Unconventional Yeasts Isolated from Chilean Honey: A Probiotic and Phenotypic Characterization
Abstract
1. Introduction
2. Materials and Methods
2.1. Samples
Isolation and Identification of Yeasts
2.2. Phenotypic Characterization of Isolated Yeasts
Analysis of Metabolites and Substrates by High Pressure Liquid Chromatography (HPLC)
2.3. Determination of Probiotic Capacity
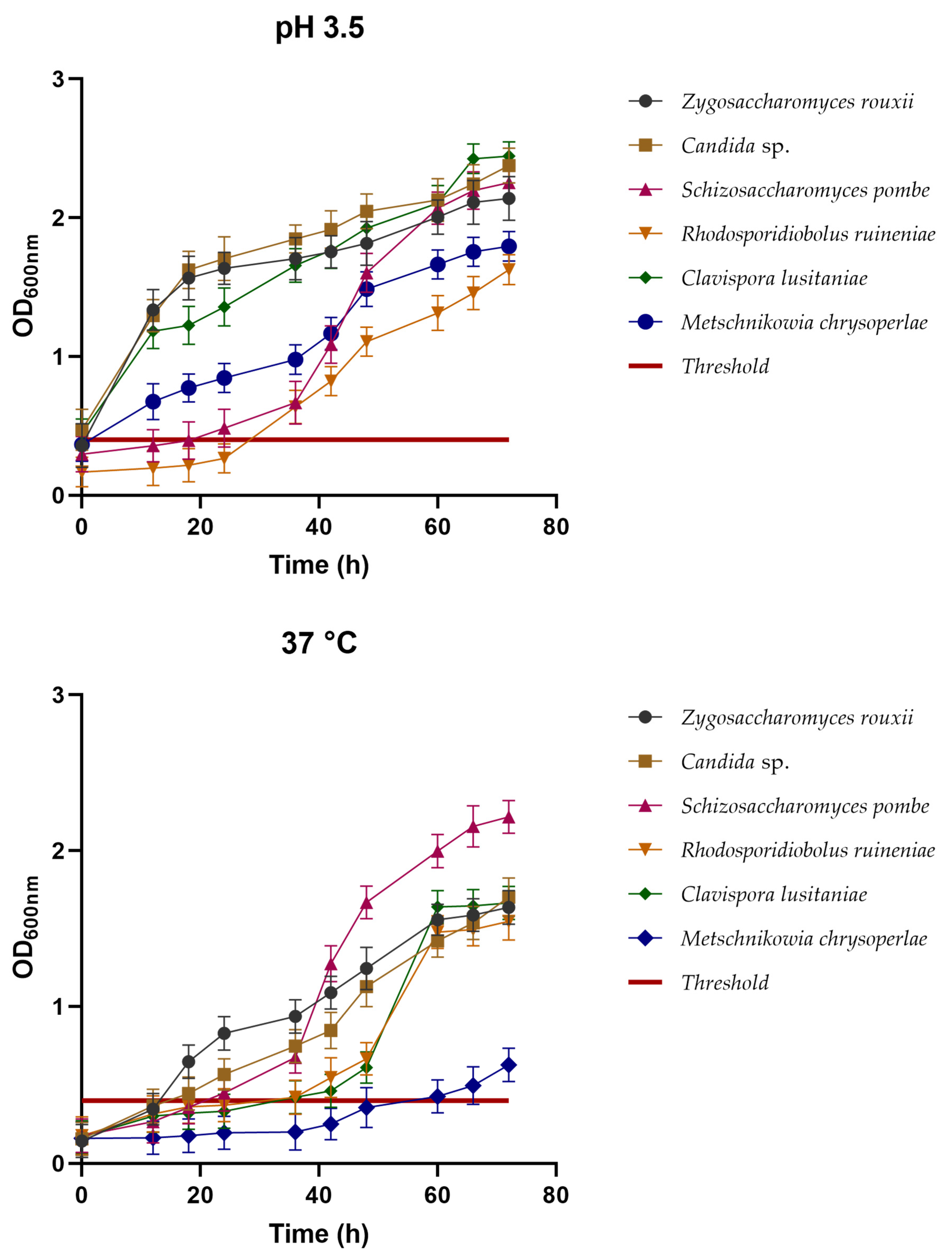
Survival at Low pH and Growth at 37 °C
2.4. Aggregation Capacity
2.5. In Vitro Digestion
2.5.1. Preparation of Simulated Digestive Fluids
2.5.2. Oral Phase of In Vitro Digestion
Curves of Change in Gastric pH
2.5.3. In Vitro Gastric Digestion
2.5.4. In Vitro Intestinal Digestion
2.6. Antimicrobial Activity Assays
2.7. Statistical Analysis and Plotting
3. Results and Discussion
3.1. Identification of Yeasts Isolated from Honey Samples
3.2. Phenotypic Characterization of Isolated Yeasts
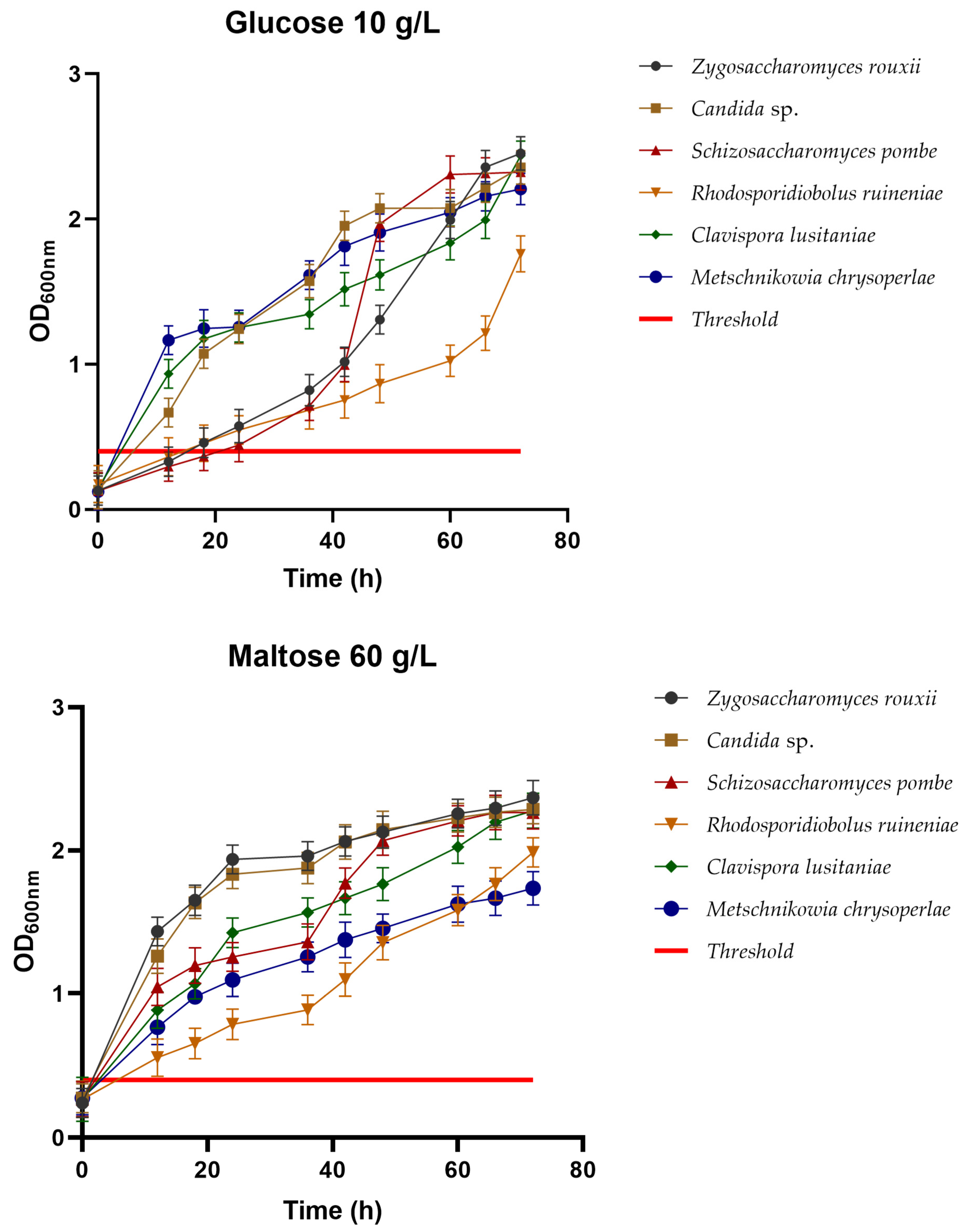
3.2.1. Carbohydrate Utilization
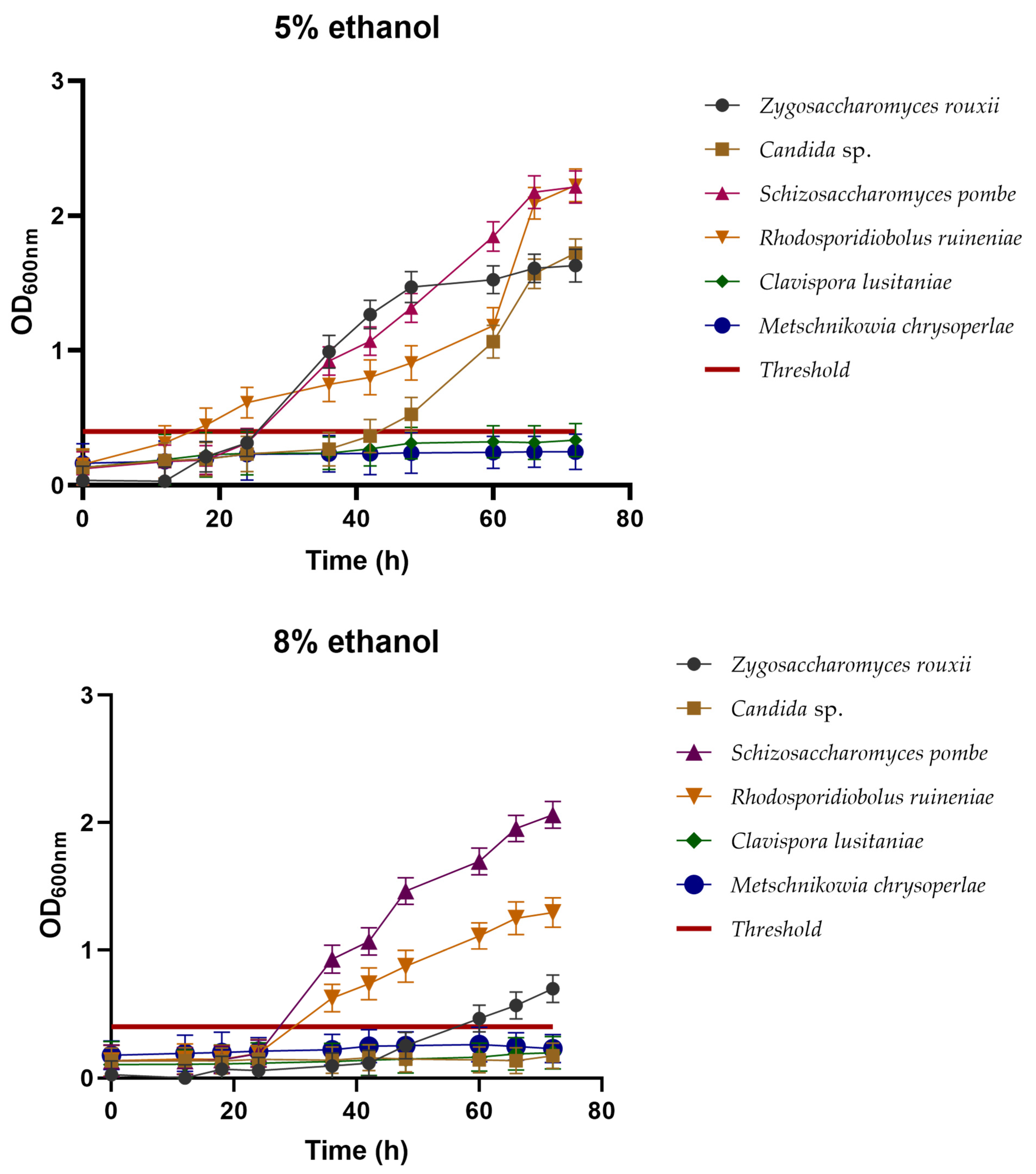
3.2.2. Ethanol Tolerance
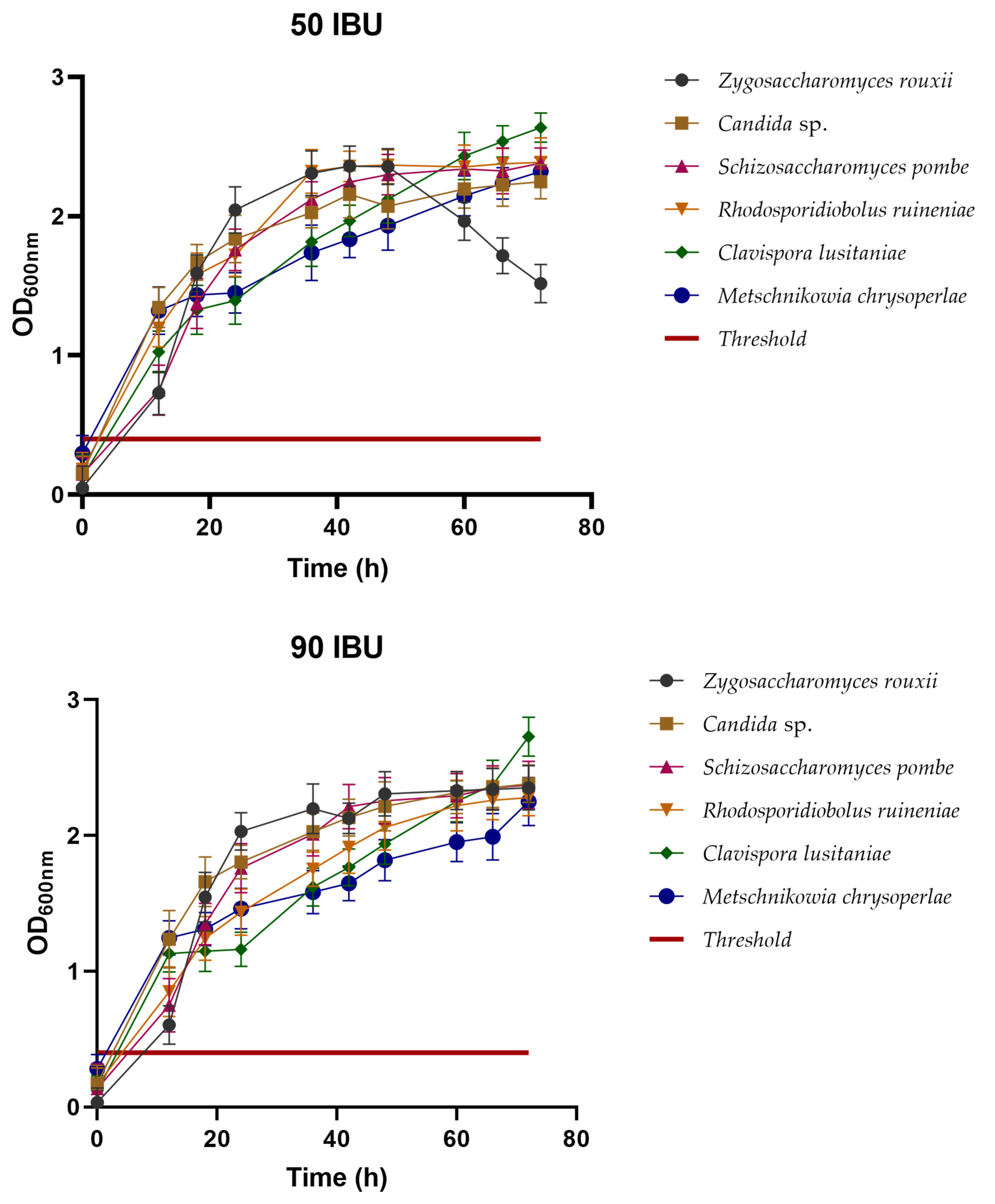
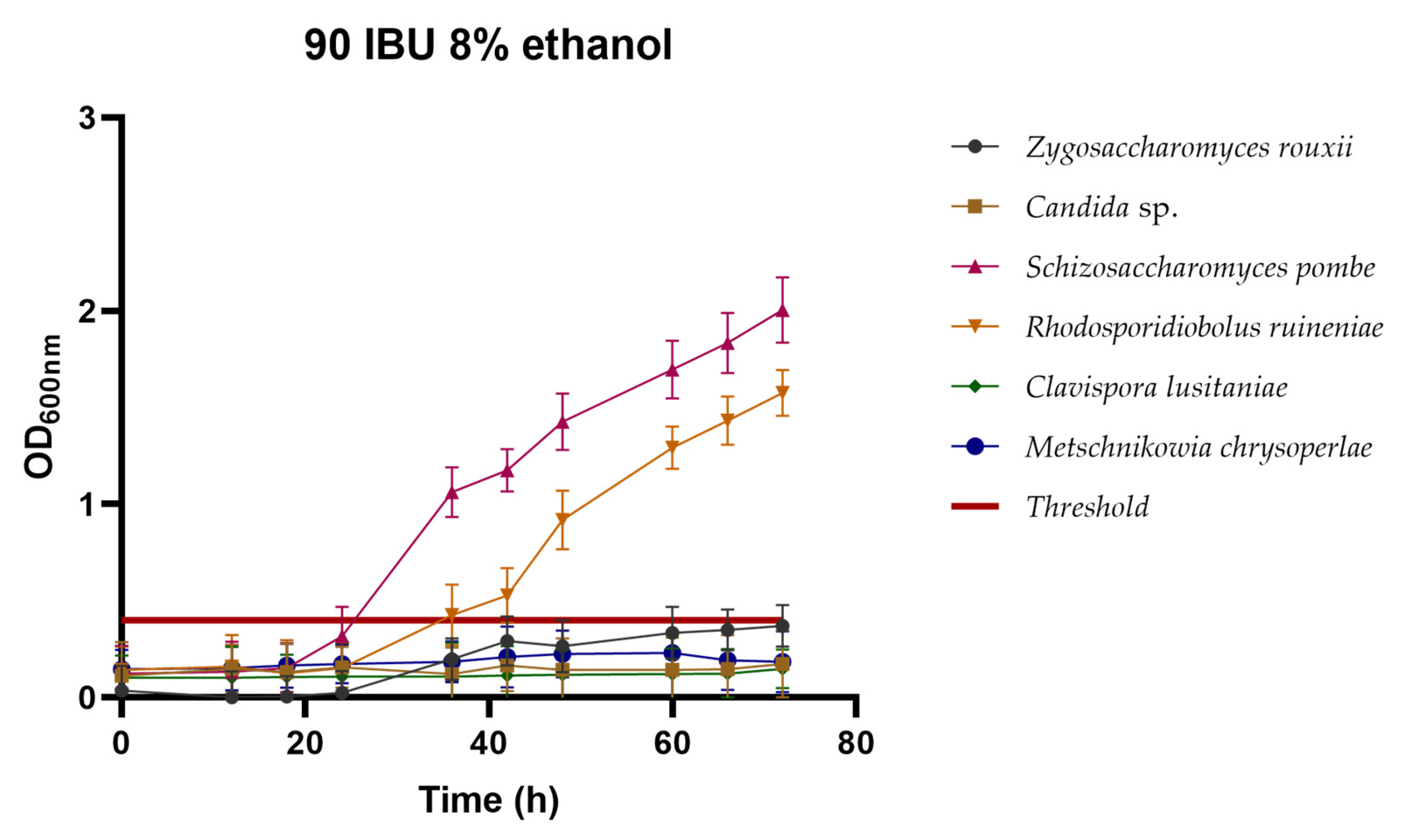
3.2.3. Hop Tolerance
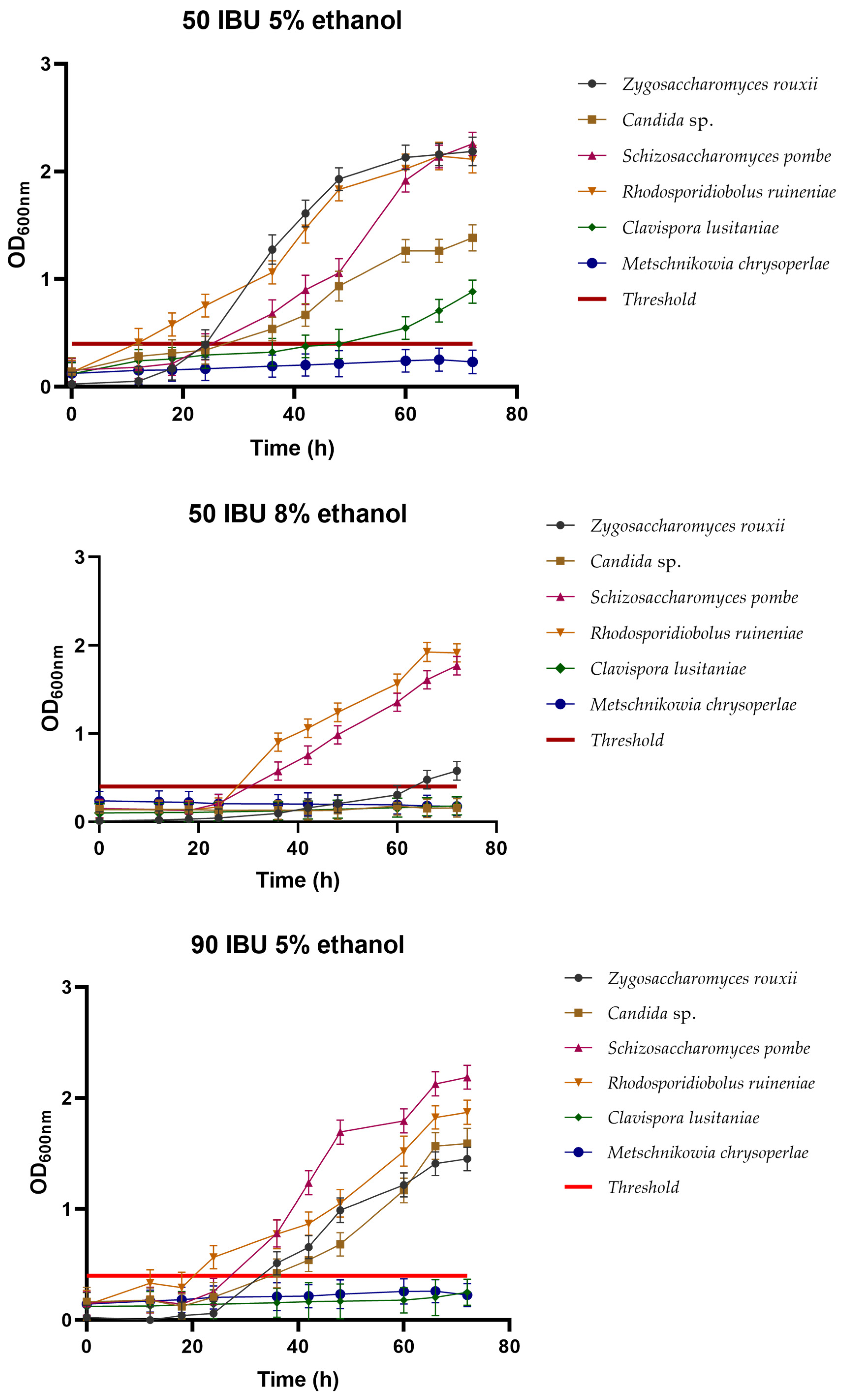
3.2.4. Cross-Tolerance between Ethanol and Hops
3.3. Probiotic Potential
3.3.1. Survival at Low pH and Growth at 37 °C
3.3.2. Auto-Aggregation Capacity
3.3.3. Antimicrobial Activity
3.3.4. Survival in Simulated In Vitro Digestion
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Araya, M.; Morelli, L.; Reid, G.; Sanders, M.; Stanton, C.; Pineiro, M.; Ben Embarek, P. Joint FAO/WHO Working Group Report on Drafting Guidelines for the Evaluation of Probiotics in Food; Fao Food and Nutrition Paper; World Health Organization, Food and Agriculture Organization of the United Nations: London, ON, Canada, 2002; Volume 30. [Google Scholar]
- Salmerón, I. Fermented cereal beverages: From probiotic, prebiotic and synbiotic towards Nanoscience designed healthy drinks. Lett. Appl. Microbiol. 2017, 65, 114–124. [Google Scholar] [CrossRef] [PubMed]
- Gharbi, Y.; Fhoula, I.; Ruas-Madiedo, P.; Afef, N.; Boudabous, A.; Gueimonde, M.; Ouzari, H.-I. In-Vitro characterization of potentially probiotic Lactobacillus strains isolated from human microbiota: Interaction with pathogenic bacteria and the enteric cell line HT29. Ann. Microbiol. 2019, 69, 61–72. [Google Scholar] [CrossRef]
- Illiano, P.; Brambilla, R.; Parolini, C. The mutual interplay of gut microbiota, diet and human disease. FEBS J. 2020, 287, 833–855. [Google Scholar] [CrossRef] [PubMed]
- Czerucka, D.; Piche, T.; Rampal, P. yeast as probiotics–Saccharomyces boulardii. Aliment. Pharmacol. Ther. 2007, 26, 767–778. [Google Scholar] [CrossRef] [PubMed]
- Otles, S.; Cagindi, O. Kefir: A probiotic dairy-composition, nutritional and therapeutic aspects. Pak. J. Nutr. 2003, 2, 54–59. [Google Scholar] [CrossRef]
- Ogunremi, O.; Sanni, A.; Agrawal, R. Probiotic potentials of yeasts isolated from some cereal-based Nigerian traditional fermented food products. J. Appl. Microbiol. 2015, 119, 797–808. [Google Scholar] [CrossRef]
- Menezes, A.G.T.; Ramos, C.L.; Dias, D.R.; Schwan, R.F. Combination of probiotic yeast and lactic acid bacteria as starter culture to produce maize-based beverages. Food Res. Int. 2018, 111, 187–197. [Google Scholar] [CrossRef]
- Nagpal, R.; Kumar, A.; Kumar, M.; Behare, P.V.; Jain, S.; Yadav, H. Probiotics, their health benefits and applications for developing healthier foods: A review. FEMS Microbiol. Lett. 2012, 334, 1–15. [Google Scholar] [CrossRef]
- Rai, A.K.; Pandey, A.; Sahoo, D. Biotechnological potential of yeasts in functional food industry. Trends Food Sci. Technol. 2019, 83, 129–137. [Google Scholar] [CrossRef]
- Fadahunsi, I.F.; Olubodun, S. Antagonistic pattern of yeast species against some selected food-borne pathogens. Bull. Natl. Res. Cent. 2021, 45, 34. [Google Scholar] [CrossRef]
- Ferronato, G.; Prandini, A. Dietary supplementation of inorganic, organic, and fatty acids in pig: A review. Animals 2020, 10, 1740. [Google Scholar] [CrossRef]
- Nicholson, J.K.; Holmes, E.; Kinross, J.; Burcelin, R.; Gibson, G.; Jia, W.; Pettersson, S. Host-gut microbiota metabolic interactions. Science 2012, 336, 1262–1267. [Google Scholar] [CrossRef]
- Zahoor, F.; Sooklim, C.; Songdech, P.; Duangpakdee, O.; Soontorngun, N. Selection of potential yeast probiotics and a cell factory for xylitol or acid production from honeybee samples. Metabolites 2021, 11, 312. [Google Scholar] [CrossRef]
- Silva, M.S.; Rabadzhiev, Y.; Eller, M.R.; Iliev, I.; Ivanova, I.; Santana, W.C. Microorganisms in honey. Honey Anal. 2017, 500, 233–257. [Google Scholar] [CrossRef]
- Mohan, A.; Quek, S.-Y.; Gutierrez-Maddox, N.; Gao, Y.; Shu, Q. Effect of honey in improving the gut microbial balance. Food Qual. Saf. 2017, 1, 107–115. [Google Scholar] [CrossRef]
- García, S.; Troncoso, J.M.; Rondanelli-Reyes, M. Study of honey according to botanical origin and physicochemical parameters in the Biobío Region, Chile. Chil. J. Agric. Res. 2020, 80, 675–685. [Google Scholar] [CrossRef]
- Sinacori, M.; Francesca, N.; Alfonzo, A.; Cruciata, M.; Sannino, C.; Settanni, L.; Moschetti, G. Cultivable microorganisms associated with honeys of different geographical and botanical origin. Food Microbiol. 2014, 38, 284–294. [Google Scholar] [CrossRef] [PubMed]
- Kurtzman, C.; Robnett, C. Identification of clinically important ascomycetous yeasts based on nucleotide divergence in the 5′ end of the large-subunit (26S) ribosomal DNA gene. J. Clin. Microbiol. 1997, 35, 1216–1223. [Google Scholar] [CrossRef] [PubMed]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Benson, D.A.; Karsch-Mizrachi, I.; Lipman, D.J.; Ostell, J.; Rapp, B.A.; Wheeler, D.L. GenBank. Nucleic Acids Res 2000, 28, 15–18. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Schwartz, S.; Wagner, L.; Miller, W. A greedy algorithm for aligning DNA sequences. J. Comput. Biol. 2000, 7, 203–214. [Google Scholar] [CrossRef]
- McFeeters, R.F. Single-injection HPLC analysis of acids, sugars, and alcohols in cucumber fermentations. J. Agric. Food Chem. 1993, 41, 1439–1443. [Google Scholar] [CrossRef]
- Pedersen, L.L.; Owusu-Kwarteng, J.; Thorsen, L.; Jespersen, L. Biodiversity and probiotic potential of yeasts isolated from Fura, a West African spontaneously fermented cereal. Int. J. Food Microbiol. 2012, 159, 144–151. [Google Scholar] [CrossRef] [PubMed]
- Gil-Rodríguez, A.M.; Carrascosa, A.V.; Requena, T. Yeasts in foods and beverages: In Vitro characterisation of probiotic traits. LWT—Food Sci. Technol. 2015, 64, 1156–1162. [Google Scholar] [CrossRef]
- Brodkorb, A.; Egger, L.; Alminger, M.; Alvito, P.; Assunção, R.; Ballance, S.; Bohn, T.; Bourlieu-Lacanal, C.; Boutrou, R.; Carrière, F.; et al. INFOGEST static in vitro simulation of gastrointestinal food digestion. Nat. Protoc. 2019, 14, 991–1014. [Google Scholar] [CrossRef] [PubMed]
- Mulet-Cabero, A.-I.; Egger, L.; Portmann, R.; Ménard, O.; Marze, S.; Minekus, M.; Le Feunteun, S.; Sarkar, A.; Grundy, M.M.L.; Carrière, F.; et al. A standardised semi-dynamic in vitro digestion method suitable for food—An international consensus. Food Funct. 2020, 11, 1702–1720. [Google Scholar] [CrossRef] [PubMed]
- Hoebler, C.; Lecannu, G.; Belleville, C.; Devaux, M.F.; Popineau, Y.; Barry, J.L. Development of an in vitro system simulating bucco-gastric digestion to assess the physical and chemical changes of food. Int. J. Food Sci. Nutr. 2002, 53, 389–402. [Google Scholar] [CrossRef] [PubMed]
- Krul, C.; Luiten-Schuite, A.; Baandagger, R.; Verhagen, H.; Mohn, G.; Feron, V.; Havenaar, R. Application of a dynamic in vitro gastrointestinal tract model to study the availability of food mutagens, using heterocyclic aromatic amines as model compounds. Food Chem. Toxicol. 2000, 38, 783–792. [Google Scholar] [CrossRef]
- Chelliah, R.; Ramakrishnan, S.R.; Prabhu, P.R.; Antony, U. Evaluation of antimicrobial activity and probiotic properties of wild-strain Pichia kudriavzevii isolated from frozen idli batter. Yeast 2016, 33, 385–401. [Google Scholar] [CrossRef]
- Palande, V.; Meora, R.; Sonavale, R.M.; Makashir, M.; Modak, M.S.; Kapse, N.; Dhakephalkar, P.K.; Ranjekar, P.K.; Kunchiraman, B.N. Inhibition of pathogenic strains of Candida albicans and non-albicans by Bacillus species isolated from traditional Indian fermented food preparations. Int. J. Curr. Microbiol. Appl. Sci. 2015, 4, 691–699. [Google Scholar]
- Carvalho, C.M.; Meirinho, S.; Estevinho, M.L.F.; Choupina, A. Yeast species associated with honey: Different identification methods. In Archivos de Zootecnia; Universidad de Córdoba: Córdoba, Andalucía, España, 2010; Volume 59, pp. 103–113. [Google Scholar]
- Wen, Y.; Wang, L.; Jin, Y.; Zhang, J.; Su, L.; Zhang, X.; Zhou, J.; Li, Y. The microbial community dynamics during the vitex honey ripening process in the honeycomb. Front. Microbiol. 2017, 8, 1649. [Google Scholar] [CrossRef]
- Lievens, B.; Hallsworth, J.E.; Pozo, M.I.; Belgacem, Z.B.; Stevenson, A.; Willems, K.A.; Jacquemyn, H. Microbiology of sugar-rich environments: Diversity, ecology and system constraints. Environ. Microbiol. 2015, 17, 278–298. [Google Scholar] [CrossRef]
- Matraxia, M.; Alfonzo, A.; Prestianni, R.; Francesca, N.; Gaglio, R.; Todaro, A.; Alfeo, V.; Perretti, G.; Columba, P.; Settanni, L. Non-conventional yeasts from fermented honey by-products: Focus on Hanseniaspora uvarum strains for craft beer production. Food Microbiol. 2021, 99, 103806. [Google Scholar] [CrossRef]
- Nandal, P.; Sharma, S.; Arora, A. Bioprospecting non-conventional yeasts for ethanol production from rice straw hydrolysate and their inhibitor tolerance. Renew. Energy 2020, 147, 1694–1703. [Google Scholar] [CrossRef]
- Čadež, N.; Fülöp, L.; Dlauchy, D.; Péter, G. Zygosaccharomyces favi sp. nov., an obligate osmophilic yeast species from bee bread and honey. Antonie Van Leeuwenhoek 2015, 107, 645–654. [Google Scholar] [CrossRef] [PubMed]
- Prestianni, R.; Matraxia, M.; Naselli, V.; Pirrone, A.; Badalamenti, N.; Ingrassia, M.; Gaglio, R.; Settanni, L.; Columba, P.; Maggio, A.; et al. Use of sequentially inoculation of Saccharomyces cerevisiae and Hanseniaspora uvarum strains isolated from honey by-products to improve and stabilize the quality of mead produced in Sicily. Food Microbiol. 2022, 107, 104064. [Google Scholar] [CrossRef] [PubMed]
- Benito, S.; Palomero, F.; Morata, A.; Calderón, F.; Palmero, D.; Suárez-Lepe, J.A. Physiological features of Schizosaccharomyces pombe of interest in making of white wines. Eur. Food Res. Technol. 2013, 236, 29–36. [Google Scholar] [CrossRef]
- Benito, Á.; Calderón, F.; Benito, S. Schizosaccharomyces pombe Biotechnological Applications in Winemaking. In Schizosaccharomyces pombe: Methods and Protocols; Singleton, T.L., Ed.; Springer: New York, NY, USA, 2018; pp. 217–226. [Google Scholar] [CrossRef]
- Benito, S. Combined use of Lachancea thermotolerans and Schizosaccharomyces pombe in winemaking: A review. Microorganisms 2020, 8, 655. [Google Scholar] [CrossRef]
- García, M.; Crespo, J.; Cabellos, J.M.; Arroyo, T. Growth of Non-Saccharomyces Native Strains under Different Fermentative Stress Conditions. Fermentation 2021, 7, 124. [Google Scholar] [CrossRef]
- Benito, Á.; Calderón, F.; Palomero, F.; Benito, S. Combine use of selected Schizosaccharomyces pombe and Lachancea thermotolerans yeast strains as an alternative to the traditional malolactic fermentation in red wine production. Molecules 2015, 20, 9510–9523. [Google Scholar] [CrossRef]
- Benito, S.; Palomero, F.; Morata, A.; Calderón, F.; Suárez-Lepe, J.A. New applications for Schizosaccharomyces pombe in the alcoholic fermentation of red wines. Int. J. Food Sci. Technol. 2012, 47, 2101–2108. [Google Scholar] [CrossRef]
- Benito, Á.; Jeffares, D.; Palomero, F.; Calderón, F.; Bai, F.-Y.; Bähler, J.; Benito, S. Selected Schizosaccharomyces pombe strains have characteristics that are beneficial for winemaking. PLoS ONE 2016, 11, e0151102. [Google Scholar] [CrossRef] [PubMed]
- Yao, S.; Hao, L.; Zhou, R.; Jin, Y.; Huang, J.; Wu, C. Co-culture with Tetragenococcus halophilus improved the ethanol tolerance of Zygosaccharomyces rouxii by maintaining cell surface properties. Food Microbiol. 2021, 97, 103750. [Google Scholar] [CrossRef] [PubMed]
- Fan, G.; Liu, P.; Chang, X.; Yin, H.; Cheng, L.; Teng, C.; Gong, Y.; Li, X. Isolation and Identification of a High-Yield Ethyl Caproate-Producing Yeast From Daqu and Optimization of Its Fermentation. Front. Microbiol. 2021, 12, 663744. [Google Scholar] [CrossRef]
- Boro, N.; Narzary, D. Amylolytic Fungi in the Ethnic Beer Starter “emao” and Their Beer-Producing Attributes. Front. Sustain. Food Syst. 2022, 6, 869430. [Google Scholar] [CrossRef]
- Michel, M.; Kopecká, J.; Meier-Dörnberg, T.; Zarnkow, M.; Jacob, F.; Hutzler, M. Screening for new brewing yeasts in the non-Saccharomyces sector with Torulaspora delbrueckii as model. Yeast 2016, 33, 129–144. [Google Scholar] [CrossRef]
- Petruzzi, L.; Rosaria Corbo, M.; Sinigaglia, M.; Bevilacqua, A. Brewer’s yeast in controlled and uncontrolled fermentations, with a focus on novel, nonconventional, and superior strains. Food Rev. Int. 2016, 32, 341–363. [Google Scholar] [CrossRef]
- Callejo, M.J.; García Navas, J.J.; Alba, R.; Escott, C.; Loira, I.; González, M.C.; Morata, A. Wort fermentation and beer conditioning with selected non-Saccharomyces yeasts in craft beers. Eur. Food Res. Technol. 2019, 245, 1229–1238. [Google Scholar] [CrossRef]
- Liu, J.; Arneborg, N.; Toldam-Andersen, T.B.; Zhang, S.; Petersen, M.A.; Bredie, W.L.P. Impact of sequential co-culture fermentations on flavour characters of Solaris wines. Eur. Food Res. Technol. 2017, 243, 437–445. [Google Scholar] [CrossRef]
- Li, S.; Chen, X.; Gao, Z.; Zhang, Z.; Bi, P.; Guo, J. Enhancing antioxidant activity and fragrant profile of low-ethanol kiwi wine via sequential culture of indigenous Zygosaccharomyces rouxii and Saccharomyces cerevisiae. Food Biosci. 2023, 51, 102210. [Google Scholar] [CrossRef]
- Allert, S.; Förster, T.M.; Svensson, C.-M.; Richardson, J.P.; Pawlik, T.; Hebecker, B.; Rudolphi, S.; Juraschitz, M.; Schaller, M.; Blagojevic, M. Candida albicans-induced epithelial damage mediates translocation through intestinal barriers. MBio 2018, 9, e00915-18. [Google Scholar] [CrossRef]
- Pfaller, M.A.; Diekema, D.J.; Turnidge, J.D.; Castanheira, M.; Jones, R.N. Twenty years of the SENTRY antifungal surveillance program: Results for Candida species from 1997–2016. In Open Forum Infectious Diseases; Oxford University Press: Oxford, UK, 2019; Volume 6, pp. S79–S94. [Google Scholar] [CrossRef]
- Kumla, J.; Nundaeng, S.; Suwannarach, N.; Lumyong, S. Evaluation of Multifarious Plant Growth Promoting Trials of Yeast Isolated from the Soil of Assam Tea (Camellia sinensis var. assamica) Plantations in Northern Thailand. Microorganisms 2020, 8, 1168. [Google Scholar] [CrossRef]
- Jansen, M.; Veurink, J.H.; Euverink, G.-J.W.; Dijkhuizen, L. Growth of the salt-tolerant yeast Zygosaccharomyces rouxii in microtiter plates: Effects of NaCl, pH and temperature on growth and fusel alcohol production from branched-chain amino acids. FEMS Yeast Res. 2003, 3, 313–318. [Google Scholar] [CrossRef] [PubMed]
- Ranjan, K.; Sahay, S. Clavispora lusitaniae produces pH and temperature tolerant extracellular amylase. Point J. Bot. Microbiol. Res. 2015, 1, 07–14. [Google Scholar]
- Merchán, A.V.; Benito, M.J.; Galván, A.I.; Ruiz-Moyano Seco de Herrera, S. Identification and selection of yeast with functional properties for future application in soft paste cheese. LWT—Food Sci. Technol. 2020, 124, 109173. [Google Scholar] [CrossRef]
- García-Cayuela, T.; Korany, A.M.; Bustos, I.; Gómez de Cadiñanos, L.P.; Requena, T.; Peláez, C.; Martínez-Cuesta, M.C. Adhesion abilities of dairy Lactobacillus plantarum strains showing an aggregation phenotype. Food Res. Int. 2014, 57, 44–50. [Google Scholar] [CrossRef]
- Goossens, K.; Willaert, R. Flocculation protein structure and cell–cell adhesion mechanism in Saccharomyces cerevisiae. Biotechnol. Lett. 2010, 32, 1571–1585. [Google Scholar] [CrossRef] [PubMed]
- Van Mulders, S.E.; Ghequire, M.; Daenen, L.; Verbelen, P.J.; Verstrepen, K.J.; Delvaux, F.R. Flocculation gene variability in industrial brewer’s yeast strains. Appl. Microbiol. Biotechnol. 2010, 88, 1321–1331. [Google Scholar] [CrossRef]
- Chan, M.Z.A.; Liu, S.-Q. Fortifying foods with synbiotic and postbiotic preparations of the probiotic yeast, Saccharomyces boulardii. Curr. Opin. Food Sci. 2022, 43, 216–224. [Google Scholar] [CrossRef]
- Pais, P.; Almeida, V.; Yılmaz, M.; Teixeira, M.C. Saccharomyces boulardii: What Makes It Tick as Successful Probiotic? J. Fungi 2020, 6, 78. [Google Scholar] [CrossRef]
- Savini, V. Chapter 10—Bacillus cereus Biocontrol Properties. In The Diverse Faces of Bacillus cereus; Clinical Microbiology and Virology, Laboratory of Bacteriology and Mycology, Civic Hospital of Pescara: Pescara, Italy, 2016; pp. 117–127. [Google Scholar] [CrossRef]
- Sisti, M.; Savini, V. Antifungal properties of the human Metschnikowia strain IHEM 25107. Folia Microbiol. 2014, 59, 263–266. [Google Scholar] [CrossRef] [PubMed]
- Savini, V.; Hendrickx, M.; Sisti, M.; Masciarelli, G.; Favaro, M.; Fontana, C.; Pitzurra, L.; Arzeni, D.; Astolfi, D.; Catavitello, C.; et al. An atypical, pigment-producing Metschnikowia strain from a leukaemia patient. Med. Mycol. 2013, 51, 438–443. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Likotrafiti, E.; Tuohy, K.M.; Gibson, G.R.; Rastall, R.A. Development of antimicrobial synbiotics using potentially-probiotic faecal isolates of Lactobacillus fermentum and Bifidobacterium longum. Anaerobe 2013, 20, 5–13. [Google Scholar] [CrossRef]
- Staniszewski, A.; Kordowska-Wiater, M. Probiotic and Potentially Probiotic Yeasts—Characteristics and Food Application. Foods 2021, 10, 1306. [Google Scholar] [CrossRef] [PubMed]
- Ok, T. Method of Utilization of Zygosaccharomyces rouxii. U.S. Patent Application No 10/153,209, 27 November 2003. [Google Scholar]
| Treatments | Value |
|---|---|
| Glucose added | 10 g/L |
| Maltose added | 60 g/L |
| pH | 3.5 |
| Temperature | 37 °C |
| Ethanol | 5% (v/v) and 8% (v/v) |
| Hop | 50 IBU and 90 IBU |
| Ethanol + Hops | 50 IBU and 5% (v/v) |
| Ethanol + Hops | 50 IBU and 8% (v/v) |
| Ethanol + Hops | 90 IBU and 5% (v/v) |
| Ethanol + Hops | 90 IBU and 8% (v/v) |
| Components | Oral Stock Solution (SSO) | Gastric Stock Solution (SSG) | Intestinal Stock Solution (SSI) | |||
|---|---|---|---|---|---|---|
| Amount in Solution | Percentage (%) | Amount in Solution | Percentage (%) | Amount in Solution | Percentage (%) | |
| KCl | 3.75 | 3.75 | 15.525 | 1.725 | 17 | 1.7 |
| KH2 PO4 | 0.925 | 0.925 | 2.025 | 0.225 | 2 | 0.2 |
| NaHCO3 | 1.7 | 1.7 | 28.125 | 3.125 | 106.25 | 10.625 |
| NaCl | - | - | 26.55 | 2.95 | 24 | 2.4 |
| MgCl2 (H2O)6 | 0.125 | 0.125 | 0.9 | 0.1 | 2.75 | 0.275 |
| (NH4) 2CO3 | 0.01 | 0.01 | 1.125 | 0.125 | - | - |
| CaCl2 | 0.007 | 0.007 | 0.011 | 0.001 | 0.11 | 0.011 |
| H2O | 93.483 | 93.483 | 825.738 | 91.748 | 847.89 | 84.789 |
| Total | 100 | 100 | 900 | 100 | 1000 | 100 |
| Components | Simulated Oral Fluid (SOF) | Simulated Gastric Fluid (SGF) | Simulated Intestinal Fluid (SIF) | |||
|---|---|---|---|---|---|---|
| Quantity in Solution | Percent (%) | Quantity in Solution | Percent (%) | Quantity in Solution | Percent (%) | |
| Stock solution | 8 | 80 | 101.76 | 84.8 | 100 | 40 |
| CaCl2 | 0.05 | 15 | 0.06 | 0.05 | 0.5 | 0.2 |
| H2O | 0.45 | 0.5 | 5376 | 4.48 | 39.5 | 15.8 |
| α-amylase | 1.5 | 4.5 | - | - | - | - |
| Pepsin | - | 8.004 | 6.67 | - | - | |
| HCL (1N) | - | 4.8 | 4 | - | - | |
| Pancreatin | - | - | - | 62.5 | 25 | |
| NaOH (1N) | - | - | 10 | 4 | ||
| Bile | - | - | - | 37.5 | 15 | |
| Total | 10 | 100 | 120 | 100 | 250 | 100 |
| Yeasts | GeneBank Accession Number | Isolation Source |
|---|---|---|
| Zygosaccharomyces rouxii | OR392815 | Ulmo Honey |
| Candida sp. | OR392816 | Ulmo Honey |
| Schizosaccharomyces pombe | OR392817 | Ulmo Honey |
| Rhodosporidiobolus ruineniae | OR392818 | Mountain Honey |
| Clavispora lusitaniae | OR392819 | Mountain Honey |
| Metschnikowia chrysoperlae | OR392820 | Quillay Honey |
| Yeasts | Glucose (g/L) | Ethanol (% v/v) | Lactic Acid (g/L) | Acetic Acid (g/L) | Glycerol (g/L) |
|---|---|---|---|---|---|
| Z. rouxii | 0.45 ± 0.002 b | 1.56 ± 0.02 b | 0.07 ± 0.009 b | 0.32 ± 0.02 de | 1.68 ± 0.03 a |
| Candida sp. | ND | 0.77 ± 0.01 d | 0.05 ± 0.006 bc | 0.34 ± 0.07 d | 0.62 ± 0.005 c |
| S. pombe | ND | 2.26 ± 0.04 a | 0.23 ± 0.01 a | 0.59 ± 0.20 b | 1.56 ± 0.06 b |
| R. ruineniae | 6.52 ± 0.20 a | 0.07 ± 0.008 e | 0.04 ± 0.004 cd | 0.04 ± 0.20 a | 0.35 ± 0.006 d |
| C. lusitaniae | ND | 1.12 ± 0.10 c | 0.03 ± 0.002 d | 0.53 ± 0.01 c | 0.21 ± 0.002 e |
| M. chrysoperlae | ND | 0.66 ± 0.04 d | 0.04 ± 0.006 cd | 0.24 ± 0.02 e | 0.41 ± 0.008 d |
| Yeasts | Glucose (g/L) | Fructose (g/L) | Sucrose (g/L) | Ethanol (% v/v) | Lactic Acid (g/L) | Acetic Acid (g/L) | Glycerol (g/L) |
|---|---|---|---|---|---|---|---|
| Z. rouxii | 0.54 ± 0.003 a | 0.21 ± 0.01 a | 4.36 ± 0.06 e | 1.17 ± 0.02 b | 0.13 ± 0.01 b | 0.48 ± 0.05 c | 0.66 ± 0.02 b |
| Candida sp. | ND | ND | 24.51 ± 0.01 b | 0.08 ± 0.007 e | 0.05 ± 0.006 c | 0.45 ± 0.05 c | ND |
| S. pombe | ND | ND | 0.35 ± 0.04 f | 1.62 ± 0.06 a | 0.27 ± 0.01 a | 0.74 ± 0.30 ab | 1.17 ± 0.06 a |
| R. ruineniae | ND | ND | 15.73 ± 0.03 d | 0.62 ± 0.02 c | 0.23 ± 0.02 a | 0.28 ± 0.04 c | ND |
| C. lusitaniae | ND | ND | 17.97 ± 0.30 c | 0.6 ± 0.04 c | 0.03 ± 0.001 c | 0.17 ± 0.03 b | ND |
| M. chrysoperlae | ND | ND | 25.91 ± 0.01 a | 0.27 ± 0.01 d | 0.04 ± 0.005 c | 0.59 ± 0.05 a | ND |
| Yeasts | % Auto-Aggregation |
|---|---|
| Z. rouxii | 89.8 ± 0.05% |
| S. pombe | 100 ± 0.0% |
| M. chrysoperlae | 94 ± 0.01% |
| Yeasts | Escherichia coli | Salmonella enteritidis | Staphylococcus aureus | |||
|---|---|---|---|---|---|---|
| P (mm) | S (mm) | P (mm) | S (mm) | P (mm) | S (mm) | |
| Z. rouxii | 9 ± 0.10 | 9 ± 0.0 | - | - | 9.33 ± 0.51 | 10.66 ± 0.52 |
| S. pombe | 9 ± 0.0 | 9.33 ± 0.50 | 9.66 ± 0.50 | - | 9.66 ± 1.10 | - |
| M. chrysoperlae | 10.33 ± 0.50 | 10 ± 0.0 | 9.33 ± 1.10 | 9 ± 1.05 | 11 ± 0.0 | 10 ± 0.10 |
| Yeasts | Cell Concentration (CFU/mL) | ||
|---|---|---|---|
| Oral Phase | Gastric Phase | Intestinal Phase | |
| Z. rouxii | 1 × 109 ± 0.65 | 1 × 109 ± 0.37 | 1 × 106 ± 0.63 |
| S. pombe | 1 × 109 ± 0.32 | 1 × 109 ± 0.52 | 1 × 107 ± 0.12 |
| M. chrysoperlae | 1 × 109 ± 0.56 | 1 × 108 ± 0.71 | 1 × 107 ± 0.25 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rodríguez Machado, A.; Caro, C.M.; Hurtado-Murillo, J.J.; Gomes Lobo, C.J.; Zúñiga, R.N.; Franco, W. Unconventional Yeasts Isolated from Chilean Honey: A Probiotic and Phenotypic Characterization. Foods 2024, 13, 1582. https://doi.org/10.3390/foods13101582
Rodríguez Machado A, Caro CM, Hurtado-Murillo JJ, Gomes Lobo CJ, Zúñiga RN, Franco W. Unconventional Yeasts Isolated from Chilean Honey: A Probiotic and Phenotypic Characterization. Foods. 2024; 13(10):1582. https://doi.org/10.3390/foods13101582
Chicago/Turabian StyleRodríguez Machado, Adrian, Camila Mella Caro, John J. Hurtado-Murillo, Cristian J. Gomes Lobo, Rommy N. Zúñiga, and Wendy Franco. 2024. "Unconventional Yeasts Isolated from Chilean Honey: A Probiotic and Phenotypic Characterization" Foods 13, no. 10: 1582. https://doi.org/10.3390/foods13101582
APA StyleRodríguez Machado, A., Caro, C. M., Hurtado-Murillo, J. J., Gomes Lobo, C. J., Zúñiga, R. N., & Franco, W. (2024). Unconventional Yeasts Isolated from Chilean Honey: A Probiotic and Phenotypic Characterization. Foods, 13(10), 1582. https://doi.org/10.3390/foods13101582







