Near-Infrared Spectroscopy Analysis of the Phytic Acid Content in Fuzzy Cottonseed Based on Machine Learning Algorithms
Abstract
1. Introduction
2. Materials and Methods
2.1. Samples and Preparation
2.2. Near-Infrared Spectroscopy Acquisition
2.2.1. Collection of the NIR Spectra of Fuzzy Cottonseeds
2.2.2. Collection of the NIR Spectra of the Delinted Cottonseeds
2.3. Determination of the PA Content in the Samples
2.4. Construction and Evaluation of the Calibration Model
3. Results
3.1. Statistical Analysis of the PA Content
3.2. NIR Spectra and Pre-Treatment
3.3. Development and Interpretation of Full-Spectrum Models
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Alford, B.B.; Liepa, G.U.; Vanbeber, A.D. Cottonseed protein: What does the future hold? Plant Foods Hum. Nutr. 1996, 49, 1–11. [Google Scholar] [CrossRef]
- Sawan, Z.M.; Elfarra, A.A.; Ellatif, S.A. Cottonseed, protein and oil yields, and oil properties as affected by nitrogen and phosphorus fertilization and growth-regulators. J. Agron. Crop Sci.-Z. Acker Pflanzenbau 1988, 161, 50–56. [Google Scholar] [CrossRef]
- Ahmad, B.; Hussain, S.M.; Ali, S.; Arsalan , M.Z.-U.-H.; Tabassum, S.; Sharif, A. Efficacy of acidified phytase supplemented cottonseed meal based diets on growth performance and proximate composition of Labeo rohita fingerlings. Braz. J. Biol. 2023, 83, e247791. [Google Scholar] [CrossRef]
- Kumar, M.; Zhang, B.; Potkule, J.; Sharma, K.; Radha; Hano, C.; Sheri, V.; Chandran, D.; Dhumal, S.; Dey, A.; et al. Cottonseed Oil: Extraction, Characterization, Health Benefits, Safety Profile, and Application. Food Anal. Methods 2023, 16, 266–280. [Google Scholar] [CrossRef]
- Riaz, T.; Iqbal, M.W.; Mahmood, S.; Yasmin, I.; Leghari, A.A.; Rehman, A.; Mushtaq, A.; Ali, K.; Azam, M.; Bilal, M. Cottonseed oil: A review of extraction techniques, physicochemical, functional, and nutritional properties. Crit. Rev. Food Sci. Nutr. 2021, 63, 1219–1237. [Google Scholar] [CrossRef]
- Zhou, J.-Z.; Zhang, H.; Gao, L.; Wang, L.; Qian, H.-F. Influence of pH and ionic strength on heat-induced formation and rheological properties of cottonseed protein gels. Food Bioprod. Process. 2015, 96, 27–34. [Google Scholar] [CrossRef]
- Kumar, M.; Tomar, M.; Punia, S.; Grasso, S.; Arrutia, F.; Choudhary, J.; Singh, S.; Verma, P.; Mahapatra, A.; Patil, S.; et al. Cottonseed: A sustainable contributor to globalprotein requirements. Trends Food Sci. Technol. 2021, 111, 100–113. [Google Scholar] [CrossRef]
- Swiatkiewicz, S.; Arczewska-Wlosek, A.; Jozefiak, D. The use of cottonseed meal as aprotein source for poultry: An updated review. World’s Poult. Sci. J. 2016, 72, 473–483. [Google Scholar] [CrossRef]
- Wang, H.; Hu, X.; Zheng, Y.; Chen, J.; Tan, B.; Shi, L.; Zhang, S. Effects of replacing fish meal with cottonseed protein concentrate on the growth, immune responses, digestive ability and intestinal microbial flora in Litopenaeus vannamei. Fish Shellfish Immunol. 2022, 128, 91–100. [Google Scholar] [CrossRef]
- Silva, V.M.; Ferrari Putti, F.; White, P.J.; dos Reis, A.R. Phytic acid accumulation in plants: Biosynthesis pathway regulation and role in human diet. Plant Physiol. Biochem. 2021, 164, 132–146. [Google Scholar] [CrossRef]
- Raboy, V. Approaches and challenges to engineering seed phytate and total phosphorus. Plant Sci. 2009, 177, 281–296. [Google Scholar] [CrossRef]
- Handa, V.; Sharma, D.; Kaur, A.; Arya, S.K. Biotechnological applications of microbial phytase and phytic acid in food and feed industries. Biocatal. Agric. Biotechnol. 2020, 25, 101600. [Google Scholar] [CrossRef]
- Raboy, V. Low phytic acid crops: Observations based on four decades of research. Plants 2020, 9, 140. [Google Scholar] [CrossRef] [PubMed]
- Brinch-Pedersen, H.; Sorensen, L.D.; Holm, P.B. Engineering crop plants: Getting a handle on phosphate. Trends Plant Sci. 2002, 7, 118–125. [Google Scholar] [CrossRef]
- Humer, E.; Schwarz, C.; Schedle, K. Phytate in pig and poultry nutrition. J. Anim. Physiol. Anim. Nutr. 2015, 99, 605–625. [Google Scholar] [CrossRef] [PubMed]
- Gupta, R.K.; Gangoliya, S.S.; Singh, N.K. Reduction of phytic acid and enhancement of bioavailable micronutrients in food grains. J. Food Sci. Technol. 2015, 52, 676–684. [Google Scholar] [CrossRef] [PubMed]
- Tangendjaja, B.; Buckle, K.A.; Wootton, M. Analysis of phytic acid by high-performance liquid-chromatography. J. Chromatogr. 1980, 197, 274–277. [Google Scholar] [CrossRef]
- Macho, S.; Larrechi, M.S. Near-infrared spectroscopy and multivariate calibration for the quantitative determination of certain properties in the petrochemical industry. Trac Trends Anal. Chem. (Regul. Ed.) 2002, 21, 799–806. [Google Scholar] [CrossRef]
- Zhang, W.; Kasun, L.C.; Wang, Q.J.; Zheng, Y.; Lin, Z. A Review of Machine Learning for Near-Infrared Spectroscopy. Sensors 2022, 22, 9764. [Google Scholar] [CrossRef]
- Chen, Y.; Bin, J.; Zou, C.; Ding, M. Discrimination of Fresh Tobacco Leaves with Different Maturity Levels by Near-Infrared (NIR) Spectroscopy and Deep Learning. J. Anal. Methods Chem. 2021, 2021, 1–11. [Google Scholar] [CrossRef]
- Li, C.; Zhao, T.; Li, C.; Mei, L.; Yu, E.; Dong, Y.; Chen, J.; Zhu, S. Determination of gossypol content in cottonseeds by near infrared spectroscopy based on Monte Carlo uninformative variable elimination and nonlinear calibration methods. Food Chem. 2017, 221, 990–996. [Google Scholar] [CrossRef] [PubMed]
- Quampah, A.; Huang, Z.R.; Wu, J.G.; Liu, H.Y.; Li, J.R.; Zhu, S.J.; Shi, C.H. Estimation of oil content and fatty acid composition in cottonseed kernel powder using near infrared reflectance spectroscopy. J. Am. Oil Chem. Soc. 2012, 89, 567–575. [Google Scholar] [CrossRef]
- Tian, J.; Chen, X.; Liang, Z.; Qi, W.; Zheng, X.; Lu, D.; Chen, B. Application of nir spectral standardization based on principal component score evaluation in wheat flour crude protein model sharing. J. Food Qual. 2022, 2022, 1–10. [Google Scholar] [CrossRef]
- Hu, M.L.; Zhi, C.Y.; Xue, X.M.; Wu, J.; Wang, J.; Yan, L.Y.; Wang, X.; Chen, Y.N.; Kang, Y.P.; Wang, Z.H.; et al. Establishment of near-infrared reflectance spectroscopy model for predicting sucrose content of single seed in peanut. Acta Agron. Sin. 2023, 49, 2498–2504. [Google Scholar]
- Daba, S.D.; Honigs, D.; McGee, R.J.; Kiszonas, A.M. Prediction of Protein Concentration in Pea (Pisum sativum L.) Using Near-Infrared Spectroscopy (NIRS) Systems. Foods 2022, 11, 3701. [Google Scholar] [CrossRef]
- Jiang, X.; Zhu, M.; Yao, J.; Zhang, Y.; Liu, Y. Study on the effect of apple size difference on soluble solids content model based on near-infrared (nir) spectroscopy. J. Spectrosc. 2022, 2022, 1–10. [Google Scholar] [CrossRef]
- Amuah, C.L.Y.; Teye, E.; Lamptey, F.P.; Nyandey, K.; Opoku-Ansah, J.; Adueming, P.O.-W. Feasibility study of the use of handheld nir spectrometer for simultaneous authentication and quantification of quality parameters in intact pineapple fruits. J. Spectrosc. 2019, 2019, 1–9. [Google Scholar] [CrossRef]
- Mishra, P.; Passos, D. A synergistic use of chemometrics and deep learning improved the predictive performance of near-infrared spectroscopy models for dry matter prediction in mango fruit. Chemom. Intell. Lab. Syst. 2021, 212, 104287. [Google Scholar] [CrossRef]
- Zhu, M.-Z.; Wen, B.; Wu, H.; Li, J.; Lin, H.; Li, Q.; Li, Y.; Huang, J.; Liu, Z. The Quality Control of Tea by Near-Infrared Reflectance (NIR) Spectroscopy and Chemometrics. J. Spectrosc. 2019, 2019, 1–11. [Google Scholar] [CrossRef]
- Yang, J.; Wang, J.; Lu, G.; Fei, S.; Yan, T.; Zhang, C.; Lu, X.; Yu, Z.; Li, W.; Tang, X. TeaNet: Deep learning on near-infrared spectroscopy (nir) data for the assurance of tea quality. Comput. Electron. Agric. 2021, 190, 106431. [Google Scholar] [CrossRef]
- Zhang, Z.-Y.; Wang, Y.-J.; Yan, H.; Chang, X.-W.; Zhou, G.-S.; Zhu, L.; Liu, P.; Guo, S.; Dong, T.T.X.; Duan, J.-A. Rapid geographical origin identification and quality assessment of angelicae sinensis radix by ft-nir spectroscopy. J. Anal. Methods Chem. 2021, 2021, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.; Bian, X.; Zheng, M.; Liu, P.; Lin, L.; Tan, X. Rapid determination of hemoglobin concentration by a novel ensemble extreme learning machine method combined with near-infrared spectroscopy. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2021, 263, 120138. [Google Scholar] [CrossRef]
- Yang, Y.; Zhang, X.; Yin, J.; Yu, X. Rapid and nondestructive on-site classification method for consumer-grade plastics based on portable nir spectrometer and machine learning. J. Spectrosc. 2020, 2020, 6631234. [Google Scholar] [CrossRef]
- Chen, H.; Xu, L.; Ai, W.; Lin, B.; Feng, Q.; Cai, K. Kernel functions embedded in support vector machine learning models for rapid water pollution assessment via near-infrared spectroscopy. Sci. Total Environ. 2020, 714, 136765. [Google Scholar] [CrossRef] [PubMed]
- Xu, Z.; Zhao, X.; Guo, X.; Guo, J. Deep Learning Application for Predicting Soil Organic Matter Content by VIS-NIR Spectroscopy. Comput. Intell. Neurosci. 2019, 2019, 3563761. [Google Scholar] [CrossRef]
- Parrish, F.W.; Madacsi, J.P.; Phillippy, B.Q.; Wilfred, A.G.; Buco, S.M. Determination of phytic acid in cottonseed by near-infrared reflectance spectroscopy. J. Agric. Food Chem. 1990, 38, 407–409. [Google Scholar] [CrossRef]
- Zhao, R.; Xu, X.; Li, J.; Li, C.; Chen, J.; Liu, Y.; Zhu, S. Rapid determination of phytic acid content in cottonseed meal via near infrared spectroscopy. J. Near Infrared Spectrosc. 2017, 25, 188–195. [Google Scholar] [CrossRef]
- Suykens, J.; Vandewalle, J. Least squares support vector machine classifiers. Neural Process. Lett. 1999, 9, 293–300. [Google Scholar] [CrossRef]
- Zhao, R. Optimization of Near Infrared Spectroscopy Nondestructive Analysis Technology for Phytic Acid Contents in Cottonseeds and Its Application in the Researches of Their Genotypic and Environmental Effects. Ph.D. Thesis, Zhejiang University, Hangzhou, China, 2017. [Google Scholar]
- Rinnan, A.; Berg FV, D.; Engelsen, S.B. Review of the most common pre-processing techniques for near-infrared spectra. TrAC Trends Anal. Chem. 2009, 28, 1201–1222. [Google Scholar] [CrossRef]
- Barnes, R.J.; Dhanoa, M.S.; Lister, S.J. Standard normal variate transformation and de-trending of near-infrared diffuse reflectance spectra. Appl. Spectrosc. 1989, 43, 772–777. [Google Scholar] [CrossRef]
- Savitzky, A.; Golay, M. Smoothing + differentiation of data by simplified least squares procedures. Anal. Chem. 1964, 36, 1627. [Google Scholar] [CrossRef]
- Hopke, P.K. The evolution of chemometrics. Anal. Chim. Acta 2003, 500, 365–377. [Google Scholar] [CrossRef]
- Kennard, R.W.; Stone, L.A. Computer aided design of experiments. Technometrics 1969, 11, 137. [Google Scholar] [CrossRef]
- Haaland, D.M.; Thomas, E.V. Partial least-squares methods for spectral analyses.1. Relation to other quantitative calibration methods and the extraction of qualitative information. Anal. Chem. 1988, 60, 1193–1202. [Google Scholar] [CrossRef]
- Nie, Z.; Han, J.; Liu, T.; Liu, X. Hot topic: Application of support vector machine method in prediction of alfalfa protein fractions by near infrared reflectance spectroscopy. J. Dairy Sci. 2008, 91, 2361–2369. [Google Scholar] [CrossRef]
- Breiman, L. Random forests. Mach. Learn. 2001, 45, 5–32. [Google Scholar] [CrossRef]
- Li, C.; Su, B.; Zhao, T.; Li, C.; Chen, J.; Zhu, S. Feasibility study on the use of near-infrared spectroscopy for rapid and nondestructive determination of gossypol content in intact cottonseeds. J. Cotton Res. 2021, 4, 13. [Google Scholar] [CrossRef]
- Batten, G.D. Phosphorus fractions in the grain of diploid, tetraploid, and hexaploid wheat grown with contrasting phosphorus supplies. Cereal Chem. 1986, 63, 384–387. [Google Scholar]
- Zagorodni, A.A.; Kotova, D.L.; Selemenev, V.F. Infrared spectroscopy of ion exchange resins: Chemical deterioration of the resins. React. Funct. Polym. 2002, 53, 157–171. [Google Scholar] [CrossRef]
- Rose, T.J.; Liu, L.; Wissuwa, M. Improving phosphorus efficiency in cereal crops: Is breeding for reduced grain phosphorus concentration part of the solution? Front. Plant Sci. 2013, 4, 444. [Google Scholar] [CrossRef]
- Zhao, Y. QTL Mapping and Genetic Analysis of Main Components in Upland Cottonseeds. Ph.D. Thesis, Zhejiang University, Hangzhou, China, 2020. [Google Scholar]
- Zhuang, T.; Xin, M.; Wang, Q.K.; Wang, Y.M.; Saeed, M.; Xing, H.X.; Zhang, H.J.; Zhang, Y.A.; Deng, Y.Y.; Zhang, G.H.; et al. Determination of protein and fatty acid composition of shell-intact upland cottonseed using near-infrared reflectance spectroscopy. Ind. Crops Prod. 2023, 191, 115909. [Google Scholar] [CrossRef]
- Nicolaï, B.M.; Beullens, K.; Bobelyn, E.; Peirs, A.; Saeys, W.; Theron, K.I.; Lammertyn, J. Nondestructive measurement of fruit and vegetable quality by means of NIR spectroscopy: A review. Postharvest Biol. Technol. 2007, 46, 99–118. [Google Scholar] [CrossRef]
- Breiman, L. Statistical modeling: The two cultures. Stat. Sci. 2001, 16, 199–215. [Google Scholar] [CrossRef]
- Hu, C.; Xu, H.; Fu, Z.; Wu, B.; Zhang, R.; Zhi, C. Non-destructive Identification of the geographical origin of red jujube by near-infrared spectroscopy and fuzzy clustering methods. Int. J. Food Prop. 2023, 26, 3275–3290. [Google Scholar] [CrossRef]
- Cozzolino, D. Advantages and limitations of using near infrared spectroscopy in plant phenomics applications. Comput. Electron. Agric. 2023, 212, 108078. [Google Scholar] [CrossRef]
- Anjali; Jena, A.; Bamola, A.; Mishra, S.; Jain, I.; Pathak, N.; Sharma, N.; Joshi, N.; Pandey, R.; Kaparwal, S.; et al. State-of-the-art non-destructive approaches for maturity index determination in fruits and vegetables: Principles, applications, and future directions. Food Prod. Process. Nutr. 2024, 6, 56. [Google Scholar] [CrossRef]
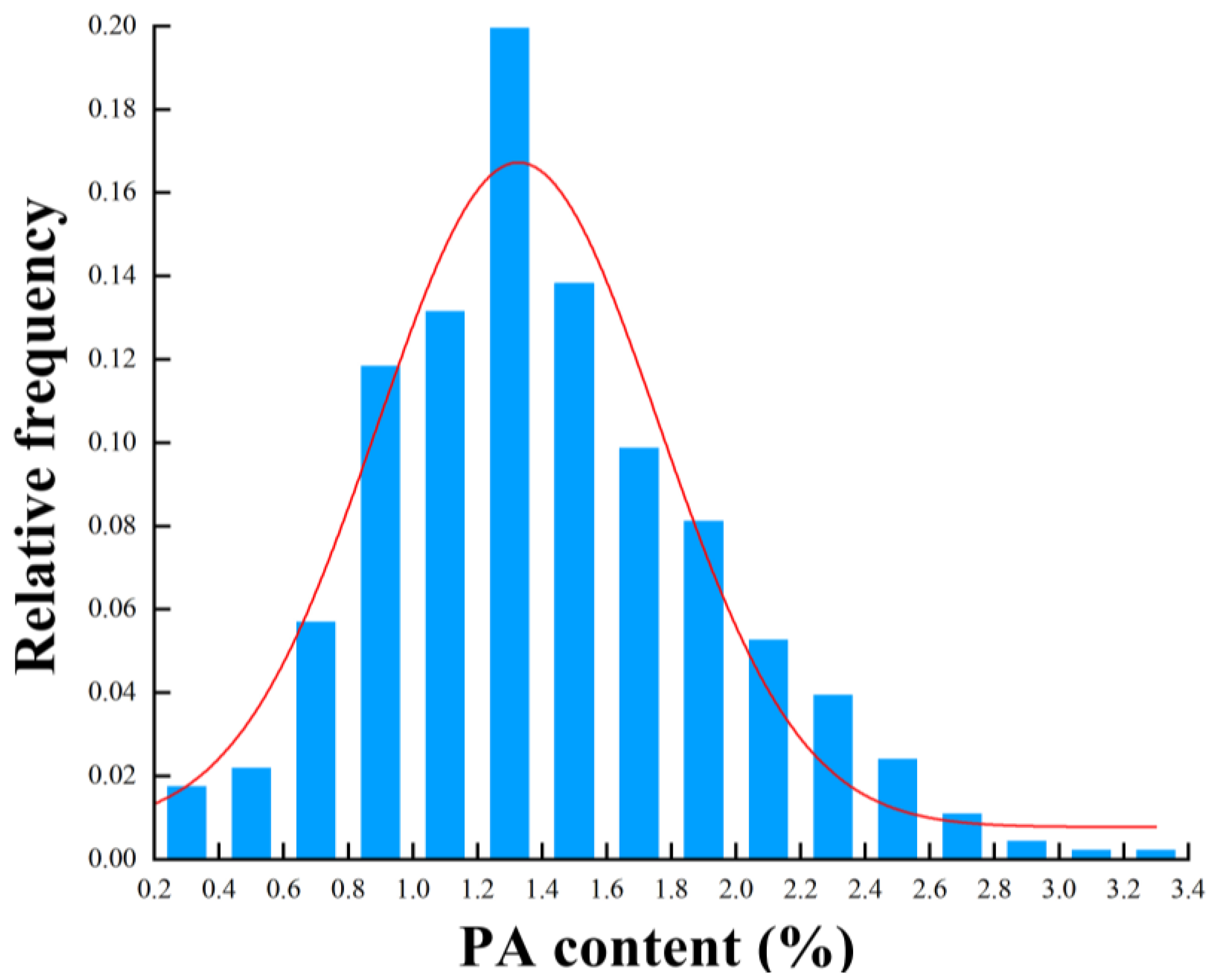
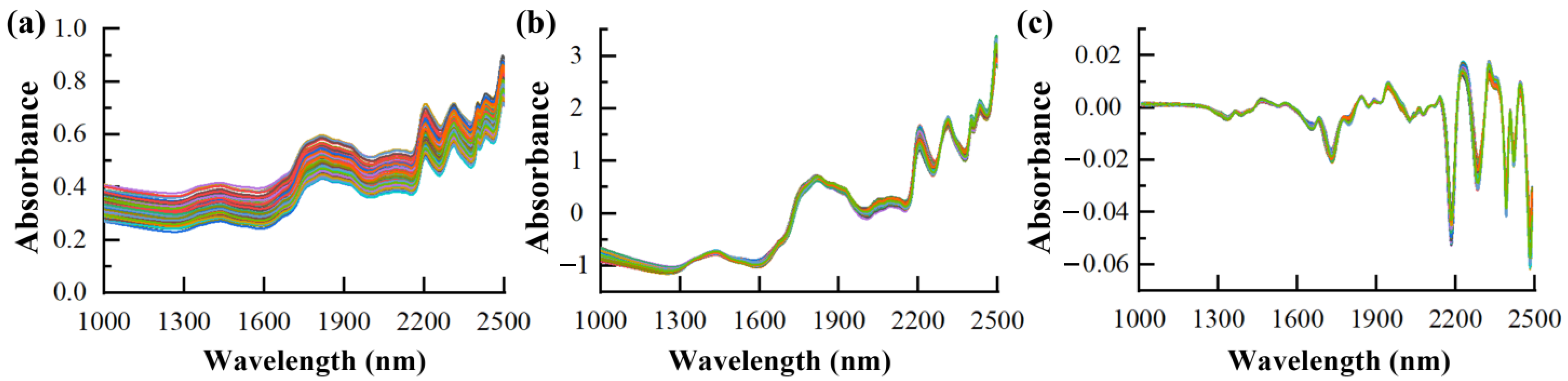
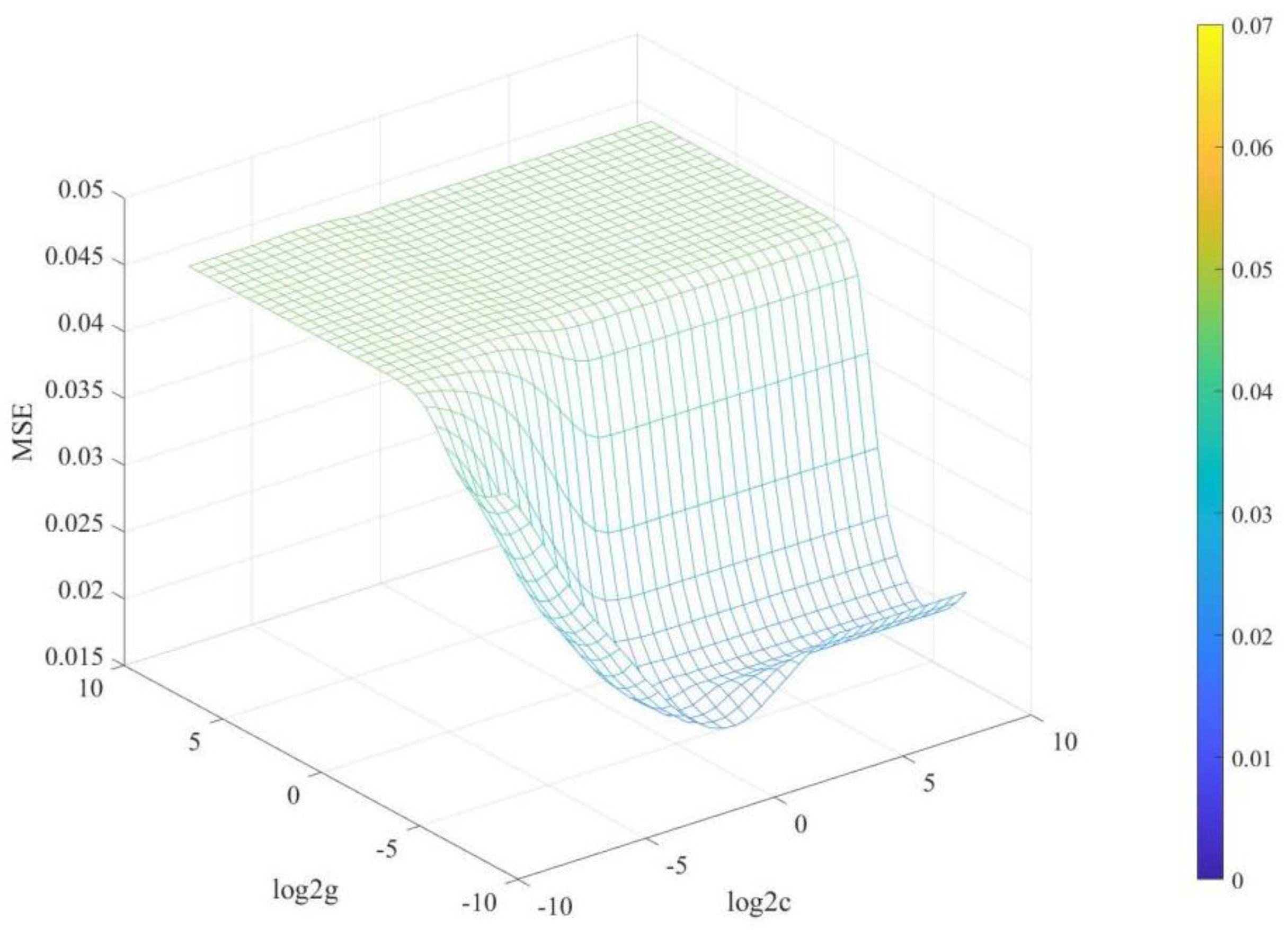

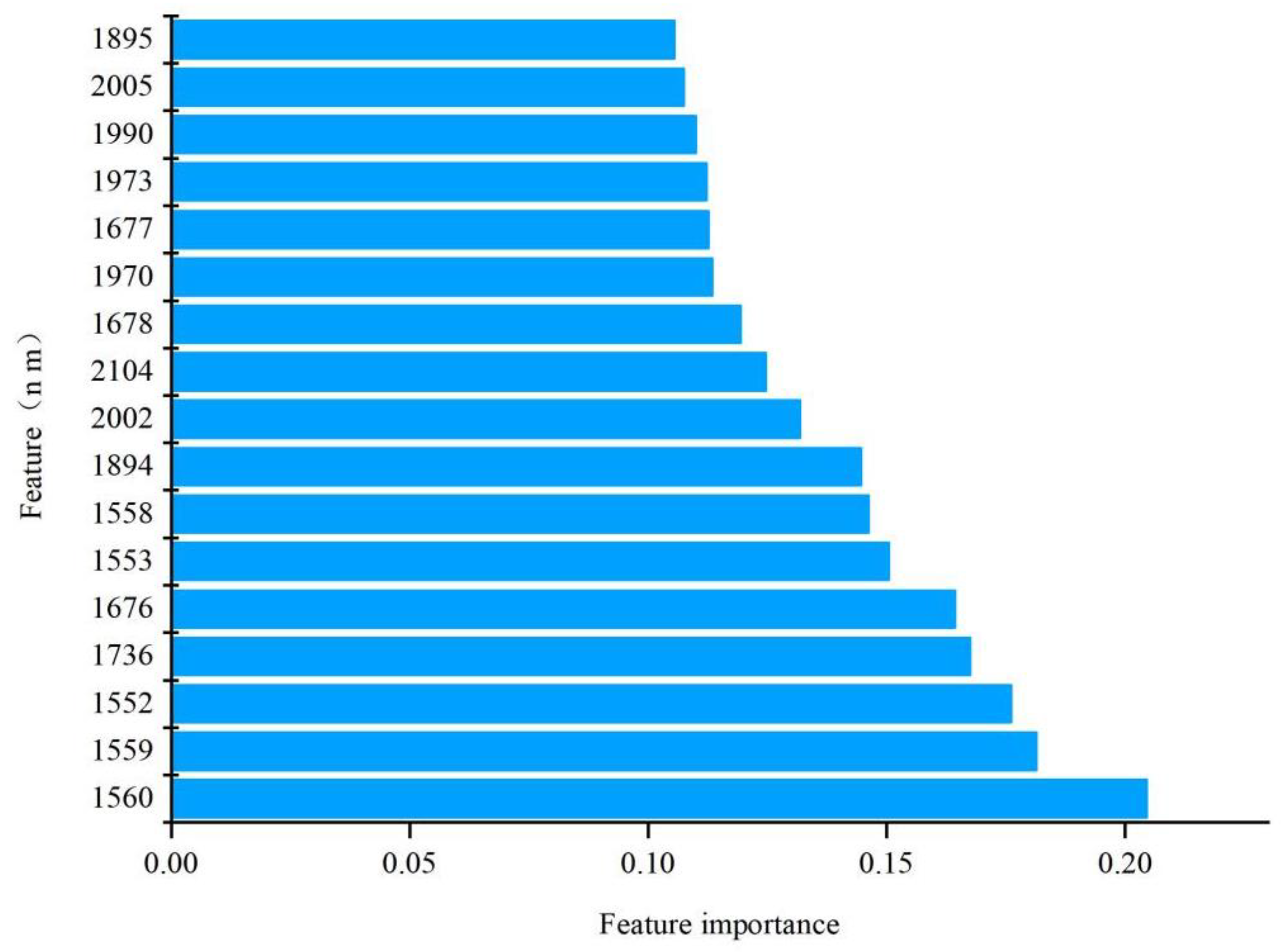
| Datasets | Min | Max | Average | SD |
|---|---|---|---|---|
| All samples | 0.7029 | 3.3267 | 1.9188 | 0.5473 |
| Calibration | 0.7029 | 3.3267 | 1.9390 | 0.5642 |
| Prediction | 0.8825 | 3.1411 | 1.8582 | 0.4930 |
| Methods | Model Parameter | |||
|---|---|---|---|---|
| RMSECV | RMSEP | RPD | R2p | |
| CK | 0.324 | 0.2785 | 1.7143 | 0.6886 |
| SG | 0.3203 | 0.2753 | 1.7457 | 0.6968 |
| 1D | 0.3252 | 0.256 | 1.7825 | 0.7426 |
| SNV | 0.3433 | 0.2456 | 1.8839 | 0.746 |
| MSC | 0.3316 | 0.2614 | 1.7999 | 0.7017 |
| SG+1D | 0.3394 | 0.2549 | 1.8261 | 0.7326 |
| SG+SNV | 0.3369 | 0.2652 | 1.7952 | 0.7139 |
| SG+MSC | 0.34 | 0.282 | 1.7139 | 0.6794 |
| SNV+1D | 0.3637 | 0.2409 | 2.1276 | 0.7865 |
| SG+SNV+1D | 0.3198 | 0.3049 | 1.4833 | 0.6353 |
| SG+MSC+1D | 0.3304 | 0.3046 | 1.4848 | 0.6309 |
| Models | Model Parameter | |||
|---|---|---|---|---|
| RMSECV | RMSEP | RPD | R2p | |
| PLS | 0.3637 | 0.2409 | 2.1276 | 0.7865 |
| SVM | 0.1274 | 0.2207 | 2.3141 | 0.8142 |
| RF | 0.0747 | 0.1294 | 3.9828 | 0.9114 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yin, H.; Mo, W.; Li, L.; Ma, Y.; Chen, J.; Zhu, S.; Zhao, T. Near-Infrared Spectroscopy Analysis of the Phytic Acid Content in Fuzzy Cottonseed Based on Machine Learning Algorithms. Foods 2024, 13, 1584. https://doi.org/10.3390/foods13101584
Yin H, Mo W, Li L, Ma Y, Chen J, Zhu S, Zhao T. Near-Infrared Spectroscopy Analysis of the Phytic Acid Content in Fuzzy Cottonseed Based on Machine Learning Algorithms. Foods. 2024; 13(10):1584. https://doi.org/10.3390/foods13101584
Chicago/Turabian StyleYin, Hong, Wenlong Mo, Luqiao Li, Yiting Ma, Jinhong Chen, Shuijin Zhu, and Tianlun Zhao. 2024. "Near-Infrared Spectroscopy Analysis of the Phytic Acid Content in Fuzzy Cottonseed Based on Machine Learning Algorithms" Foods 13, no. 10: 1584. https://doi.org/10.3390/foods13101584
APA StyleYin, H., Mo, W., Li, L., Ma, Y., Chen, J., Zhu, S., & Zhao, T. (2024). Near-Infrared Spectroscopy Analysis of the Phytic Acid Content in Fuzzy Cottonseed Based on Machine Learning Algorithms. Foods, 13(10), 1584. https://doi.org/10.3390/foods13101584







