Fresh Meat Classification Using Laser-Induced Breakdown Spectroscopy Assisted by LightGBM and Optuna
Abstract
1. Introduction
2. Experiments
2.1. Sample Preparation
2.2. Experimental Setup and Measurement
3. Method
3.1. LightGBM
3.2. Optuna
- Define-by-run style API: Optuna provides a flexible API (application programming interface) that allows defining and optimizing hyperparameters within the code, making it easy to incorporate into workflows.
- Efficient sampling and pruning mechanism: Optuna employs efficient sampling and pruning techniques to explore the hyperparameter search space effectively and eliminate unpromising trials, thus improving efficiency.
- Easy to setup: Optuna is designed to be user-friendly and easy to set up, enabling users to quickly get started with hyperparameter optimization.
- (1)
- Define an objective function that takes a set of hyperparameters as input and returns the metrics representing model performance (such as the accuracy of the validation set, root mean square error (RMSE), and multi-loss). Additionally, specify the range of hyperparameters that need to be adjusted, including the distribution type and value range for each hyperparameter.
- (2)
- Create an Optuna study to minimize or maximize the objective function and set the number of trials in a study. In each trial, Optuna finds a set of hyperparameters and passes them into the objective function. The sampling methods were used to traverse the hyperparameter space.
- (3)
- Get the result, the best hyperparameter combination at the end of all trials. The “plot_optimization_history (study) API” can be used to observe the trend of the objective function’s value increasing or decreasing.
- (4)
- Apply the optimal hyperparameters to the classification model and test it on the test set and determine if further optimization is needed.
4. Result and Discussion
4.1. Classification with SVM
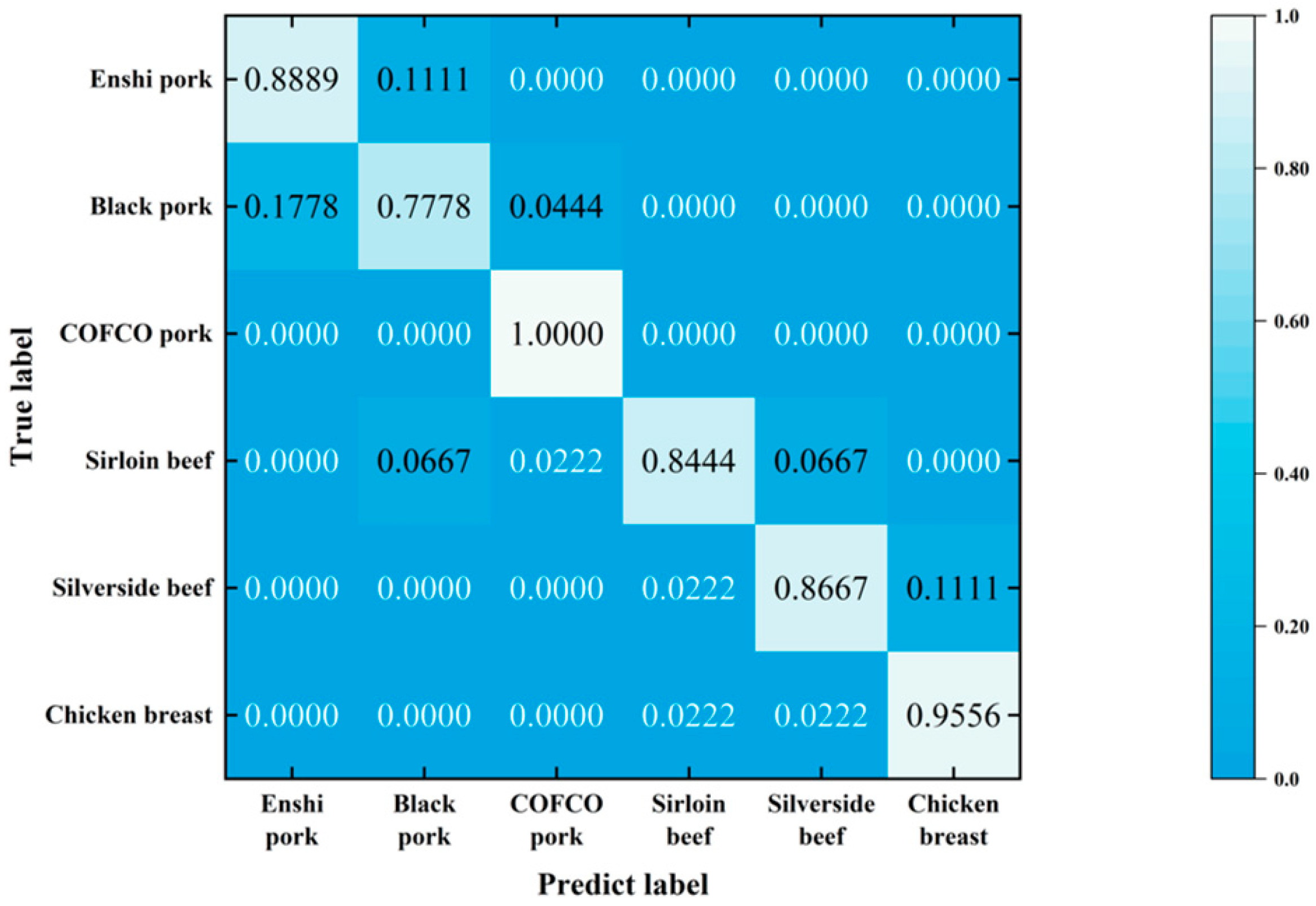
4.2. Classification with LightGBM
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Boyacı, İ.H.; Temiz, H.T.; Uysal, R.S.; Velioğlu, H.M.; Yadegari, R.J.; Rishkan, M.M. A novel method for discrimination of beef and horsemeat using Raman spectroscopy. Food Chem. 2014, 148, 37–41. [Google Scholar] [CrossRef] [PubMed]
- Vallejo-Cordoba, B.; González-Córdova, A.F.; Mazorra-Manzano, M.A.; Rodríguez-Ramírez, R. Caillary electrophoresis for the analysis of meat authenticity. J. Sep. Sci. 2005, 28, 826–836. [Google Scholar] [CrossRef] [PubMed]
- Rahman, M.M.; Ali, M.E.; Hamid, S.B.A.; Mustafa, S.; Hashim, U.; Hanapi, U.K. Polymerase chain reaction assay targeting cytochrome b gene for the detection of dog meat adulteration in meatball formulation. Meat Sci. 2014, 97, 404–409. [Google Scholar] [CrossRef] [PubMed]
- Nurjuliana, M.; Che Man, Y.B.; Mat Hashim, D.; Mohamed, A.K.S. Rapid identification of pork for halal authentication using the electronic nose and gas chromatography mass spectrometer with headspace analyzer. Meat Sci. 2011, 88, 638–644. [Google Scholar] [CrossRef] [PubMed]
- Floren, C.; Wiedemann, I.; Brenig, B.; Schütz, E.; Beck, J. Species identification and quantification in meat and meat products using droplet digital PCR (ddPCR). Food Chem. 2015, 173, 1054–1058. [Google Scholar] [CrossRef] [PubMed]
- Hellberg, R.S.; Hernandez, B.C.; Hernandez, E.L. Identification of meat and poultry species in food products using DNA barcoding. Food Control 2017, 80, 23–28. [Google Scholar] [CrossRef]
- Ivanova, B. Special Issue with Research Topics on “Recent Analysis and Applications of Mass Spectra on Biochemistry”. Int. J. Mol. Sci. 2024, 25, 1995. [Google Scholar] [CrossRef] [PubMed]
- Windarsih, A.; Suratno; Warmiko, H.D.; Indrianingsih, A.W.; Rohman, A.; Ulumuddin, Y.I. Untargeted metabolomics and proteomics approach using liquid chromatography-Orbitrap high resolution mass spectrometry to detect pork adulteration in Pangasius hypopthalmus meat. Food Chem. 2022, 386, 132856. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Liu, M.; Wang, S.; Kang, C.; Zhang, M.; Li, Y. Identification and quantification of fox meat in meat products by liquid chromatography–tandem mass spectrometry. Food Chem. 2022, 372, 131336. [Google Scholar] [CrossRef]
- Erasmus, S.W.; Muller, M.; Alewijn, M.; Koot, A.H.; van Ruth, S.M.; Hoffman, L.C. Proton-transfer reaction mass spectrometry (PTR-MS) for the authentication of regionally unique South African lamb. Food Chem. 2017, 233, 331–342. [Google Scholar] [CrossRef]
- Pu, K.; Qiu, J.; Li, J.; Huang, W.; Lai, X.; Liu, C.; Lin, Y.; Ng, K.-M. MALDI-TOF MS Protein Profiling Combined with Multivariate Analysis for Identification and Quantitation of Beef Adulteration. Food Anal. Methods 2023, 16, 132–142. [Google Scholar] [CrossRef]
- Zou, Z.; Deng, Y.; Hu, J.; Jiang, X.; Hou, X. Recent trends in atomic fluorescence spectrometry towards miniaturized instrumentation-A review. Anal. Chim. Acta 2018, 1019, 25–37. [Google Scholar] [CrossRef] [PubMed]
- Vu, D.M.; Auxier, J.D.; Judge, E.J.; Aldrich, K.E.; Gifford, B.J.; Saumon, D.; Neukirch, A.J.; Auxier, J.P.; Barefield, J.E.; Clegg, S.M.; et al. A data analysis method to rapidly characterize gallium concentration in plutonium matrices using LIBS. Spectrochim. Acta Part B At. Spectrosc. 2023, 203, 106650. [Google Scholar] [CrossRef]
- Pagnotta, S.; Lezzerini, M.; Ripoll-Seguer, L.; Hidalgo, M.; Grifoni, E.; Legnaioli, S.; Lorenzetti, G.; Poggialini, F.; Palleschi, V. Micro-Laser-Induced Breakdown Spectroscopy (Micro-LIBS) Study on Ancient Roman Mortars. Appl. Spectrosc. 2017, 71, 721–727. [Google Scholar] [CrossRef]
- Girón, D.; Delgado, T.; Ruiz, J.; Cabalín, L.M.; Laserna, J.J. In-situ monitoring and characterization of airborne solid particles in the hostile environment of a steel industry using stand-off LIBS. Measurement 2018, 115, 1–10. [Google Scholar] [CrossRef]
- Guo, L.B.; Zhu, Z.H.; Li, J.M.; Tang, Y.; Tang, S.S.; Hao, Z.Q.; Li, X.Y.; Lu, Y.F.; Zeng, X.Y. Determination of boron with molecular emission using laser-induced breakdown spectroscopy combined with laser-induced radical fluorescence. Opt. Express 2018, 26, 2634–2642. [Google Scholar] [CrossRef] [PubMed]
- Guo, Y.M.; Guo, L.B.; Hao, Z.Q.; Tang, Y.; Ma, S.X.; Zeng, Q.D.; Tang, S.S.; Li, X.Y.; Lu, Y.F.; Zeng, X.Y. Accuracy improvement of iron ore analysis using laser-induced breakdown spectroscopy with a hybrid sparse partial least squares and least-squares support vector machine model. J. Anal. At. Spectrom. 2018, 33, 1330–1335. [Google Scholar] [CrossRef]
- Garcia, J.A.; da Silva, J.R.A.; Pereira-Filho, E.R. LIBS as an alternative method to control an industrial hydrometallurgical process for the recovery of Cu in waste from electro-electronic equipment (WEEE). Microchem. J. 2021, 164, 106007. [Google Scholar] [CrossRef]
- Silva, T.V.; Milori, D.M.B.P.; Neto, J.A.G.; Ferreira, E.J.; Ferreira, E.C. Prediction of black, immature and sour defective beans in coffee blends by using Laser-Induced Breakdown Spectroscopy. Food Chem. 2019, 278, 223–227. [Google Scholar] [CrossRef]
- Tian, Y.; Chen, Q.; Lin, Y.; Lu, Y.; Li, Y.; Lin, H. Quantitative determination of phosphorus in seafood using laser-induced breakdown spectroscopy combined with machine learning. Spectrochim. Acta Part B At. Spectrosc. 2021, 175, 106027. [Google Scholar] [CrossRef]
- Yang, P.; Zhou, R.; Zhang, W.; Yi, R.; Tang, S.; Guo, L.; Hao, Z.; Li, X.; Lu, Y.; Zeng, X. High-sensitivity determination of cadmium and lead in rice using laser-induced breakdown spectroscopy. Food Chem. 2019, 272, 323–328. [Google Scholar] [CrossRef]
- Viana, L.F.; Súarez, Y.R.; Cardoso, C.A.L.; Lima, S.M.; Andrade, L.H.d.C.; Lima-Junior, S.E. Use of fish scales in environmental monitoring by the application of Laser-Induced Breakdown Spectroscopy (LIBS). Chemosphere 2019, 228, 258–263. [Google Scholar] [CrossRef] [PubMed]
- Gaudiuso, R.; Ewusi-Annan, E.; Melikechi, N.; Sun, X.; Liu, B.; Campesato, L.F.; Merghoub, T. Using LIBS to diagnose melanoma in biomedical fluids deposited on solid substrates: Limits of direct spectral analysis and capability of machine learning. Spectrochim. Acta Part B At. Spectrosc. 2018, 146, 106–114. [Google Scholar] [CrossRef]
- Skalny, A.V.; Korobeinikova, T.V.; Aschner, M.; Baranova, O.V.; Barbounis, E.G.; Tsatsakis, A.; Tinkov, A.A. Medical application of laser-induced breakdown spectroscopy (LIBS) for assessment of trace element and mineral in biosamples: Laboratory and clinical validity of the method. J. Trace Elem. Med. Biol. 2023, 79, 127241. [Google Scholar] [CrossRef] [PubMed]
- Beck, P.; Meslin, P.Y.; Fau, A.; Forni, O.; Gasnault, O.; Lasue, J.; Cousin, A.; Schröder, S.; Maurice, S.; Rapin, W.; et al. Detectability of carbon with ChemCam LIBS: Distinguishing sample from Mars atmospheric carbon, and application to Gale crater. Icarus 2024, 408, 115840. [Google Scholar] [CrossRef]
- Bilge, G.; Velioglu, H.M.; Sezer, B.; Eseller, K.E.; Boyaci, I.H. Identification of meat species by using laser-induced breakdown spectroscopy. Meat Sci. 2016, 119, 118–122. [Google Scholar] [CrossRef] [PubMed]
- Casado-Gavalda, M.P.; Dixit, Y.; Geulen, D.; Cama-Moncunill, R.; Cama-Moncunill, X.; Markiewicz-Keszycka, M.; Cullen, P.J.; Sullivan, C. Quantification of copper content with laser induced breakdown spectroscopy as a potential indicator of offal adulteration in beef. Talanta 2017, 169, 123–129. [Google Scholar] [CrossRef] [PubMed]
- Sezer, B.; Durna, S.; Bilge, G.; Berkkan, A.; Yetisemiyen, A.; Boyaci, I.H. Identification of milk fraud using laser-induced breakdown spectroscopy (LIBS). Int. Dairy J. 2018, 81, 1–7. [Google Scholar] [CrossRef]
- Chu, Y.W.; Tang, S.S.; Ma, S.X.; Ma, Y.Y.; Hao, Z.Q.; Guo, Y.M.; Guo, L.B.; Lu, Y.F.; Zeng, X.Y. Accuracy and stability improvement for meat species identification using multiplicative scatter correction and laser-induced breakdown spectroscopy. Opt. Express 2018, 26, 10119–10127. [Google Scholar] [CrossRef]
- Velioglu, H.M.; Sezer, B.; Bilge, G.; Baytur, S.E.; Boyaci, I.H. Identification of offal adulteration in beef by laser induced breakdown spectroscopy (LIBS). Meat Sci. 2018, 138, 28–33. [Google Scholar] [CrossRef]
- Chen, C.; Zhang, Q.; Ma, Q.; Yu, B. LightGBM-PPI: Predicting protein-protein interactions through LightGBM with multi-information fusion. Chemom. Intell. Lab. Syst. 2019, 191, 54–64. [Google Scholar] [CrossRef]
- Sezer, B.; Bjelak, A.; Velioglu, H.M.; Boyaci, I.H. Protein based evaluation of meat species by using laser induced breakdown spectroscopy. Meat Sci. 2021, 172, 108361. [Google Scholar] [CrossRef] [PubMed]
- Wei, J.; Li, Z.; Pinker, R.T.; Wang, J.; Sun, L.; Xue, W.; Li, R.; Cribb, M. Himawari-8-derived diurnal variations in ground-level PM2.5 pollution across China using the fast space-time Light Gradient Boosting Machine (LightGBM). Atmos. Chem. Phys. 2021, 21, 7863–7880. [Google Scholar] [CrossRef]
- Ke, G.; Meng, Q.; Finley, T.; Wang, T.; Chen, W.; Ma, W.; Ye, Q.; Liu, T.-Y. LightGBM: A highly efficient gradient boosting decision tree. In Proceedings of the 31st International Conference on Neural Information Processing Systems, Long Beach, CA, USA, 4–9 December 2017; pp. 3149–3157. [Google Scholar]
- Akiba, T.; Sano, S.; Yanase, T.; Ohta, T.; Koyama, M. Optuna: A Next-generation Hyperparameter Optimization Framework. In Proceedings of the 25th ACM SIGKDD International Conference on Knowledge Discovery & Data Mining, Anchorage, AK, USA, 4–8 August 2019; pp. 2623–2631. [Google Scholar] [CrossRef]






| Parameters | Interpretation |
|---|---|
| num_leaves | This parameter determines the number of leaves per tree. |
| learning_rate | This parameter controls the speed of iterations in the training process. |
| max_depth | This parameter sets the maximum depth of the tree. |
| min_data | This parameter defines the minimum number of records that a leaf node should have. |
| feature_fraction | This parameter specifies the fraction of features to be selected at each iteration. |
| bagging_fraction | This parameter determines the fraction of data to be used for each iteration through bagging. |
| Parameters | Optimal Values |
|---|---|
| num_leaves | 34 |
| learning_rate | 0.05343712612981269 |
| max_depth | 8 |
| min_data_in_leaf | 26 |
| feature_fraction | 0.13 |
| bagging_fraction | 0.64 |
| max_bin | 213 |
| lambda_l1 | 0 |
| lambda_l2 | 0 |
| min_gain_to_split | 0.005296940015136468 |
| Model | Kappa | Macro-F1 | Accuracy |
|---|---|---|---|
| SVM | 0.8666 | 0.8881 | 88.88% |
| LightGBM | 0.9244 | 0.9364 | 93.70% |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mo, K.; Tang, Y.; Zhu, Y.; Li, X.; Li, J.; Peng, X.; Liao, P.; Zou, P. Fresh Meat Classification Using Laser-Induced Breakdown Spectroscopy Assisted by LightGBM and Optuna. Foods 2024, 13, 2028. https://doi.org/10.3390/foods13132028
Mo K, Tang Y, Zhu Y, Li X, Li J, Peng X, Liao P, Zou P. Fresh Meat Classification Using Laser-Induced Breakdown Spectroscopy Assisted by LightGBM and Optuna. Foods. 2024; 13(13):2028. https://doi.org/10.3390/foods13132028
Chicago/Turabian StyleMo, Kaifeng, Yun Tang, Yining Zhu, Xiangyou Li, Jingfeng Li, Xuxiang Peng, Ping Liao, and Penghui Zou. 2024. "Fresh Meat Classification Using Laser-Induced Breakdown Spectroscopy Assisted by LightGBM and Optuna" Foods 13, no. 13: 2028. https://doi.org/10.3390/foods13132028
APA StyleMo, K., Tang, Y., Zhu, Y., Li, X., Li, J., Peng, X., Liao, P., & Zou, P. (2024). Fresh Meat Classification Using Laser-Induced Breakdown Spectroscopy Assisted by LightGBM and Optuna. Foods, 13(13), 2028. https://doi.org/10.3390/foods13132028






