Optimization of Protease Treatment Conditions for Chlorella pyrenoidosa Protein Extraction and Investigation of Its Potential as an Alternative Protein Source
Abstract
1. Introduction
2. Materials and Methods
2.1. Materials and Chemicals
2.2. Enzymatic Hydrolysis of C. pyrenoidosa
2.3. Determination of Protein Extraction Yield
2.4. Experimental Design and Optimization for Enzymatic Hydrolysis Conditions
2.5. Characteristic Analysis of CP and CPE
2.5.1. Amino Acid Composition
2.5.2. Sodium Dodecyl Sulfate–Polyacrylamide Gel Electrophoresis (SDS–PAGE) Analysis
2.5.3. Antioxidant Properties
2.5.4. Total Phenolic Content (TPC)
2.6. Statistical Analysis
3. Results
3.1. Selection of Enzyme for Optimization
3.2. Results of Single-Factor Analysis and Experimental Design
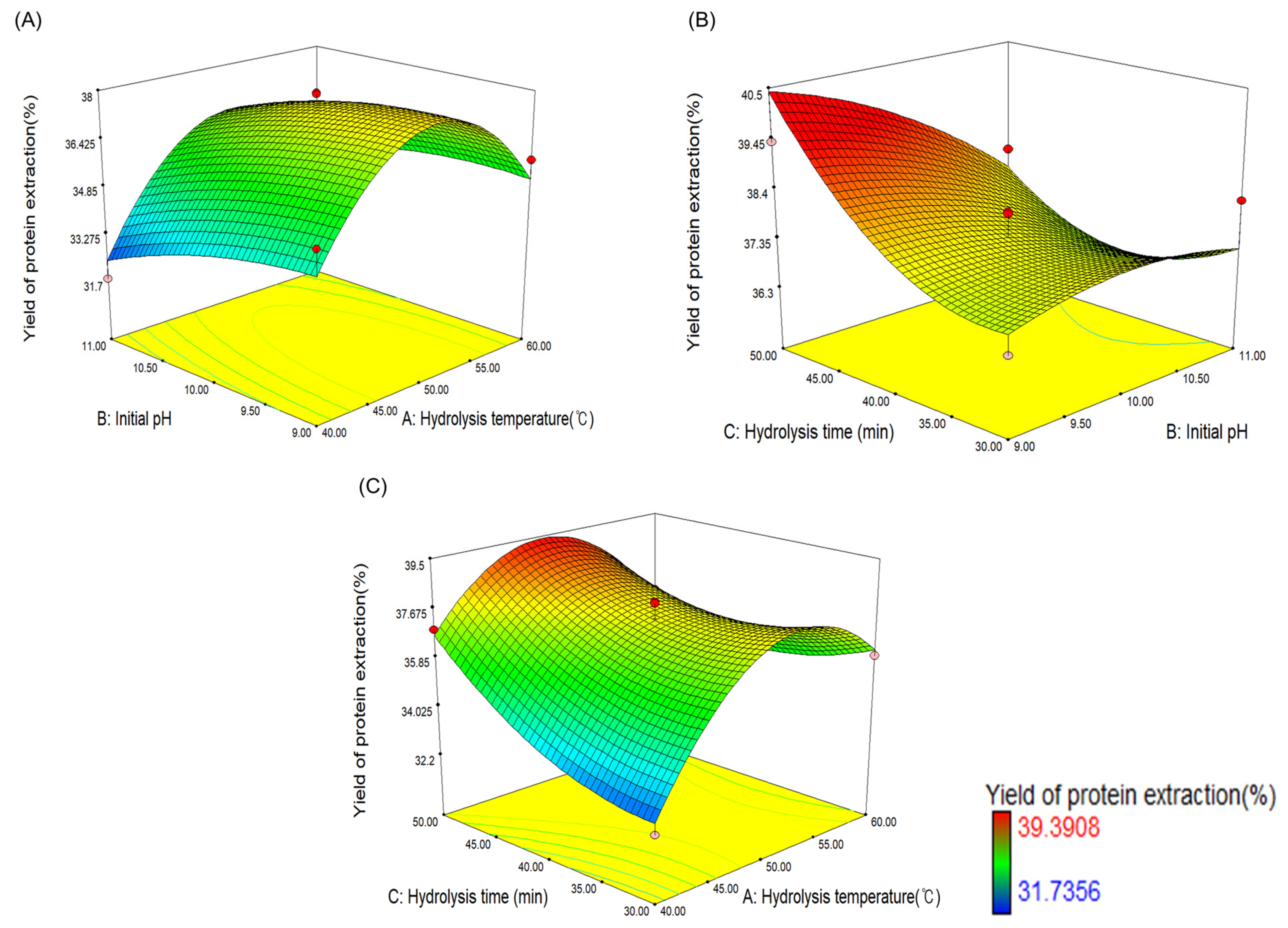
3.3. Optimization of the Enzymatic Hydrolysis Conditions
3.4. Validation of the Optimized Microbial Protease Treatment Condition
3.5. Amino Acids Analysis of CPE
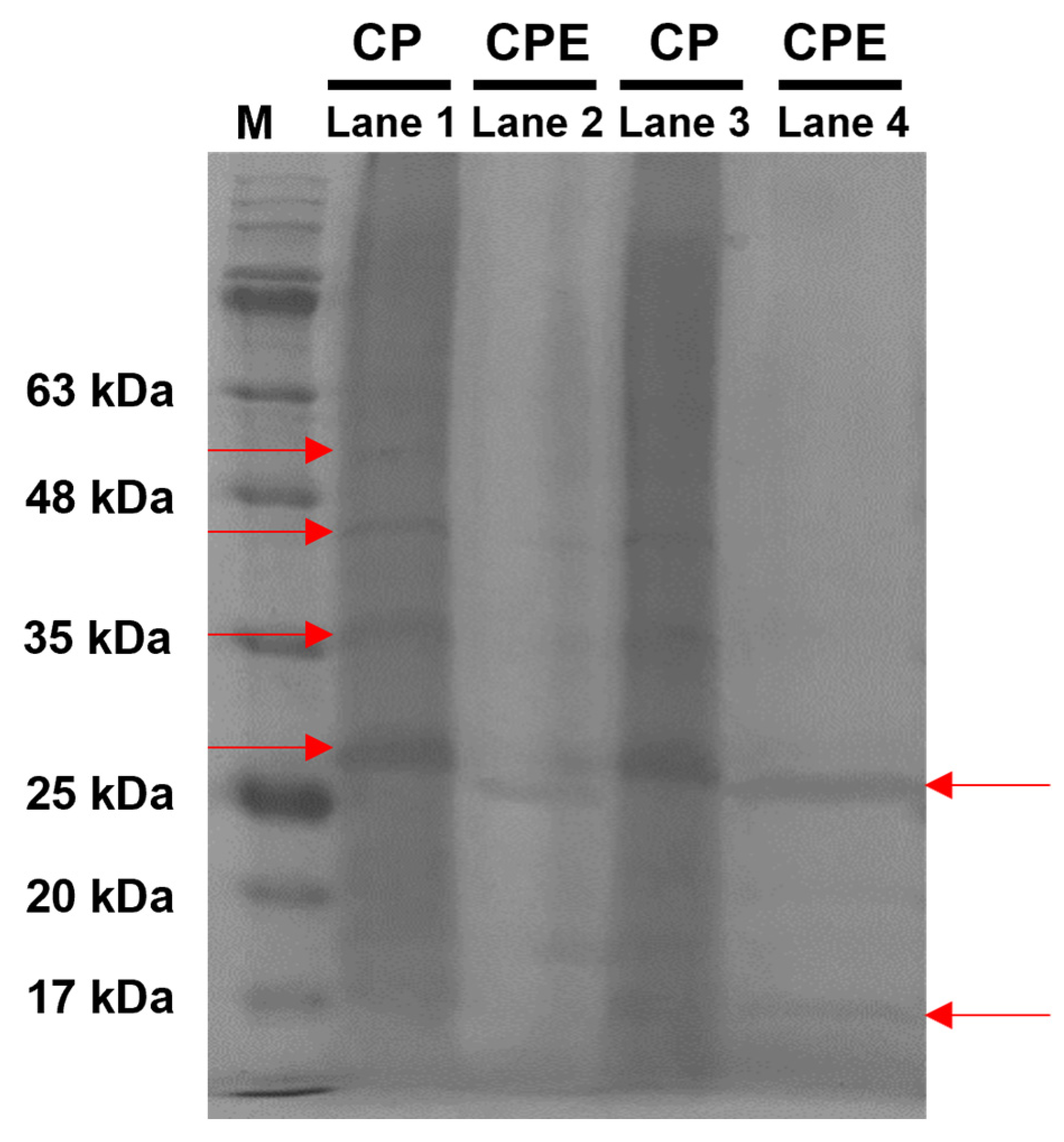
3.6. Protein Profiles of CPE
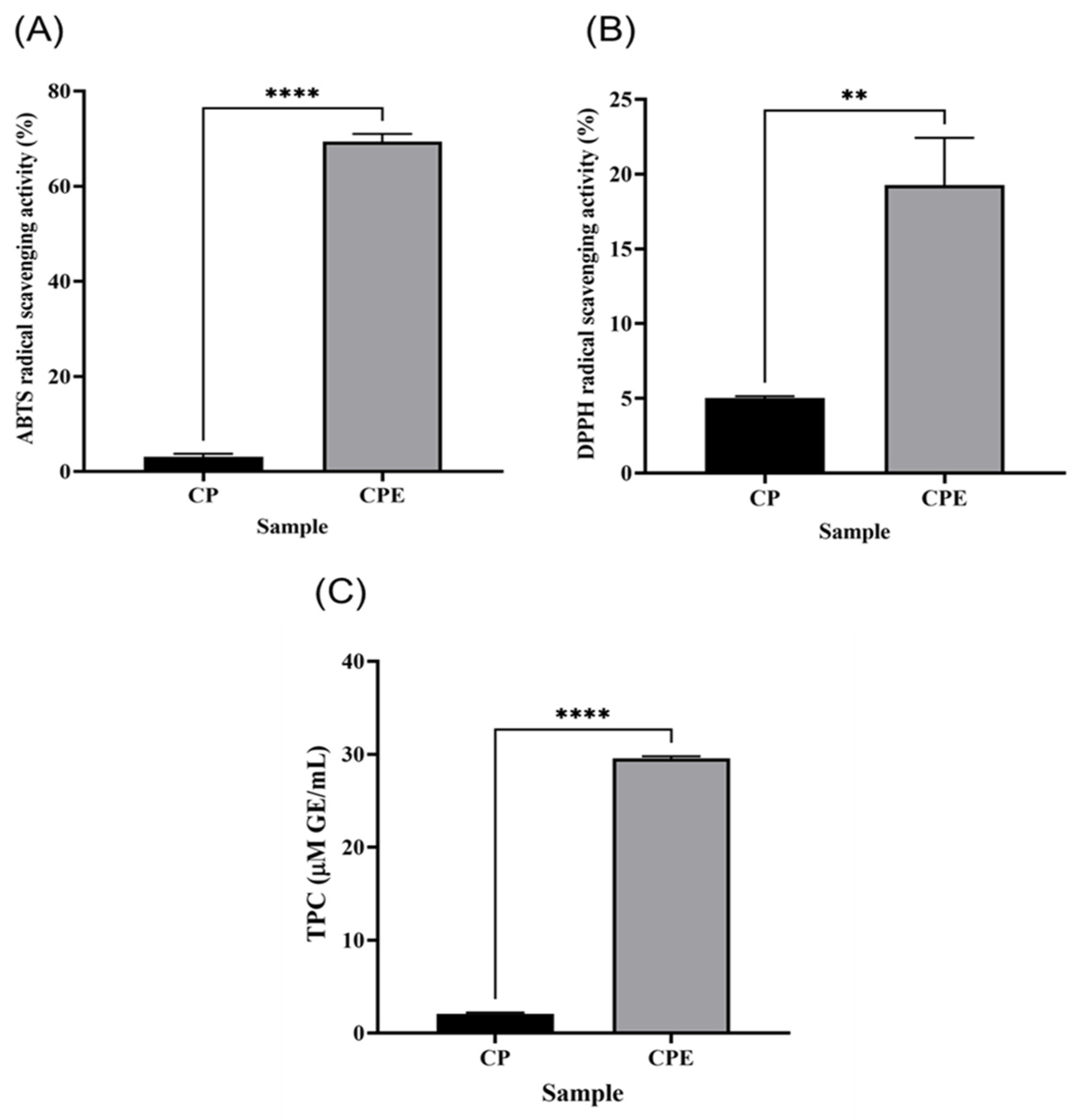
3.7. Antioxidant Activity and Phenolic Content of CPE
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bernaerts, T.M.M.; Verstreken, H.; Dejonghe, C.; Gheysen, L.; Foubert, I.; Grauwet, T.; Van Loey, A.M. Cell disruption of Nannochloropsis sp. improves in vitro bioaccessibility of carotenoids and ω3-LC-PUFA. J. Funct. Foods 2020, 65, 103770. [Google Scholar] [CrossRef]
- Sarma, S.; Sharma, S.; Patel, A.; Upadhyay, J.; Rathod, V.; Narra, M. Statistical optimization of microalgal biodiesel production and protein extraction from Chlorella sorokiniana cultivated in dairy effluent. Mater. Today Proc. 2023, 72, 2731–2740. [Google Scholar] [CrossRef]
- Katsimichas, A.; Karveli, I.; Dimopoulos, G.; Giannakourou, M.; Taoukis, P. Kinetics of high pressure homogenization assisted protein extraction from Chlorella pyrenoidosa. Innov. Food Sci. Emerg. Technol. 2023, 88, 103438. [Google Scholar] [CrossRef]
- Ramos-Romero, S.; Torrella, J.R.; Pagès, T.; Viscor, G.; Torres, J.L. Edible microalgae and their bioactive compounds in the prevention and treatment of metabolic alterations. Nutrients 2021, 13, 563. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Zhou, Z.; Wang, Z.; Hao, S.; Xue, N.; Liu, B. Cultivation of Chlorella for biomass production and nutrient removal from unsterilized human urine containing washing powder wastewater. Energy Sources Part A Recovery Util. Environ. Eff. 2023, 45, 12899–12908. [Google Scholar] [CrossRef]
- Gerde, J.A.; Montalbo-Lomboy, M.; Yao, L.; Grewell, D.; Wang, T. Evaluation of microalgae cell disruption by ultrasonic treatment. Bioresour. Technol. 2012, 125, 175–181. [Google Scholar] [CrossRef]
- Lee, J.-Y.; Yoo, C.; Jun, S.-Y.; Ahn, C.-Y.; Oh, H.-M. Comparison of several methods for effective lipid extraction from microalgae. Bioresour. Technol. 2010, 101, S75–S77. [Google Scholar] [CrossRef]
- Zheng, H.; Yin, J.; Gao, Z.; Huang, H.; Ji, X.; Dou, C. Disruption of Chlorella vulgaris cells for the release of biodiesel-producing lipids: A comparison of grinding, ultrasonication, bead milling, enzymatic lysis, and microwaves. Appl. Biochem. Biotechnol. 2011, 164, 1215–1224. [Google Scholar] [CrossRef]
- Mendes-Pinto, M.; Raposo, M.; Bowen, J.; Young, A.; Morais, R. Evaluation of different cell disruption processes on encysted cells of Haematococcus pluvialis: Effects on astaxanthin recovery and implications for bio-availability. J. Appl. Phycol. 2001, 13, 19–24. [Google Scholar] [CrossRef]
- Lian, H.; Wen, C.; Zhang, J.; Feng, Y.; Duan, Y.; Zhou, J.; He, Y.; Zhang, H.; Ma, H. Effects of simultaneous dual-frequency divergent ultrasound-assisted extraction on the structure, thermal and antioxidant properties of protein from Chlorella pyrenoidosa. Algal Res. 2021, 56, 102294. [Google Scholar] [CrossRef]
- Wu, Q.; Ma, Y.; Zhang, L.; Han, J.; Lei, Y.; Le, Y.; Huang, C.; Kan, J.; Fu, C. Extraction, functionality, and applications of Chlorella pyrenoidosa protein/peptide. Curr. Res. Food Sci. 2023, 7, 100621. [Google Scholar] [CrossRef] [PubMed]
- Qi, X.; Sun, Y.; Xiong, S. A single freeze-thawing cycle for highly efficient solubilization of inclusion body proteins and its refolding into bioactive form. Microb. Cell Factories 2015, 14, 24. [Google Scholar] [CrossRef] [PubMed]
- Kleekayai, T.; Khalesi, M.; Amigo-Benavent, M.; Cermeño, M.; Harnedy-Rothwell, P.; FitzGerald, R.J. Enzyme-Assisted Extraction of Plant Proteins. In Green Protein Processing Technologies from Plants: Novel Extraction and Purification Methods for Product Development; Springer: Cham, Switzerland, 2023; pp. 131–178. [Google Scholar] [CrossRef]
- Alavijeh, R.S.; Karimi, K.; Wijffels, R.H.; van den Berg, C.; Eppink, M. Combined bead milling and enzymatic hydrolysis for efficient fractionation of lipids, proteins, and carbohydrates of Chlorella vulgaris microalgae. Bioresour. Technol. 2020, 309, 123321. [Google Scholar] [CrossRef] [PubMed]
- Al-Zuhair, S.; Ashraf, S.; Hisaindee, S.; Darmaki, N.A.; Battah, S.; Svistunenko, D.; Reeder, B.; Stanway, G.; Chaudhary, A. Enzymatic pre-treatment of microalgae cells for enhanced extraction of proteins. Eng. Life Sci. 2017, 17, 175–185. [Google Scholar] [CrossRef] [PubMed]
- Deng, A.; Wu, J.; Zhang, Y.; Zhang, G.; Wen, T. Purification and characterization of a surfactant-stable high-alkaline protease from Bacillus sp. B001. Bioresour. Technol. 2010, 101, 7100–7106. [Google Scholar] [CrossRef] [PubMed]
- Babalola, B.A.; Akinwande, A.I.; Gboyega, A.E.; Otunba, A.A. Extraction, purification and characterization of papain cysteine-proteases from the leaves of Carica papaya. Sci. Afr. 2023, 19, e01538. [Google Scholar] [CrossRef]
- Manzoor, Z.; Nawaz, A.; Mukhtar, H.; Haq, I. Bromelain: Methods of extraction, purification and therapeutic applications. Braz. Arch. Biol. Technol. 2016, 59. [Google Scholar] [CrossRef]
- Darabzadeh, N.; Hamidi-Esfahani, Z.; Hejazi, P. Improvement of cellulase production and its characteristics by inducing mutation on Trichoderma reesei 2414 under solid state fermentation on rice by-products. Appl. Food Biotechnol. 2018, 5, 11–18. [Google Scholar] [CrossRef]
- Guan, X.; Yao, H. Optimization of Viscozyme L-assisted extraction of oat bran protein using response surface methodology. Food Chem. 2008, 106, 345–351. [Google Scholar] [CrossRef]
- Krieg, R.C.; Dong, Y.; Schwamborn, K.; Knuechel, R. Protein quantification and its tolerance for different interfering reagents using the BCA-method with regard to 2D SDS PAGE. J. Biochem. Biophys. Methods 2005, 65, 13–19. [Google Scholar] [CrossRef]
- Pekkoh, J.; Ruangrit, K.; Pumas, C.; Duangjan, K.; Chaipoot, S.; Phongphisutthinant, R.; Jeerapan, I.; Sawangrat, K.; Pathom-aree, W.; Srinuanpan, S. Transforming microalgal Chlorella biomass into cosmetically and nutraceutically protein hydrolysates using high-efficiency enzymatic hydrolysis approach. Biomass Convers. Biorefinery 2023, 13, 6299–6315. [Google Scholar] [CrossRef]
- Manns, J.M. SDS-polyacrylamide gel electrophoresis (SDS-PAGE) of proteins. Curr. Protoc. Microbiol. 2011, 22, A.3M.1–A.3M.13. [Google Scholar] [CrossRef]
- Jo, D.-M.; Song, M.-R.; Park, S.-K.; Choi, J.-H.; Oh, D.K.; Kim, D.H.; Kim, Y.-M. Potential application of lactic acid bacteria for controlling discoloration in tuna (Thunnus orientalis). Food Biosci. 2023, 54, 102856. [Google Scholar] [CrossRef]
- Madjirebaye, P.; Xiao, M.; Mahamat, B.; Xiong, S.; Mueed, A.; Wei, B.; Huang, T.; Peng, F.; Xiong, T.; Peng, Z. In vitro characteristics of lactic acid bacteria probiotics performance and antioxidant effect of fermented soymilk. Food Biosci. 2022, 49, 101952. [Google Scholar] [CrossRef]
- Tsai, T.-H.; Tsai, T.-H.; Chien, Y.-C.; Lee, C.-W.; Tsai, P.-J. In vitro antimicrobial activities against cariogenic streptococci and their antioxidant capacities: A comparative study of green tea versus different herbs. Food Chem. 2008, 110, 859–864. [Google Scholar] [CrossRef]
- Mahdy, A.; Mendez, L.; Blanco, S.; Ballesteros, M.; González-Fernández, C. Protease cell wall degradation of Chlorella vulgaris: Effect on methane production. Bioresour. Technol. 2014, 171, 421–427. [Google Scholar] [CrossRef]
- Xu, Y.; Galanopoulos, M.; Sismour, E.; Ren, S.; Mersha, Z.; Lynch, P.; Almutaimi, A. Effect of enzymatic hydrolysis using endo-and exo-proteases on secondary structure, functional, and antioxidant properties of chickpea protein hydrolysates. J. Food Meas. Charact. 2020, 14, 343–352. [Google Scholar] [CrossRef]
- Cunha, S.A.; Coscueta, E.R.; Nova, P.; Silva, J.L.; Pintado, M.M. Bioactive hydrolysates from Chlorella vulgaris: Optimal process and bioactive properties. Molecules 2022, 27, 2505. [Google Scholar] [CrossRef]
- Lee, S.Y.; Show, P.L.; Ling, T.C.; Chang, J.-S. Single-step disruption and protein recovery from Chlorella vulgaris using ultrasonication and ionic liquid buffer aqueous solutions as extractive solvents. Biochem. Eng. J. 2017, 124, 26–35. [Google Scholar] [CrossRef]
- Callejo-López, J.; Ramírez, M.; Bolívar, J.; Cantero, D. Main variables affecting a chemical-enzymatic method to obtain protein and amino acids from resistant microalgae. J. Chem. 2019, 2019, 1390463. [Google Scholar] [CrossRef]
- Shi, X.; Karachi, A.; Hosseini, M.; Yazd, M.S.; Kamyab, H.; Ebrahimi, M.; Parsaee, Z. Ultrasound wave assisted removal of Ceftriaxone sodium in aqueous media with novel nano composite g-C3N4/MWCNT/Bi2WO6 based on CCD-RSM model. Ultrason. Sonochemistry 2020, 68, 104460. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, T.V.T.; Unpaprom, Y.; Manmai, N.; Whangchai, K.; Ramaraj, R. Impact and significance of pretreatment on the fermentable sugar production from low-grade longan fruit wastes for bioethanol production. Biomass Convers. Biorefin. 2020, 12, 1605–1617. [Google Scholar] [CrossRef]
- Panjiara, D.; Pramanik, H. Optimization of process parameters using response surface methodology (RSM) for power generation via electrooxidation of glycerol in T-Shaped air breathing microfluidic fuel cell (MFC). Int. J. Hydrogen Energy 2020, 45, 33968–33979. [Google Scholar] [CrossRef]
- Said, N.S.; Sarbon, N.M. Response surface methodology (RSM) of chicken skin gelatin based composite films with rice starch and curcumin incorporation. Polym. Test. 2020, 81, 106161. [Google Scholar] [CrossRef]
- Nath, A.; Chattopadhyay, P. Optimization of oven toasting for improving crispness and other quality attributes of ready to eat potato-soy snack using response surface methodology. J. Food Eng. 2007, 80, 1282–1292. [Google Scholar] [CrossRef]
- Chiavelli, L.U.R.; Godoy, A.C.; Silveira, R.d.; Santos, P.D.S.; Lopes, T.A.; Santos, O.O.; Visentainer, J.V. Optimization of milk sample cleanup using response surface methodology. Food Anal. Methods 2020, 13, 166–175. [Google Scholar] [CrossRef]
- Hildebrand, G.; Poojary, M.M.; O’Donnell, C.; Lund, M.N.; Garcia-Vaquero, M.; Tiwari, B.K. Ultrasound-assisted processing of Chlorella vulgaris for enhanced protein extraction. J. Appl. Phycol. 2020, 32, 1709–1718. [Google Scholar] [CrossRef]
- Vieira, E.F.; Soares, C.; Machado, S.; Correia, M.; Ramalhosa, M.J.; Oliva-Teles, M.T.; Carvalho, A.P.; Domingues, V.F.; Antunes, F.; Oliveira, T.A.C. Seaweeds from the Portuguese coast as a source of proteinaceous material: Total and free amino acid composition profile. Food Chem. 2018, 269, 264–275. [Google Scholar] [CrossRef]
- Brestenský, M.; Nitrayová, S.; Patras, P.; Heger, J.; Nitray, J. Branched chain amino acids and their importance in nutrition. J. Microbiol. Biotechnol. Food Sci. 2015, 5, 197. [Google Scholar] [CrossRef]
- Moayedi, A.; Mora, L.; Aristoy, M.-C.; Hashemi, M.; Safari, M.; Toldrá, F. ACE-inhibitory and antioxidant activities of peptide fragments obtained from tomato processing by-products fermented using Bacillus subtilis: Effect of amino acid composition and peptides molecular mass distribution. Appl. Biochem. Biotechnol. 2017, 181, 48–64. [Google Scholar] [CrossRef]
- Kurt, H.; Hosoglu, M.I.; Guneser, O.; Karagul-Yuceer, Y. Influence of different bacteria species in chemical composition and sensory properties of fermented Spirulina. Food Chem. 2023, 400, 133994. [Google Scholar] [CrossRef] [PubMed]
- Guo, W.; Zhang, Y.; Long, Z.; Fu, X.; Ren, K. Study on the taste active compounds in Douchi using metabolomics method. Food Chem. 2023, 412, 135343. [Google Scholar] [CrossRef] [PubMed]
- Shao, M.; Wang, Q.; Lv, Q.; Zhang, Y.; Gao, G.; Lu, S. Advances in the research on myokine-driven regulation of bone metabolism. Heliyon 2023, 10, e22547. [Google Scholar] [CrossRef] [PubMed]
- Diez-Gutiérrez, L.; San Vicente, L.; Barrón, L.J.R.; del Carmen Villarán, M.; Chávarri, M. Gamma-aminobutyric acid and probiotics: Multiple health benefits and their future in the global functional food and nutraceuticals market. J. Funct. Foods 2020, 64, 103669. [Google Scholar] [CrossRef]
- Sharma, R.; Singh, G.P.; Sharma, V. Effects of culture conditions on growth and biochemical profile of Chlorella vulgaris. J. Plant Pathol. Microbiol. 2012, 3, 1000131. [Google Scholar] [CrossRef]
- Bachmanov, A.A.; Bosak, N.P.; Glendinning, J.I.; Inoue, M.; Li, X.; Manita, S.; McCaughey, S.A.; Murata, Y.; Reed, D.R.; Tordoff, M.G. Genetics of amino acid taste and appetite. Adv. Nutr. 2016, 7, 806S–822S. [Google Scholar] [CrossRef]
- Re, R.; Pellegrini, N.; Proteggente, A.; Pannala, A.; Yang, M.; Rice-Evans, C. Antioxidant activity applying an improved ABTS radical cation decolorization assay. Free. Radic. Biol. Med. 1999, 26, 1231–1237. [Google Scholar] [CrossRef]
- Shi, Y.; Kovacs-Nolan, J.; Jiang, B.; Tsao, R.; Mine, Y. Antioxidant activity of enzymatic hydrolysates from eggshell membrane proteins and its protective capacity in human intestinal epithelial Caco-2 cells. J. Funct. Foods 2014, 10, 35–45. [Google Scholar] [CrossRef]
- Zou, T.-B.; He, T.-P.; Li, H.-B.; Tang, H.-W.; Xia, E.-Q. The structure-activity relationship of the antioxidant peptides from natural proteins. Molecules 2016, 21, 72. [Google Scholar] [CrossRef]
- Patel, A.K.; Albarico, F.P.J.B.; Perumal, P.K.; Vadrale, A.P.; Nian, C.T.; Chau, H.T.B.; Anwar, C.; ud din Wani, H.M.; Pal, A.; Saini, R. Algae as an emerging source of bioactive pigments. Bioresour. Technol. 2022, 351, 126910. [Google Scholar] [CrossRef]

| Enzyme | Source | Enzyme Activity | Composition | Optimal Hydrolysis Conditions | Reference | |
|---|---|---|---|---|---|---|
| pH | Temperature (°C) | |||||
| Microbial protease | Bacillus sp. | ≥16 U/g | Endopeptidase | 10.0 | 50 | [16] |
| Papain | Carica papaya | ≥10 U/mg protein | Endopeptidase | 6.0 | 50 | [17] |
| Bromelain | Pineapple stem | ≥3 U/mg protein | Endopeptidase | 6.0 | 50 | [18] |
| Cellulase | Trichoderma reesei | ≥700 U/g | Cellulase | 5.0 | 50 | [19] |
| Viscozyme L | Aspergillus sp. | ≥100 U/g | Hemicellulase | 5.0 | 50 | [20] |
| Time (h) | Yield of Protein Extraction (%) | |||||
|---|---|---|---|---|---|---|
| Microbial Protease | Papain | Bromelain | Cellulase | Viscozyme L | Control | |
| 0 | 10.52 ± 0.33 Ad | 7.11 ± 0.13 Be | 5.41 ± 0.23 Cd | 6.23 ± 0.25 BCa | 6.42 ± 0.11 BCd | 5.62 ± 0.12 Cd |
| 3 | 34.54 ± 2.48 Aa | 13.78 ± 1.29 Bd | 8.37 ± 0.86 Cc | 5.31 ± 0.55 Db | 7.76 ± 0.04 Cc | 5.51 ± 0.20 Dcd |
| 6 | 32.45 ± 2.73 Aa | 16.09 ± 0.29 Bbc | 9.21 ± 0.75 Cb | 6.57 ± 0.27 Da | 9.23 ± 0.70 Ca | 5.80 ± 0.11 Dcd |
| 9 | 32.72 ± 0.07 Aa | 15.47 ± 0.61 Bc | 9.58 ± 0.67 Cab | 4.80 ± 0.30 Ec | 7.55 ± 0.15 Dc | 5.89 ± 0.32 DEbc |
| 12 | 29.48 ± 2.05 Ab | 16.94 ± 0.80 Bab | 9.34 ± 0.44 Cab | 5.63 ± 0.08 Db | 8.40 ± 0.79 Ca | 6.21 ± 0.15 Db |
| 15 | 33.60 ± 3.16 Aa | 17.53 ± 0.51 Ba | 9.93 ± 0.24 Ca | 6.52 ± 0.47 Da | 8.68 ± 0.29 Cab | 6.65 ± 0.07 Da |
| 18 | 26.47 ± 0.65 Ac | 18.22 ± 0.47 Ba | 10.02 ± 0.32 Ca | 6.48 ± 0.24 Ea | 8.52 ± 0.30 Db | 6.80 ± 0.21 Ea |
| No. | A, Hydrolysis Temperature (°C) | B, Initial pH | C, Hydrolysis Time (min) | Y, Yield of Protein Extraction (%) |
|---|---|---|---|---|
| 1 | 40 | 9 | 40 | 35.07 |
| 2 | 40 | 11 | 40 | 31.74 |
| 3 | 50 | 9 | 30 | 36.59 |
| 4 | 50 | 10 | 40 | 37.88 |
| 5 | 60 | 10 | 50 | 36.41 |
| 6 | 40 | 10 | 30 | 32.25 |
| 7 | 40 | 10 | 50 | 36.91 |
| 8 | 60 | 11 | 40 | 33.50 |
| 9 | 50 | 10 | 40 | 37.88 |
| 10 | 60 | 9 | 40 | 35.75 |
| 11 | 50 | 10 | 40 | 37.88 |
| 12 | 50 | 11 | 50 | 38.02 |
| 13 | 60 | 10 | 30 | 35.95 |
| 14 | 50 | 9 | 50 | 39.39 |
| 15 | 50 | 10 | 40 | 37.88 |
| 16 | 50 | 11 | 30 | 38.16 |
| 17 | 50 | 10 | 40 | 36.45 |
| Source | Sum of Squares | Degrees of Freedom | Mean Square | F-Value | p-Value |
|---|---|---|---|---|---|
| Model | 65.19 | 9 | 7.24 | 7.58 | 0.0071 |
| Significant | |||||
| A | 3.99 | 1 | 3.99 | 4.18 | 0.0402 |
| B | 3.62 | 1 | 3.62 | 3.79 | 0.0426 |
| C | 7.57 | 1 | 7.57 | 7.92 | 0.0260 |
| AB | 0.29 | 1 | 0.29 | 0.31 | 0.5958 |
| AC | 4.43 | 1 | 4.43 | 4.63 | 0.0384 |
| BC | 2.16 | 1 | 2.16 | 2.26 | 0.1763 |
| A2 | 39.23 | 1 | 39.23 | 41.06 | 0.0004 |
| B2 | 0.64 | 1 | 0.64 | 0.67 | 0.4394 |
| C2 | 4.00 | 1 | 4.00 | 4.19 | 0.0299 |
| Residual | 6.69 | 7 | 0.96 | ||
| Lack of Fit | 5.02 | 3 | 1.67 | 4.01 | 0.1064 |
| Not significant | |||||
| R2 | 0.9270 | ||||
| Regression equation | Y = −99.86311 + 3.27244 A + 8.71967 B + 0.57823 C + 0.027155 AB − 0.010520 AC − 0.073515 BC − 0.030525 A2 − 0.39049 B2 + 0.00975255 C2 | ||||
| Source | Predicted Value (%) |
|---|---|
| Mean | 39.96 |
| Standard deviation | 0.82 |
| Model degree | Quadratic |
| R2 | 0.9270 |
| Adequacy precision | 10.749 |
| Amino Acids | Concentration of Total Amino Acid (mg/100 g) | t-Value | p-Value | |
|---|---|---|---|---|
| CP | CPE | |||
| Aspartic acid | 3847.4 ± 26.4 | 5555.0 ± 90.3 | −25.650 | 0.002 |
| Threonine * | 2036.0 ± 32.9 | 2934.0 ± 54.4 | −18.601 | 0.003 |
| Serine | 1748.6 ± 35.1 | 2433.5 ± 85.1 | −10.077 | 0.010 |
| Glutamic acid | 5032.5 ± 26.8 | 7104.2 ± 61.9 | −79.657 | 0.000 |
| Glycine | 2539.1 ± 58.9 | 3353.6 ± 41.1 | −52.056 | 0.000 |
| Alanine | 3763.1 ± 36.0 | 5591.8 ± 80.8 | −42.933 | 0.001 |
| Cystine | 533.4 ± 13.7 | 760.9 ± 16.4 | −67.131 | 0.000 |
| Valine * | 2813.2 ± 40.6 | 3524.0 ± 49.7 | −42.430 | 0.001 |
| Methionine * | 945.9 ± 11.1 | 1363.4 ± 38.4 | −21.709 | 0.002 |
| Isoleucine * | 1770.8 ± 44.3 | 2193.6 ± 30.5 | −15.417 | 0.004 |
| Leucine * | 4161.3 ± 61.1 | 5223.7 ± 34.7 | −54.192 | 0.000 |
| Tyrosine | 1586.0 ± 16.2 | 1932.3 ± 18.3 | −41.479 | 0.001 |
| Phenylalanine * | 2473.6 ± 42.0 | 2927.1 ± 37.1 | −10.223 | 0.009 |
| Lysine * | 3717.1 ± 22.5 | 3472.4 ± 17.4 | 10.996 | 0.008 |
| Histidine * | 923.4 ± 10.5 | 1210.9 ± 12.1 | −313.688 | 0.000 |
| Arginine | 2801.0 ± 47.4 | 3474.2 ± 41.9 | −16.966 | 0.003 |
| Proline | 2342.4 ± 43.4 | 3341.7 ± 55.7 | −18.615 | 0.003 |
| ∑EAA | 18,841.3 ± 129.2 | 22,849.1 ± 194.9 | −103.755 | 0.000 |
| ∑NEAA | 24,193.5 ± 80.6 | 33,547.2 ± 340.2 | −41.255 | 0.001 |
| TAA | 43,034.8 ± 69.8 | 56,396.3 ± 517.7 | −51.196 | 0.000 |
| Amino Acids | Concentration of Total Amino Acid (mg/100 g) | t-Value | p-Value | |
|---|---|---|---|---|
| CP | CPE | |||
| Aspartic acid | 13.6 ± 0.2 | 25.8 ± 0.6 | −48.478 | 0.000 |
| Threonine * | 50.1 ± 1.3 | 96.0 ± 1.8 | −58.641 | 0.000 |
| Serine | 17.3 ± 0.8 | 50.6 ± 1.3 | −46.381 | 0.000 |
| Glutamic acid | 288.9 ± 7.9 | 541.4 ± 10.6 | −71.645 | 0.000 |
| Glycine | 23.0 ± 2.7 | 61.8 ± 0.6 | −30.563 | 0.001 |
| Alanine | 164.5 ± 6.9 | 357.4 ± 5.5 | −118.647 | 0.000 |
| Cystine | 1.9 ± 0.5 | 183.2 ± 5.3 | −60.848 | 0.000 |
| Valine * | 30.4 ± 1.0 | 102.4 ± 1.2 | −59.216 | 0.000 |
| Methionine * | 8.1 ± 0.3 | 74.4 ± 2.8 | −38.866 | 0.001 |
| Isoleucine * | 11.8 ± 0.5 | 183.6 ± 6.4 | −43.774 | 0.001 |
| Leucine * | 17.0 ± 0.7 | 47.7 ± 0.7 | −45.102 | 0.000 |
| Tyrosine | 6.9 ± 0.3 | 21.7 ± 0.8 | −36.414 | 0.001 |
| Phenylalanine * | 22.9 ± 1.5 | 102.1 ± 1.8 | −114.135 | 0.000 |
| Lysine * | 72.7 ± 2.3 | 139.3 ± 12.6 | −10.943 | 0.008 |
| Arginine | 35.6 ± 1.4 | 68.7 ± 4.0 | −14.871 | 0.004 |
| Proline | 121.3 ± 3.0 | 217.1 ± 7.0 | −30.673 | 0.001 |
| β-amino isobutyric acid | ND | 650.8 ± 23.6 | −47.725 | 0.000 |
| γ-amino-n-butyric acid | 9.9 ± 0.3 | 117.2 ± 5.5 | −34.831 | 0.001 |
| ∑EAA | 213.0 ± 4.4 | 745.5 ± 23.1 | −40.604 | 0.001 |
| ∑NEAA | 683.0 ± 20.3 | 2295.6 ± 54.2 | −59.654 | 0.000 |
| TAA | 896.0 ± 24.7 | 3041.2 ± 74.7 | −53.824 | 0.000 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cho, K.-J.; Kim, M.-U.; Jeong, G.-J.; Khan, F.; Jo, D.-M.; Kim, Y.-M. Optimization of Protease Treatment Conditions for Chlorella pyrenoidosa Protein Extraction and Investigation of Its Potential as an Alternative Protein Source. Foods 2024, 13, 366. https://doi.org/10.3390/foods13030366
Cho K-J, Kim M-U, Jeong G-J, Khan F, Jo D-M, Kim Y-M. Optimization of Protease Treatment Conditions for Chlorella pyrenoidosa Protein Extraction and Investigation of Its Potential as an Alternative Protein Source. Foods. 2024; 13(3):366. https://doi.org/10.3390/foods13030366
Chicago/Turabian StyleCho, Kyung-Jin, Min-Ung Kim, Geum-Jae Jeong, Fazlurrahman Khan, Du-Min Jo, and Young-Mog Kim. 2024. "Optimization of Protease Treatment Conditions for Chlorella pyrenoidosa Protein Extraction and Investigation of Its Potential as an Alternative Protein Source" Foods 13, no. 3: 366. https://doi.org/10.3390/foods13030366
APA StyleCho, K.-J., Kim, M.-U., Jeong, G.-J., Khan, F., Jo, D.-M., & Kim, Y.-M. (2024). Optimization of Protease Treatment Conditions for Chlorella pyrenoidosa Protein Extraction and Investigation of Its Potential as an Alternative Protein Source. Foods, 13(3), 366. https://doi.org/10.3390/foods13030366







