Characterization of a Ligilactobacillus salivarius Strain Isolated from a Cheese Seal Which Was Last Used in 1936
Abstract
1. Introduction
2. Materials and Methods
2.1. In Search for the Seal
2.2. Collection of Biological Material from the Seal
2.3. ELISA Detection of Ovine- and Bovine-Specific Caseins and β-Lactoglobulins
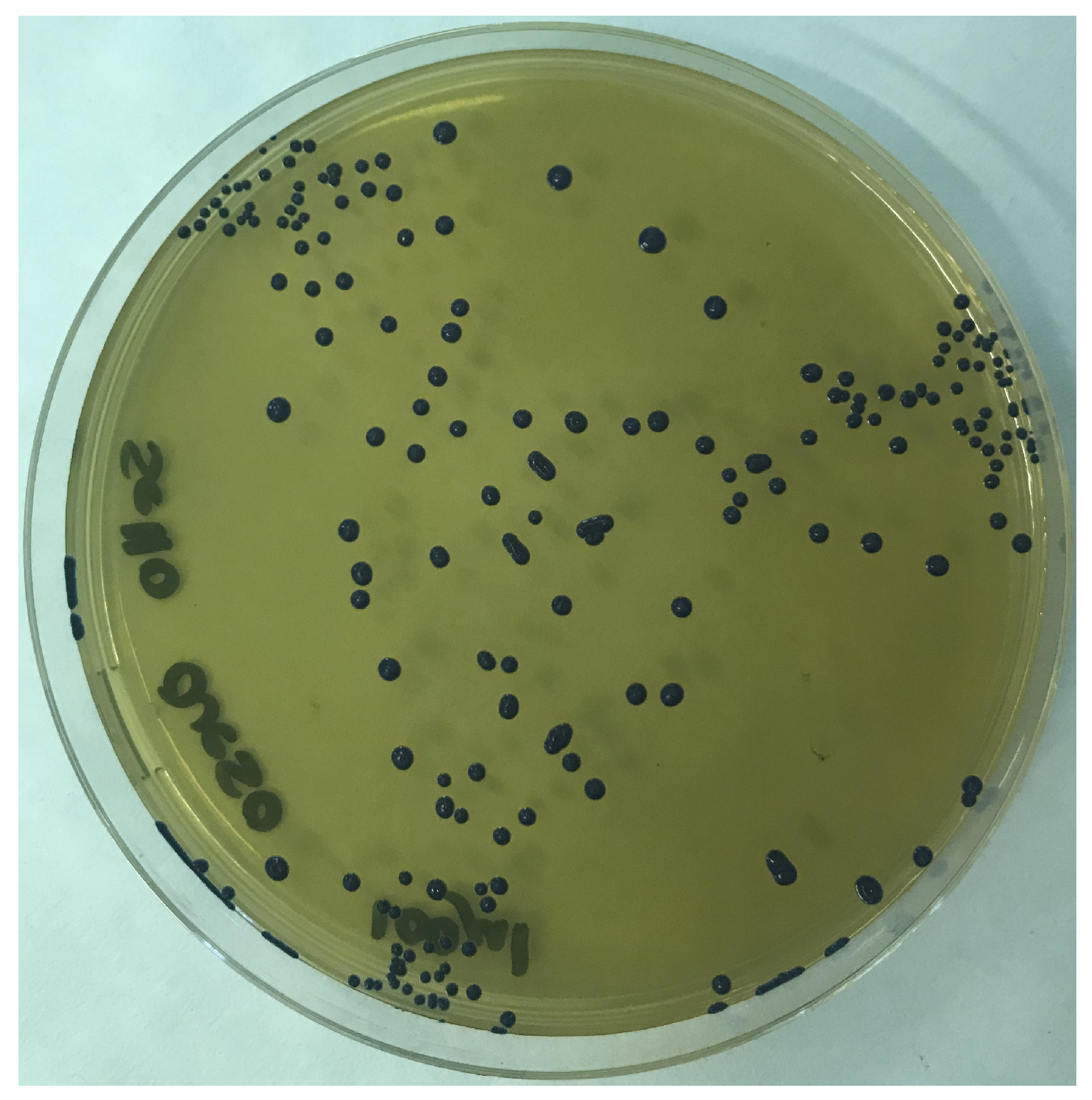
2.4. Culture of the Biological Material
2.5. In Vitro Characterization of L. salivarius SP36
2.6. Metataxonomic Analysis of the Milky Crust of the Seal
2.7. Whole Genome Sequencing and Mining of L. salivarius SP36
3. Results
3.1. ELISA Detection of Ovine- and Bovine-Specific Caseins and β-Lactoglobulins
3.2. Isolation and In Vitro Characterization of L. salivarius SP36
3.3. Metataxonomic Analysis of the Milky Crust of the Seal
3.4. Genome Sequencing and Mining of L. salivarius SP36
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Salque, M.; Bogucki, P.I.; Pyzel, J.; Sobkowiak-Tabaka, I.; Grygiel, R.; Szmyt, M.; Evershed, R.P. Earliest evidence for cheese making in the sixth millennium BC in northern Europe. Nature 2013, 493, 522–525. [Google Scholar] [CrossRef]
- Krausmann, F. Milk, manure and muscular power. Livestock and the industrialization of Agriculture. Hum. Ecol. 2004, 32, 735–773. [Google Scholar] [CrossRef]
- McClure, S.B.; Magill, C.; Podrug, E.; Moore, A.M.T.; Harper, T.K.; Culleton, B.J.; Kennett, D.J.; Freeman, K.H. Fatty acid specific δ13C values reveal earliest Mediterranean cheese production 7200 years ago. PLoS ONE 2018, 13, e0202807. [Google Scholar] [CrossRef] [PubMed]
- Dias, C.; Mendes, L. Protected Designation of Origin (PDO), Protected Geographical Indication (PGI) and Traditional Speciality Guaranteed (TSG): A bibiliometric analysis. Food Res. Int. 2018, 103, 492–508. [Google Scholar] [CrossRef]
- Muchnik, J.; Biénabe, E.; Cerdan, C. Food identity/food quality: Insights from the “coalho” cheese in the Northeast of Brazil. Anthropol. Food 2005, 4. [Google Scholar] [CrossRef]
- Bale, M.J.; Bennett, P.M.; Beringer, J.E.; Hinton, M. The survival of bacteria exposed to desiccation on surfaces associated with farm buildings. J. Appl. Bacteriol. 1993, 75, 519–528. [Google Scholar] [CrossRef] [PubMed]
- Beuchat, L.; Komitopoulou, E. Persistence and Survival of Pathogens in Dry Foods and Dry Food Processing Environments; ILSI Europe Report Series; ILSI Europe: Brussels, Belgium, 2011; pp. 1–48. [Google Scholar]
- Juma, N.A.; Manning, G.; Forsythe, S.J. Desiccation survival of Acinetobacter spp. in infant formula. Food Control 2016, 68, 162–166. [Google Scholar] [CrossRef]
- Chen, G.; Lin, M.; Chen, Y.; Xu, W.; Zhang, H. Induction of a viable but nonculturable state, thermal and sanitizer tolerance, and gene expression correlation with desiccation-adapted biofilm and planktonic Salmonella in powdered infant formula. J. Food Prot. 2021, 84, 1194–1201. [Google Scholar] [CrossRef]
- Martín, V.; Mediano, P.; Del Campo, R.; Rodríguez, J.M.; Marín, M. Streptococcal diversity of human milk and comparison of different methods for the taxonomic identification of streptococci. J. Hum. Lact. 2016, 32, NP84–NP94. [Google Scholar] [CrossRef]
- Marín, M.; Arroyo, R.; Espinosa-Martos, I.; Fernández, L.; Rodríguez, J.M. Identification of emerging human mastitis pathogens by MALDI-TOF and assessment of their antibiotic resistance patterns. Front. Microbiol. 2017, 8, 1258. [Google Scholar] [CrossRef]
- Martín, V.; Cárdenas, N.; Ocaña, S.; Marín, M.; Arroyo, R.; Beltrán, D.; Badiola, C.; Fernández, L.; Rodríguez, J.M. Rectal and vaginal eradication of Streptococcus agalactiae (GBS) in pregnant women by using Lactobacillus salivarius CECT 9145, a target-specific probiotic strain. Nutrients 2019, 11, 810. [Google Scholar] [CrossRef]
- Cárdenas, N.; Martín, V.; Arroyo, R.; López, M.; Carrera, M.; Badiola, C.; Jiménez, E.; Rodríguez, J.M. Prevention of recurrent acute otitis media in children through the use of Lactobacillus salivarius PS7, a target-specific probiotic strain. Nutrients 2019, 11, 376. [Google Scholar] [CrossRef] [PubMed]
- Magnusson, J.; Schnürer, J. Lactobacillus coryniformis subsp. coryniformis strain Si3 produces a broad-spectrum proteinaceous antifungal compound. Appl. Environ. Microbiol. 2001, 67, 1–5. [Google Scholar] [CrossRef] [PubMed]
- Martín, R.; Jiménez, E.; Olivares, M.; Marín, M.L.; Fernández, L.; Xaus, J.; Rodríguez, J.M. Lactobacillus salivarius CECT 5713, a potential probiotic strain isolated from infant feces and breast milk of a mother-child pair. Int. J. Food Microbiol. 2006, 112, 35–43. [Google Scholar] [CrossRef] [PubMed]
- Song, Y.L.; Kato, N.; Matsumiya, Y.; Liu, C.X.; Kato, H.; Watanabe, K. Identification of and hydrogen peroxide production by fecal and vaginal lactobacilli isolated from Japanese women and newborn infants. J. Clin. Microbiol. 1999, 37, 3062–3064. [Google Scholar] [CrossRef] [PubMed]
- Yap, P.S.; Gilliland, S.E. Comparison of newly isolated strains of Lactobacillus delbrueckii subsp. lactis for hydrogen peroxide production at 5 degrees C. J. Dairy Sci. 2000, 83, 628–632. [Google Scholar] [CrossRef]
- Cohen, P.S.; Laux, D.C. Bacterial adhesion to and penetration of intestinal mucus in vitro. Methods Enzymol. 1995, 253, 309–314. [Google Scholar] [CrossRef] [PubMed]
- Marteau, P.; Minekus, M.; Havenaar, R.; Huis in’t Veld, J.H. Survival of lactic acid bacteria in a dynamic model of the stomach and small intestine: Validation and the effects of bile. J. Dairy Sci. 1997, 80, 1031–1037. [Google Scholar] [CrossRef] [PubMed]
- EFSA; Rychen, G.; Aquilina, G.; Azimonti, G.; Bampidis, V.; Bastos, M.L.; Bories, G.; Chesson, A.; Cocconcelli, P.S.; Flachowsky, G.; et al. Guidance on the characterisation of microorganisms used as feed additives or as production organisms. EFSA J. 2018, 16, e05206. [Google Scholar]
- Bover-Cid, S.; Holzapfel, W.H. Improved screening procedure for biogenic amine production by lactic acid bacteria. Int. J. Food Microbiol. 1999, 53, 33–41. [Google Scholar] [CrossRef]
- Zhou, J.S.; Gopal, P.K.; Hill, H.S. Potential probiotic lactic acid bacteria Lactobacillus rhamnosus (HN001), Lactobacillus acidophilus (HN017) and Bifidobacterium lactis (HN019) do not degrade gastric mucin in vitro. Int. J. Food Microbiol. 2001, 63, 81–90. [Google Scholar] [CrossRef] [PubMed]
- Lackey, K.A.; Williams, J.E.; Meehan, C.L.; Zachek, J.A.; Benda, E.D.; Price, W.J.; Foster, J.A.; Sellen, D.W.; Kamau-Mbuthia, E.W.; Kamundia, E.W.; et al. What’s normal? Microbiomes in human milk and infant feces are related to each other but vary geographically: The INSPIRE study. Front. Nutr. 2019, 6, 45. [Google Scholar] [CrossRef] [PubMed]
- Klindworth, A.; Pruesse, E.; Schweer, T.; Peplies, J.; Quast, C.; Horn, M.; Glöckner, F.O. Evaluation of general 16S ribosomal RNA gene PCR primers for classical and next-generation sequencing-based diversity studies. Nucleic Acids Res. 2013, 41, e1. [Google Scholar] [CrossRef] [PubMed]
- Palau, R.; Bloomfield, S.J.; Jenkins, C.; Greig, D.R.; Jorgensen, F.; Mather, A.E. Yersinia enterocolitica biovar 1A: An underappreciated potential pathogen in the food chain. Int. J. Food Microbiol. 2024, 412, 110554. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 2018, 34, i884–i890. [Google Scholar] [CrossRef] [PubMed]
- Wood, D.E.; Salzberg, S.L. Kraken: Ultrafast metagenomic sequence classification using exact alignments. Genome Biol. 2014, 15, R46. [Google Scholar] [CrossRef] [PubMed]
- Seemann, T. Shovill: Faster SPAdes Assembly of Illumina Reads. Available online: https://github.com/tseemann/shovill (accessed on 14 March 2024).
- Parks, D.H.; Imelfort, M.; Skennerton, C.T.; Hugenholtz, P.; Tyson, G.W. CheckM: Assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Res. 2015, 25, 1043–1055. [Google Scholar] [CrossRef] [PubMed]
- Krawczyk, P.S.; Lipinski, L.; Dziembowski, A. PlasFlow: Predicting plasmid sequences in metagenomic data using genome signatures. Nucleic Acids Res. 2018, 46, e35. [Google Scholar] [CrossRef] [PubMed]
- Seemann, T. ABRicate: Mass Screening of Contigs for Antiobiotic Resistance Genes. Available online: https://github.com/tseemann/abricate (accessed on 14 March 2024).
- Gurevich, A.; Saveliev, V.; Vyahhi, N.; Tesler, G. QUAST: Quality assessment tool for genome assemblies. Bioinformatics 2013, 29, 1072–1075. [Google Scholar] [CrossRef]
- Starikova, E.V.; Tikhonova, P.O.; Prianichnikov, N.A.; Rands, C.M.; Zdobnov, E.M.; Ilina, E.N.; Govorun, V.M. Phigaro: High-throughput prophage sequence annotation. Bioinformatics 2020, 36, 3882–3884. [Google Scholar] [CrossRef]
- Alvarez-Sieiro, P.; Montalbán-López, M.; Mu, D.; Kuipers, O.P. Bacteriocins of lactic acid bacteria: Extending the family. Appl. Microbiol. Biotechnol. 2016, 100, 2939–2951. [Google Scholar] [CrossRef] [PubMed]
- Urbach, E.; Daniels, B.; Salama, M.S.; Sandine, W.E.; Giovannoni, S.J. The ldh phylogeny for environmental isolates of Lactococcus lactis is consistent with rRNA genotypes but not with phenotypes. Appl. Environ. Microbiol. 1997, 63, 694–702. [Google Scholar] [CrossRef]
- Karaseva, O.; Ozhegov, G.; Khusnutdinova, D.; Siniagina, M.; Anisimova, E.; Akhatova, F.; Fakhrullin, R.; Yarullina, D. Whole genome sequencing of the novel probiotic strain Lactiplantibacillus plantarum FCa3L. Microorganisms 2023, 11, 1234. [Google Scholar] [CrossRef]
- Bhukya, K.K.; Bhukya, B. Unraveling the probiotic efficiency of bacterium Pediococcus pentosaceus OBK05 isolated from buttermilk: An in vitro study for cholesterol assimilation potential and antibiotic resistance status. PLoS ONE 2021, 16, e0259702. [Google Scholar] [CrossRef] [PubMed]
- Foley, M.H.; O’Flaherty, S.; Allen, G.; Rivera, A.J.; Stewart, A.K.; Barrangou, R.; Theriot, C.M. Lactobacillus bile salt hydrolase substrate specificity governs bacterial fitness and host colonization. Proc. Natl. Acad. Sci. USA 2021, 118, e2017709118. [Google Scholar] [CrossRef]
- Langa, S.; Maldonado-Barragán, A.; Delgado, S.; Martín, R.; Martín, V.; Jiménez, E.; Ruíz-Barba, J.L.; Mayo, B.; Connor, R.I.; Suárez, J.E.; et al. Characterization of Lactobacillus salivarius CECT 5713. A strain isolated from human milk: From genotype to phenotype. Appl. Microbiol. Biotechnol. 2012, 94, 1279–1287. [Google Scholar] [CrossRef]
- Derunets, A.S.; Selimzyanova, A.I.; Rykov, S.V.; Kuznetsov, A.E.; Berezina, O.V. Strategies to enhance stress tolerance in lactic acid bacteria across diverse stress conditions. World J. Microbiol. Biotechnol. 2024, 40, 126. [Google Scholar] [CrossRef] [PubMed]
- Fernández, M.; Zúñiga, M. Amino acid catabolic pathways of lactic acid bacteria. Crit. Rev. Microbiol. 2006, 32, 155–183. [Google Scholar] [CrossRef]
- Abdul Hakim, B.N.; Xuan, N.J.; Oslan, S.N.H. A Comprehensive review of bioactive compounds from lactic acid bacteria: Potential functions as functional food in dietetics and the food industry. Foods 2023, 12, 2850. [Google Scholar] [CrossRef]
- Smit, G.; Smit, B.A.; Engels, W.J. Flavour formation by lactic acid bacteria and biochemical flavour profiling of cheese products. FEMS Microbiol. Rev. 2005, 29, 591–610. [Google Scholar] [CrossRef]
- Zheng, J.; Wittouck, S.; Salvetti, E.; Franz, C.M.A.P.; Harris, H.M.B.; Mattarelli, P.; O’Toole, P.W.; Pot, B.; Vandamme, P.; Walter, J.; et al. A taxonomic note on the genus Lactobacillus: Description of 23 novel genera, emended description of the genus Lactobacillus Beijerinck 1901, and union of Lactobacillaceae and Leuconostocaceae. Int. J. Syst. Evol. Microbiol. 2020, 70, 2782–2858. [Google Scholar] [CrossRef] [PubMed]
- Dréan, P.; McAuley, C.M.; Moore, S.C.; Fegan, N.; Fox, E.M. Characterization of the spore-forming Bacillus cereus sensu lato group and Clostridium perfringens bacteria isolated from the Australian dairy farm environment. BMC Microbiol. 2015, 15, 38. [Google Scholar] [CrossRef] [PubMed]
- Knuth, R.M.; Stewart, W.C.; Taylor, J.B.; Bisha, B.; Yeoman, C.J.; Van Emon, M.L.; Murphy, T.W. Relationships among intramammary health, udder and teat characteristics, and productivity of extensively managed ewes. J. Anim. Sci. 2021, 99, skab059. [Google Scholar] [CrossRef] [PubMed]
- Ozturkoglu-Budak, S.; Wiebenga, A.; Bron, P.A.; de Vries, R.P. Protease and lipase activities of fungal and bacterial strains derived from an artisanal raw ewe’s milk cheese. Int. J. Food Microbiol. 2016, 237, 17–27. [Google Scholar] [CrossRef] [PubMed]
- Tirloni, E.; Bernardi, C.; Celandroni, F.; Mazzantini, D.; Massimino, M.; Stella, S.; Ghelardi, E. Prevalence, virulence potential, and growth in cheese of Bacillus cereus strains isolated from fresh and short-ripened cheeses sold on the Italian market. Microorganisms 2023, 11, 521. [Google Scholar] [CrossRef] [PubMed]
- Ronimus, R.S.; Rueckert, A.; Morgan, H.W. Survival of thermophilic spore-forming bacteria in a 90+ year old milk powder from Ernest Shackelton’s Cape Royds Hut in Antarctica. J. Dairy Res. 2006, 73, 235–243. [Google Scholar] [CrossRef] [PubMed]
- Balasubramanian, B.; Soundharrajan, I.; Al-Dhabi, N.A.; Vijayaraghavan, P.; Balasubramanian, K.; Valan Arasu, M.; Choi, K.C. Probiotic characteristics of Ligilactobacillus salivarius AS22 isolated from sheep dung and its application in corn-fox tail millet silage. Appl. Sci. 2021, 11, 9447. [Google Scholar] [CrossRef]
- Carbonne, C.; Chadi, S.; Kropp, C.; Molimard, L.; Chain, F.; Langella, P.; Martin, R. Ligilactobacillus salivarius CNCM I-4866, a potential probiotic candidate, shows anti-inflammatory properties in vitro and in vivo. Front. Microbiol. 2023, 14, 1270974. [Google Scholar] [CrossRef]
- Martín, R.; Delgado, S.; Maldonado, A.; Jiménez, E.; Olivares, M.; Fernández, L.; Sobrino, O.J.; Rodríguez, J.M. Isolation of lactobacilli from sow milk and evaluation of their probiotic potential. J. Dairy Res. 2009, 76, 418–425. [Google Scholar] [CrossRef]
- Diaz, M.A.; Bik, E.M.; Carlin, K.P.; Venn-Watson, S.K.; Jensen, E.D.; Jones, S.E.; Gaston, E.P.; Relman, D.A.; Versalovic, J. Identification of Lactobacillus strains with probiotic features from the bottlenose dolphin (Tursiops truncatus). J. Appl. Microbiol. 2013, 115, 1037–1051. [Google Scholar] [CrossRef]
- Quilodrán-Vega, S.; Albarracin, L.; Mansilla, F.; Arce, L.; Zhou, B.; Islam, M.A.; Tomokiyo, M.; Al Kassaa, I.; Suda, Y.; Kitazawa, H.; et al. functional and genomic characterization of Ligilactobacillus salivarius TUCO-L2 isolated from Lama glama milk: A promising immunobiotic strain to combat infections. Front. Microbiol. 2020, 11, 608752. [Google Scholar] [CrossRef] [PubMed]
- Jobby, R.; Flora, Y.; Bora, A.; Jha, P.; Kawalkar, H.; Desai, N. Exploring probiotic activity of Lactobacillus sp. isolated from indigenous breeds of cattle milk and fecal samples in Bhatan village, MH., IN. Curr. Microbiol. 2020, 77, 1184–1190. [Google Scholar] [CrossRef] [PubMed]
- Sobrino, O.J.; Alba, C.; Arroyo, R.; Pérez, I.; Sariego, L.; Delgado, S.; Fernández, L.; de María, J.; Fumanal, P.; Fumanal, A.; et al. Replacement of metaphylactic antimicrobial therapy by oral administration of Ligilactobacillus salivarius MP100 in a pig farm. Front. Vet. Sci. 2021, 8, 666887. [Google Scholar] [CrossRef] [PubMed]
- Guerrero Sanchez, M.; Passot, S.; Campoy, S.; Olivares, M.; Fonseca, F. Ligilactobacillus salivarius functionalities, applications, and manufacturing challenges. Appl. Microbiol. Biotechnol. 2022, 106, 57–80. [Google Scholar] [CrossRef] [PubMed]
- Buahom, J.; Siripornadulsil, S.; Sukon, P.; Sooksawat, T.; Siripornadulsil, W. Survivability of freeze- and spray-dried probiotics and their effects on the growth and health performance of broilers. Vet. World. 2023, 16, 1849–1865. [Google Scholar] [CrossRef] [PubMed]
- Esteban-Blanco, C.; Gutiérrez-Gil, B.; Marina, H.; Pelayo, R.; Suárez-Vega, A.; Acedo, A.; Arranz, J.J. The milk microbiota of the Spanish churra sheep breed: New insights into the complexity of the milk microbiome of dairy species. Animals 2020, 10, 1463. [Google Scholar] [CrossRef] [PubMed]
- Lysitsas, M.; Spyrou, V.; Billinis, C.; Valiakos, G. Coagulase-negative staphylococci as an etiologic agent of ovine mastitis, with a focus on subclinical forms. Antibiotics 2023, 12, 1661. [Google Scholar] [CrossRef] [PubMed]
- Zhou, M.; Huang, F.; Du, X.; Wang, C.; Liu, G. Microbial quality of donkey milk during lactation stages. Foods 2023, 12, 4272. [Google Scholar] [CrossRef]
- Zhang, J.; Liu, X.; Usman, T.; Tang, Y.; Mi, S.; Li, W.; Yang, M.; Yu, Y. Integrated analysis of transcriptome and milk metagenome in subclinical mastitic and healthy cows. Anim. Biosci. 2024, 37, 709–717. [Google Scholar] [CrossRef]
- Flórez, A.B.; Vázquez, L.; Rodríguez, J.; Mayo, B. Phenotypic and safety assessment of the cheese strain Lactiplantibacillus plantarum LL441, and sequence analysis of its complete genome and plasmidome. Int. J. Mol. Sci. 2022, 24, 605. [Google Scholar] [CrossRef]
- Apostolakos, I.; Paramithiotis, S.; Mataragas, M. Comparative genomic analysis reveals the functional traits and safety status of lactic acid bacteria retrieved from artisanal cheeses and raw sheep milk. Foods 2023, 12, 599. [Google Scholar] [CrossRef] [PubMed]
- Luo, R.; Liu, C.; Li, Y.; Liu, Q.; Su, X.; Peng, Q.; Lei, X.; Li, W.; Menghe, B.; Bao, Q.; et al. Comparative genomics analysis of habitat adaptation by Lactobacillus kefiranofaciens. Foods 2023, 12, 1606. [Google Scholar] [CrossRef] [PubMed]


| Category | Peptide or Protein-Encoding Genes | Brief Description |
| Bacteriocin production | nisA/nisI (nisin) salA/salI (salivaricin) | Antimicrobial peptides [34] |
| Lactate dehydrogenases | ldhL (l-lactate dehydrogenase) | Critical enzyme for lactic acid fermentation, affecting taste and shelf-life of fermented products [35]. |
| Peroxide biosynthesis | pox (pyruvate oxidase) nox (NADH oxidase) | Generation of hydrogen peroxide, which can serve as a defense mechanism against pathogens [36]. |
| Antibiotic resistance | mate (multi-antimicrobial extrusion protein) gyrB (DNA gyrase subunit B) gyrA (DNA gyrase subunit A) | Genes that may confer intrinsic resistance to some antibiotics [37]. |
| Bile salt hydrolases | bsh (bile salt hydrolase) | Key enzyme for bacterial survival in the gastrointestinal tract by deconjugating bile acids [38]. |
| Phage proteins | int (phage integrase) portal (phage portal protein) | Components of prophages that may impact bacterial genetics and the stability of bacterial populations in fermented foods [39]. |
| Stress response (thermal stress) | dnaK (chaperone DnaK) dnaJ (chaperone DnaJ) groEL (chaperonin GroEL) groES (co-chaperonin GroES) | Proteins that assist in the proper folding of other proteins and stress recovery, essential for survival at high temperatures [40]. |
| Stress response (oxidative stress) | gshA (glutamate-cysteine ligase) grx (glutaredoxin) | Enzymes that participate in the cellular response to oxidative stress by synthesizing antioxidants and repairing damaged proteins [40]. |
| Stress response (osmotic stress) | glpF (glycerol uptake facilitator), proP (propanediol diffusion facilitator) | Proteins that help bacteria cope with high osmolarity environments by regulating the uptake of compatible solutes [40]. |
| Proteases | glpG (Rhomboid protease) clpX (ATP-dependent Clp protease ATP-binding subunit) | Proteases that degrade misfolded proteins, influencing texture and flavor maturation in fermented products [41,42]. |
| Peptidases | pepT (Peptidase T) pepD (Dipeptidase) | Enzymes that cleave peptides into amino acids, affecting the taste profile of fermented foods by releasing flavor precursors [41,42]. |
| Aromatic pathways | aroD (3-dehydroquinate dehydratase) qsuB (quinate 5-dehydrogenase) | Enzymes involved in the synthesis of aromatic amino acids, precursors to a variety of flavor and aroma compounds in foods [43]. |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Alba, C.; Arroyo, R.; Fernández, L.; Narbad, A.; Rodríguez, J.M. Characterization of a Ligilactobacillus salivarius Strain Isolated from a Cheese Seal Which Was Last Used in 1936. Foods 2024, 13, 2005. https://doi.org/10.3390/foods13132005
Alba C, Arroyo R, Fernández L, Narbad A, Rodríguez JM. Characterization of a Ligilactobacillus salivarius Strain Isolated from a Cheese Seal Which Was Last Used in 1936. Foods. 2024; 13(13):2005. https://doi.org/10.3390/foods13132005
Chicago/Turabian StyleAlba, Claudio, Rebeca Arroyo, Leónides Fernández, Arjan Narbad, and Juan M. Rodríguez. 2024. "Characterization of a Ligilactobacillus salivarius Strain Isolated from a Cheese Seal Which Was Last Used in 1936" Foods 13, no. 13: 2005. https://doi.org/10.3390/foods13132005
APA StyleAlba, C., Arroyo, R., Fernández, L., Narbad, A., & Rodríguez, J. M. (2024). Characterization of a Ligilactobacillus salivarius Strain Isolated from a Cheese Seal Which Was Last Used in 1936. Foods, 13(13), 2005. https://doi.org/10.3390/foods13132005






