Efficient Green Extraction of Nutraceutical Compounds from Nannochloropsis gaditana: A Comparative Electrospray Ionization LC-MS and GC-MS Analysis for Lipid Profiling
Abstract
1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Combined Pretreatment Method
2.3. Extraction of Microalgal Biomass
2.3.1. Ultrasound-Assisted Extraction
2.3.2. Pressurized Liquid Extraction
2.3.3. Microwave-Assisted Extraction
2.3.4. Traditional Lipid Extraction of Microalgal Biomass
2.4. LC-MS Analysis Method and Equipment
2.5. HPLC–ELSD-DAD Analysis
2.6. Fatty Acid Composition Through GC-MS
2.7. Statistical Analysis
3. Results and Discussion
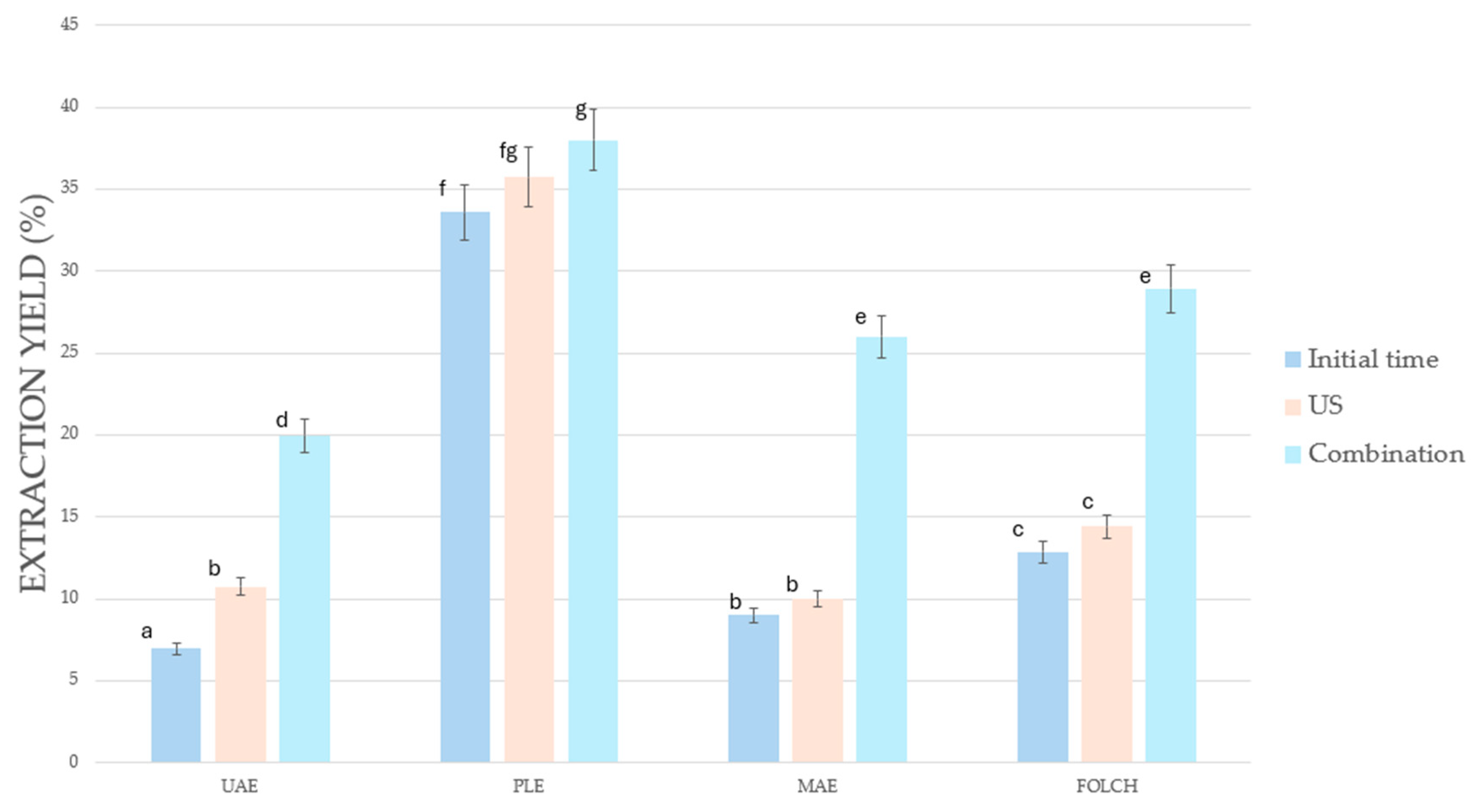
3.1. Comparison of Extraction Yield Obtained Through Different Extraction Methods After Pretreatment
3.2. Chemical Characterization of N. gaditana Extracts Through LC-MS
Comparison Between PLE, UAE, and MAE Extracts: Influence of the Pretreatment on Polar Lipids
3.3. Omega 3 PUFA Content in the Different Extracts
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Blanco-Llamero, C.; García-García, P.; Señoráns, F.J. Cross-Linked Enzyme Aggregates and Their Application in Enzymatic Pretreatment of Microalgae: Comparison Between CLEAs and Combi-CLEAs. Front. Bioeng. Biotechnol. 2021, 9, 794672. [Google Scholar] [CrossRef] [PubMed]
- Blanco-Llamero, C.; García-García, P.; Señoráns, F.J. Combination of synergic enzymes and ultrasounds as an effective pretreatment process to break microalgal cell wall and enhance algal oil extraction. Foods 2021, 10, 1928. [Google Scholar] [CrossRef]
- Mahdy, A.; Mendez, L.; Ballesteros, M.; González-Fernández, C. Enhanced methane production of Chlorella vulgaris and Chlamydomonas reinhardtii by hydrolytic enzymes addition. Energy Convers. Manag. 2014, 85, 551–557. [Google Scholar] [CrossRef]
- Gianluca, C. Use of Cell Wall Degrading Enzymes for the Recovery of Lipids from Microalgae. Ph.D. Thesis, Faculty of Civil and Industrial Engineering Chemical, Material and Environmental Engineering Department, SAPIENZA Università di Roma, Rome, Italy, 2017. Available online: https://core.ac.uk/download/pdf/80313205.pdf (accessed on 15 October 2024).
- Zuorro, A.; Maffei, G.; Lavecchia, R. Optimization of enzyme-assisted lipid extraction from Nannochloropsis microalgae. J. Taiwan Inst. Chem. Eng. 2016, 67, 106–114. [Google Scholar] [CrossRef]
- Sander, K.; Murthy, G.S. Enzymatic degradation of microalgal cell walls. In Proceedings of the American Society of Agricultural and Biological Engineers Annual International Meeting 2009, ASABE, Reno, NV, USA, 21–24 June 2009; Volume 4, pp. 2489–2500. [Google Scholar] [CrossRef]
- Coelho, D.; Lopes, P.A.; Cardoso, V.; Ponte, P.; Brás, J.; Madeira, M.S.; Alfaia, C.M.; Bandarra, N.M.; Fontes, C.M.G.A.; Prates, J.A.M. A two-enzyme constituted mixture to improve the degradation of Arthrospira platensis microalga cell wall for monogastric diets. J. Anim. Physiol. Anim. Nutr. 2020, 104, 310–321. [Google Scholar] [CrossRef] [PubMed]
- Zhang, R.; Chen, J.; Zhang, X. Extraction of intracellular protein from Chlorella pyrenoidosa using a combination of ethanol soaking, enzyme digest, ultrasonication and homogenization techniques. Bioresour. Technol. 2018, 247, 267–272. [Google Scholar] [CrossRef] [PubMed]
- Señoráns, M.; Castejón, N.; Señoráns, F.J. Advanced Extraction of Lipids with DHA from Isochrysis galbana with Enzymatic Pre-Treatment Combined with Pressurized Liquids and Ultrasound Assisted Extractions. Molecules 2020, 25, 3310. [Google Scholar] [CrossRef]
- Demuez, M.; Mahdy, A.; Tomás-Pejó, E.; González-Fernández, C.; Ballesteros, M. Enzymatic cell disruption of microalgae biomass in biorefinery processes. Biotechnol. Bioeng. 2015, 112, 1955–1966. [Google Scholar] [CrossRef] [PubMed]
- Ciudad, G.; Rubilar, O.; Azócar, L.; Toro, C.; Cea, M.; Torres, Á.; Ribera, A.; Navia, R. Performance of an enzymatic extract in Botrycoccus braunii cell wall disruption. J. Biosci. Bioeng. 2014, 117, 75–80. [Google Scholar] [CrossRef]
- Al-Zuhair, S.; Ashraf, S.; Hisaindee, S.; Al Darmaki, N.; Battah, S.; Svistunenko, D.; Reeder, B.; Stanway, G.; Chaudhary, A. Enzymatic pre-treatment of microalgae cells for enhanced extraction of proteins. Eng. Life Sci. 2017, 17, 175–185. [Google Scholar] [CrossRef] [PubMed]
- Gerken, H.G.; Donohoe, B.; Knoshau, E.P.G. Enzymatic cell wall degradation of Chlorella vulgaris and other microalgae for biofuels production. Planta 2013, 237, 239–253. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Li, Y.; Hu, X.; Su, W.; Zhong, M. Combined enzymatic and mechanical cell disruption and lipid extraction of green alga Neochloris oleoabundans. Int. J. Mol. Sci. 2015, 16, 7707–7722. [Google Scholar] [CrossRef] [PubMed]
- Sabater, C.; Corzo, N.; Olano, A.; Montilla, A. Enzymatic extraction of pectin from artichoke (Cynara scolymus L.) by-products using Celluclast®1.5L. Carbohydr. Polym. 2018, 190, 43–49. [Google Scholar] [CrossRef] [PubMed]
- Maffei, G.; Bracciale, M.P.; Broggi, A.; Zuorro, A.; Santarelli, M.L.; Lavecchia, R. Effect of an enzymatic treatment with cellulase and mannanase on the structural properties of Nannochloropsis microalgae. Bioresour. Technol. 2018, 249, 592–598. [Google Scholar] [CrossRef]
- Chen, Q.; Liu, D.; Wu, C.; Xu, A.; Xia, W.; Wang, Z.; Wen, F.; Yu, D. Influence of a facile pretreatment process on lipid extraction from: Nannochloropsis sp. through an enzymatic hydrolysis reaction. RSC Adv. 2017, 7, 53270–53277. [Google Scholar] [CrossRef]
- Chen, L.; Li, R.; Ren, X.; Liu, T. Improved aqueous extraction of microalgal lipid by combined enzymatic and thermal lysis from wet biomass of Nannochloropsis oceanica. Bioresour. Technol. 2016, 214, 138–143. [Google Scholar] [CrossRef] [PubMed]
- Castejón, N.; Señoráns, F.J. Enzymatic modification to produce health-promoting lipids from fish oil, algae and other new omega-3 sources: A review. New Biotechnol. 2020, 57, 45–54. [Google Scholar] [CrossRef]
- Jaime, L.; Mendiola, J.A.; Herrero, M.; Soler-Rivas, C.; Santoyo, S.; Señorans, F.J.; Cifuentes, A.; Ibáñez, E. Separation and characterization of antioxidants from Spirulina platensis microalga combining pressurized liquid extraction, TLC, and HPLC-DAD. J. Sep. Sci. 2005, 28, 2111–2119. [Google Scholar] [CrossRef] [PubMed]
- He, Y.; Huang, Z.; Zhong, C.; Guo, Z.; Chen, B. Pressurized liquid extraction with ethanol as a green and efficient technology to lipid extraction of Isochrysis biomass. Bioresour. Technol. 2019, 293, 122049. [Google Scholar] [CrossRef] [PubMed]
- Pühringer, M.; Rampler, E.; Castejón, N. Unwrapping the (glyco-)lipidome in the microalgae Microchloropsis gaditana: Effects of eco-friendly extraction methods. Algal Res. 2024, 79, 103480. [Google Scholar] [CrossRef]
- Castejón, N.; Marko, D. Fatty Acid Composition and Cytotoxic Activity of Lipid Extracts from Nannochloropsis gaditana Produced by Green Technologies. Molecules 2022, 27, 3710. [Google Scholar] [CrossRef] [PubMed]
- Blanco-Llamero, C.; Señoráns, F.J. Biobased Solvents for Pressurized Liquid Extraction of Nannochloropsis gaditana Omega-3 Lipids. Mar. Drugs 2021, 19, 107. [Google Scholar] [CrossRef] [PubMed]
- Chemat, F.; Vian, M.A.; Cravotto, G. Green extraction of natural products: Concept and principles. Int. J. Mol. Sci. 2012, 13, 8615–8627. [Google Scholar] [CrossRef] [PubMed]
- Araujo, R.G.; Rodríguez-Jasso, R.M.; Ruíz, H.A.; Govea-Salas, M.; Pintado, M.; Aguilar, C.N. Recovery of bioactive components from avocado peels using microwave-assisted extraction. Food Bioprod. Process. 2021, 127, 152–161. [Google Scholar] [CrossRef]
- Araújo, R.G.; Rodriguez-Jasso, R.M.; Ruiz, H.A.; Govea-Salas, M.; Pintado, M.E.; Aguilar, C.N. Process optimization of microwave-assisted extraction of bioactive molecules from avocado seeds. Ind. Crops Prod. 2020, 154, 112623. [Google Scholar] [CrossRef]
- Garoma, T.; Janda, D. Investigation of the effects of microalgal cell concentration and electroporation, microwave and ultrasonication on lipid extraction efficiency. Renew. Energy 2016, 86, 117–123. [Google Scholar] [CrossRef]
- Castejón, N.; Luna, P.; Señoráns, F.J. Alternative oil extraction methods from Echium plantagineum L. seeds using advanced techniques and green solvents. Food Chem. 2018, 244, 75–82. [Google Scholar] [CrossRef]
- Sharayei, P.; Azarpazhooh, E.; Zomorodi, S.; Einafshar, S.; Ramaswamy, H.S. Optimization of ultrasonic-assisted extraction of astaxanthin from green tiger (Penaeus semisulcatus) shrimp shell. Ultrason. Sonochem. 2021, 76, 105666. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Xie, Y.; Johnson, D.R.; Li, Y.; He, Z.; Li, H. Ultrasonic-pretreated lipase-catalyzed synthesis of medium–long–medium lipids using different fatty acids as sn-2 acyl-site donors. Food Sci. Nutr. 2019, 7, 2361–2373. [Google Scholar] [CrossRef] [PubMed]
- Castejón, N.; Luna, P.; Señorans, F.J. Ultrasonic removal of mucilage for pressurized liquid extraction of omega-3 rich oil from chia seeds (Salvia hispanica L.). J. Agric. Food Chem. 2017, 65, 2572–2579. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.Y.; Show, P.L.; Ling, T.C.; Chang, J.S. Single-step disruption and protein recovery from Chlorella vulgaris using ultrasonication and ionic liquid buffer aqueous solutions as extractive solvents. Biochem. Eng. J. 2017, 124, 26–35. [Google Scholar] [CrossRef]
- Zielinski, A.A.F.; del Pilar Sanchez-Camargo, A.; Benvenutti, L.; Ferro, D.M.; Dias, J.L.; Ferreira, S.R.S. High-pressure fluid technologies: Recent approaches to the production of natural pigments for food and pharmaceutical applications. Trends Food Sci. Technol. 2021, 118, 850–869. [Google Scholar] [CrossRef]
- Poerschmann, J.; Carlson, R. New fractionation scheme for lipid classes based on ‘in-cell fractionation’ using sequential pressurized liquid extraction. J. Chromatogr. A 2006, 1127, 18–25. [Google Scholar] [CrossRef] [PubMed]
- Villanueva-Bermejo, D.; Calvo, M.V.; Castro-Gómez, P.; Fornari, T.; Fontecha, J. Production of omega 3-rich oils from underutilized chia seeds. Comparison between supercritical fluid and pressurized liquid extraction methods. Food Res. Int. 2019, 115, 400–407. [Google Scholar] [CrossRef] [PubMed]
- Castejón, N.; Señoráns, F.J. Simultaneous extraction and fractionation of omega-3 acylglycerols and glycolipids from wet microalgal biomass of Nannochloropsis gaditana using pressurized liquids. Algal Res. 2019, 37, 74–82. [Google Scholar] [CrossRef]
- da Silva, L.C.; Souza, M.C.; Sumere, B.R.; Silva, L.G.; da Cunha, D.T.; Barbero, G.F.; Bezerra, R.M.; Rostagno, M.A. Simultaneous extraction and separation of bioactive compounds from apple pomace using pressurized liquids coupled on-line with solid-phase extraction. Food Chem. 2020, 318, 126450. [Google Scholar] [CrossRef]
- Wang, Y.; Peng, D.; Zhu, Y.; Xie, S.; Pan, Y.; Chen, D.; Tao, Y.; Yuan, Z. Establishment of pressurized liquid extraction followed by HPLC–MS/MS method for the screening of adrenergic drugs, steroids, sedatives, colorants and antioxidants in swine feed. J. Sep. Sci. 2019, 42, 1915–1929. [Google Scholar] [CrossRef] [PubMed]
- Otero, P.; Quintana, S.E.; Reglero, G.; Fornari, T.; García-Risco, M.R. Pressurized Liquid Extraction (PLE) as an innovative green technology for the effective enrichment of galician algae extracts with high quality fatty acids and antimicrobial and antioxidant properties. Mar. Drugs 2018, 16, 156. [Google Scholar] [CrossRef]
- Rombaut, R.; Camp, J.V.; Dewettinck, K. Analysis of phospho- and sphingolipids in dairy products by a new HPLC method. J. Dairy Sci. 2005, 88, 482–488. [Google Scholar] [CrossRef] [PubMed]
- Ma, H.-F.; Wei, F.; Wu, B.-F.; Yang, C.; Xie, Y.; Wu, Z.-Y.; Lv, X.; Chen, H. Profiling and quantification of aminophospholipids based on chemical derivatization coupled with HPLC-MS. J. Lipid Res. 2019, 60, 121–134. [Google Scholar] [CrossRef] [PubMed]
- Giacometti, J.; Milošević, A.; Milin, Č. Gas chromatographic determination of fatty acids contained in different lipid classes after their separation by solid-phase extraction. J. Chromatogr. A 2002, 976, 47–54. [Google Scholar] [CrossRef] [PubMed]
- Bradford, M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef] [PubMed]
- Folch, J.; Lees, M.; Stanley, G.H.S. A simple method for the isolation and purification of total lipides from animal tissues. J. Biol. Chem. 1957, 226, 497–509. [Google Scholar] [CrossRef]
- Martin, G.J.O.; Hill, D.R.A.; Olmstead, I.L.D.; Bergamin, A.; Shears, M.J.; Dias, D.A.; Kentish, S.E.; Scales, P.J.; Botté, C.Y.; Callahan, D.L. Lipid profile remodeling in response to nitrogen deprivation in the Microalgae chlorella sp. (trebouxiophyceae) and Nannochloropsis sp. (eustigmatophyceae). PLoS ONE 2014, 9, e103389. [Google Scholar] [CrossRef] [PubMed]
- Couto, D.; Conde, T.A.; Melo, T.; Neves, B.; Costa, M.; Cunha, P.; Guerra, I.; Correia, N.; Silva, J.T.; Pereira, H.; et al. Effects of outdoor and indoor cultivation on the polar lipid composition and antioxidant activity of Nannochloropsis oceanica and Nannochloropsis limnetica: A lipidomics perspective. Algal Res. 2022, 64, 102718. [Google Scholar] [CrossRef]
- Chua, E.T.; Schenk, P.M. A biorefinery for Nannochloropsis: Induction, harvesting, and extraction of EPA-rich oil and high-value protein. Bioresour. Technol. 2017, 244, 1416–1424. [Google Scholar] [CrossRef] [PubMed]
- Ho, B.C.H.; Kamal, S.M.M.; Danquah, M.K.; Harun, R. Optimization of Subcritical Water Extraction (SWE) of Lipid and Eicosapentaenoic Acid (EPA) from Nannochloropsis gaditana. BioMed Res. Int. 2018, 2018, 8273581. [Google Scholar] [CrossRef]
- Hendriks, T. Maldi-msi-lc-ms/ms workflow for single-section single step combined proteomics and quantitative lipidomics. Anal. Chem. 2024, 96, 4266–4274. [Google Scholar] [CrossRef] [PubMed]
- Vasilopoulou, C.G.; Sulek, K.; Brunner, A.-D.; Meitei, N.S.; Schweiger-Hufnagel, U.; Meyer, S.W.; Barsch, A.; Mann, M.; Meier, F. Trapped ion mobility spectrometry and pasef enable in-depth lipidomics from minimal sample amounts. Nat. Commun. 2020, 11, 331. [Google Scholar] [CrossRef]
- Arif, M.; Bai, Y.; Usman, M.; Jalalah, M.; Harraz, F.A.; Al-Assiri, M.; Li, X.; Salama, E.-S.; Zhang, C. Highest accumulated microalgal lipids (polar and non-polar) for biodiesel production with advanced wastewater treatment: Role of lipidomics. Bioresour. Technol. 2020, 298, 122299. [Google Scholar] [CrossRef]
- Scholz, M.J.; Weiss, T.L.; Jinkerson, R.E.; Jing, J.; Roth, R.; Goodenough, U.; Posewitz, M.C.; Gerken, H.G. Ultrastructure and composition of the Nannochloropsis gaditana cell wall. Eukaryot. Cell 2014, 13, 1450–1464. [Google Scholar] [CrossRef] [PubMed]
- Molino, A.; Martino, M.; Larocca, V.; Di Sanzo, G.; Spagnoletta, A.; Marino, T.; Karatza, D.; Iovine, A.; Mehariya, S.; Musmarra, D. Eicosapentaenoic acid extraction from Nannochloropsis gaditana using carbon dioxide at supercritical conditions. Mar. Drugs 2019, 17, 132. [Google Scholar] [CrossRef] [PubMed]
- Mitra, M.; Patidar, S.K.; George, B.; Shah, F.; Mishra, S. A euryhaline Nannochloropsis gaditana with potential for nutraceutical (EPA) and biodiesel production. Algal Res. 2015, 8, 161–167. [Google Scholar] [CrossRef]
- López, E.N.; Medina, A.R.; Moreno, P.A.G.; Cerdán, L.E.; Valverde, L.M.; Grima, E.M. Biodiesel production from Nannochloropsis gaditana lipids through transesterification catalyzed by Rhizopus oryzae lipase. Bioresour. Technol. 2016, 203, 236–244. [Google Scholar] [CrossRef] [PubMed]
- Mitra, M.; Mishra, S. A comparative analysis of different extraction solvent systems on the extractability of eicosapentaenoic acid from the marine eustigmatophyte Nannochloropsis oceanica. Algal Res. 2019, 38, 101387. [Google Scholar] [CrossRef]
- Lai, Y.J. Omega-3 fatty acid obtained from Nannochloropsis oceanica cultures grown under low urea protect against Abeta-induced neural damage. J. Food Sci. Technol. 2015, 52, 2982–2989. [Google Scholar] [CrossRef]
- Harris, W.; Hoem, N. Effects of Different Chemical fFrms of EPA and DHA on the Omega-3 Index: A Meta-Analysis of 16 Trials. J. Clin. Lipidol. 2020, 14, 589–590. [Google Scholar] [CrossRef]
- Yen, H.W.; Hu, I.C.; Chen, C.Y.; Ho, S.H.; Lee, D.J.; Chang, J.S. Microalgae-based biorefinery—From biofuels to natural products. Bioresour. Technol. 2013, 135, 166–174. [Google Scholar] [CrossRef]
- Jacob-Lopes, E.; Maroneze, M.M.; Deprá, M.C.; Sartori, R.B.; Dias, R.R.; Zepka, L.Q. Bioactive food compounds from microalgae: An innovative framework on industrial biorefineries. In Current Opinion in Food Science; Elsevier Ltd.: Amsterdam, The Netherlands, 2019; Volume 25, pp. 1–7. [Google Scholar] [CrossRef]
- Chew, K.W.; Yap, J.Y.; Show, P.L.; Suan, N.H.; Juan, J.C.; Ling, T.C.; Lee, D.-J.; Chang, J.-S. Microalgae biorefinery: High value products perspectives. Bioresour. Technol. 2017, 229, 53–62. [Google Scholar] [CrossRef] [PubMed]
- Vanthoor-Koopmans, M.; Wijffels, R.H.; Barbosa, M.J.; Eppink, M.H.M. Biorefinery of microalgae for food and fuel. Bioresour. Technol. 2013, 135, 142–149. [Google Scholar] [CrossRef] [PubMed]
- Katiyar, R.; Arora, A. Health promoting functional lipids from microalgae pool: A review. Algal Res. 2020, 46, 101800. [Google Scholar] [CrossRef]
| Retention Time (min) | Observed m/z | Theoretical m/z | Error (ppm) | Adduct Ion Type m/z | Type of Second Adduct | Second Adduct m/z | Compound or Lipid Type | Identified Compound | Confidence | Comments |
|---|---|---|---|---|---|---|---|---|---|---|
| 3.0 | 205.0682 | 205.0680 | 1.2 | [M+Na]+ | - | Manitol Derivative | C6H14NaO6 (sodium Manitol derivative) | High | Structure suggested by SciFinder. | |
| 3.2 | 205.0680 | 205.0680 | 0.0 | [M+Na]+ | - | Manitol Derivative | C6H14NaO6 (isomer of previous compound) | High | Identified as isomer with 91.14% score. | |
| 3.4 | 197.1171 | 197.1171 | 0.0 | [M+H]+ | [M+Na]+ | 219.0988 | Fatty Acid Derivative | C11H17O3 | High | Identified with 100% score in SciFinder. Possible fatty acid derivative. |
| 4.7 | 494.3269 | 494.3270 | −0.2 | [M+NH4]+ | - | Lipid With Phosphates | C31H44NO4, C23H50N3O4P2 | High | High confidence with 100% score. C23H50N3O4P2 also identified. | |
| 4.9 | 520.3640 | 520.3645 | −0.1 | [M+H]+ | - | Lipid With Ammonium | C27H49NO6 | Medium | Score of 73.32%. Identified as possible phospholipid derivative. | |
| 8.1 | 282.2791 | 282.2789 | 0.7 | [M+H]+ | - | Amide (Erucamide) | Octadecenamide | High | Found in blank, as well; possible contamination or common background signal. | |
| 13.8 | 338.3415 | 338.3420 | −0.2 | [M+H]+ | - | Erucamide | Erucamide | High | Typical of plastic material additive; high confidence. | |
| 17.2 | 730.5373 | 730.5370 | 0.4 | [M+H]+ | [M+Na]+ | 752.5226 | Lipid Phosphates | C34H78N5O7P2 | High | Identified with 100% score. |
| 18.1 | 804.5773 | 804.5773 | 0.0 | [M+NH4]+ | - | Lipid | C50H78NO7 | Medium | Possibility of ammonium adduct. Score of 76.08%. | |
| 19.7 | 806.5927 | 806.5925 | 0.2 | [M+NH4]+ | - | Lipid | C50H80NO7 | Medium | Similar to previous peak; ammonium adduct; possibly an isomer. | |
| 20.2 | 758.5687 | 758.5690 | −0.4 | [M+NH4]+ | - | Lipid Phosphates | C34H85N3O10PS | Low | Possible diacylglycerol phosphate, with low confidence due to fragmentation. | |
| 24.0 | 840.5619 | 840.5624 | −0.6 | [M+NH4]+ | [M+Na]+ | 845.5173 | Galactosyl Diacyl Glycerol (MGDG) | C49H74NaO10 | Low | Possibility of ammonium adduct. |
| 28.4 | 600.5194 | 600.5190 | 0.6 | [M+NH4]+ | [M+Na]+ | 605.4752 | Triglyceride Ester | C35H70NO6 | Medium | Possible triglyceride esterified with fatty acids; moderate confidence. |
| 30.5 | 548.5028 | 548.5027 | 0.2 | [M+NH4]+ | [M+Na]+ | 553.4586 | Lipid Derivative | C35H66NO3 | High | Identified with high confidence as a triglyceride ester. |
| 33.2 | 871.5726 | 871.5725 | 0.1 | [M+H]+ | - | Phytopigment | Pheophytin A | High | Identified with a score of 79.34%; good match with the literature. | |
| 34.4 | 871.5719 | 871.5720 | 0.0 | [M+H]+ | - | Phytopigment | Pheophytin A isomer | Medium | Possible isomer of Pheophytin A; could indicate slight matrix effect. | |
| 35.1 | 918.8105 | 918.8100 | 0.5 | [M+NH4]+ | - | Lipid Ester | C57H108NO7 | Low | Esterified lipid with fatty acids, identified with low confidence. | |
| 36.2 | 813.5666 | 813.5660 | 0.7 | [M+H]+ | 2 [M+H]+ | 1626.12 | Lipid | C52H77O7 | Medium | Identified as lipid ester with 54.27% score; low confidence due to fragmentation. |
| 36.8 | 818.7220 | 818.7223 | −0.4 | [M+NH4]+ | - | Lipid Phosphate | C45H97N5O5P | High | High confidence; identified as lipid with nitrogen; [M+NH4]+ clearly dominant. | |
| 37.5 | 820.7375 | 820.7380 | −0.6 | [M+NH4]+ | [M+Na]+ | 825.6935 | Lipid | C51H98NO6 | Medium | Triglyceride derivative; possible fatty acid ester; moderate confidence. |
| 38.2 | 848.7688 | 848.7685 | 0.4 | [M+NH4]+ | [M+Na]+ | 853.7249 | Lipid Ester | C53H102NO6 | Medium | Esterified lipid with high score but moderate confidence. |
| 39.0 | 902.8166 | 902.8165 | 0.1 | [M+NH4]+ | [M+Na]+ | 907.7715 | Lipid Phosphates | C56H104N4O8P | Low | Possible triglyceride with ammonium adduct; further validation needed. |
| Extraction Technique | GL (%) 1 | GL (mg) 2 | Lutein (mg) 2 | EPA (%) 1 |
|---|---|---|---|---|
| PLE | ||||
| Initial time | 22.82 a | 112.73 ± 0.93 a | 4.34 ± 0.11 a | 35.24 a |
| Combined pretreatment | 33.46 b | 186.54 ± 2.11 b | 8.95 ± 0.08 b | 37.29 b |
| UAE | ||||
| Initial time | 33.51 b | 101.01 ± 1.13 c | 7.55 ± 0.45 c | 29.38 c |
| Combined pretreatment | 55.52 c | 133.36 ± 3.01 d | 8.36 ± 0.26 c | 38.63 b |
| MAE | ||||
| Initial time | 20.47 a | 110.57 ± 1.12 a | 5.79 ± 0.17 d | 34.52 d |
| Combined pretreatment | 38.90 d | 170.38 ± 0.89 e | 8.56 ± 0.63 e | 36.46 e |
| Identified Fatty Acid | Rt (min) | %FAME |
|---|---|---|
| 14:0 | 9.70 | 4.74 |
| 16:0 | 11.77 | 16.64 |
| 16:1 | 12.50 | 23.07 |
| 18:0 | 14.59 | 0.49 |
| 18:1 | 15.59 | 4.17 |
| 18:2 | 17.12 | 3.37 |
| 20:3 | 21.90 | 0.51 |
| 20:4 | 22.69 | 7.92 |
| 20:5 | 24.61 | 37.38 |
| SFA | 21.87 | |
| MUFA | 27.24 | |
| PUFA | 49.18 | |
| n-3 | 37.89 | |
| n-6 | 11.29 | |
| n-6/n-3 | 0.30 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Blanco-Llamero, C.; García-García, P.; Señoráns, F.J. Efficient Green Extraction of Nutraceutical Compounds from Nannochloropsis gaditana: A Comparative Electrospray Ionization LC-MS and GC-MS Analysis for Lipid Profiling. Foods 2024, 13, 4117. https://doi.org/10.3390/foods13244117
Blanco-Llamero C, García-García P, Señoráns FJ. Efficient Green Extraction of Nutraceutical Compounds from Nannochloropsis gaditana: A Comparative Electrospray Ionization LC-MS and GC-MS Analysis for Lipid Profiling. Foods. 2024; 13(24):4117. https://doi.org/10.3390/foods13244117
Chicago/Turabian StyleBlanco-Llamero, Cristina, Paz García-García, and Francisco Javier Señoráns. 2024. "Efficient Green Extraction of Nutraceutical Compounds from Nannochloropsis gaditana: A Comparative Electrospray Ionization LC-MS and GC-MS Analysis for Lipid Profiling" Foods 13, no. 24: 4117. https://doi.org/10.3390/foods13244117
APA StyleBlanco-Llamero, C., García-García, P., & Señoráns, F. J. (2024). Efficient Green Extraction of Nutraceutical Compounds from Nannochloropsis gaditana: A Comparative Electrospray Ionization LC-MS and GC-MS Analysis for Lipid Profiling. Foods, 13(24), 4117. https://doi.org/10.3390/foods13244117









