Comparing the Effect of HPP on the Structure and Stability of Soluble and Membrane-Bound Polyphenol Oxidase from ‘Lijiang Snow’ Peach: Multispectroscopic and Molecular Dynamics Simulation
Abstract
1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Extraction and Purification of sPPO and mPPO
2.3. Enzyme Assay
2.4. Bioinformatics Analysis
2.5. Three-Dimensional Structure Modeling
2.6. High-Pressure Processing
2.7. CD Spectra Analysis
2.8. Fluorescence Spectra Analysis
2.9. Molecular Dynamics Simulation
2.10. Molecular Docking
2.11. Statistical Analysis
3. Results and Discussion
3.1. Bioinformatics and Three-Dimensional Structure Analysis of sPPO and mPPO
3.2. Effect of HPP on the Activity of sPPO and mPPO
3.3. Multispectral Analysis
3.4. MD Simulations of sPPO and mPPO at the Single-Molecule Level
3.4.1. RMSD and RMSF Analysis of sPPO and mPPO
3.4.2. The Secondary and Tertiary Structure Analysis of sPPO and mPPO
3.4.3. The Copper-Binding Region Analysis of sPPO and mPPO
3.4.4. The Three-Dimensional Conformational Change Analysis of sPPO and mPPO
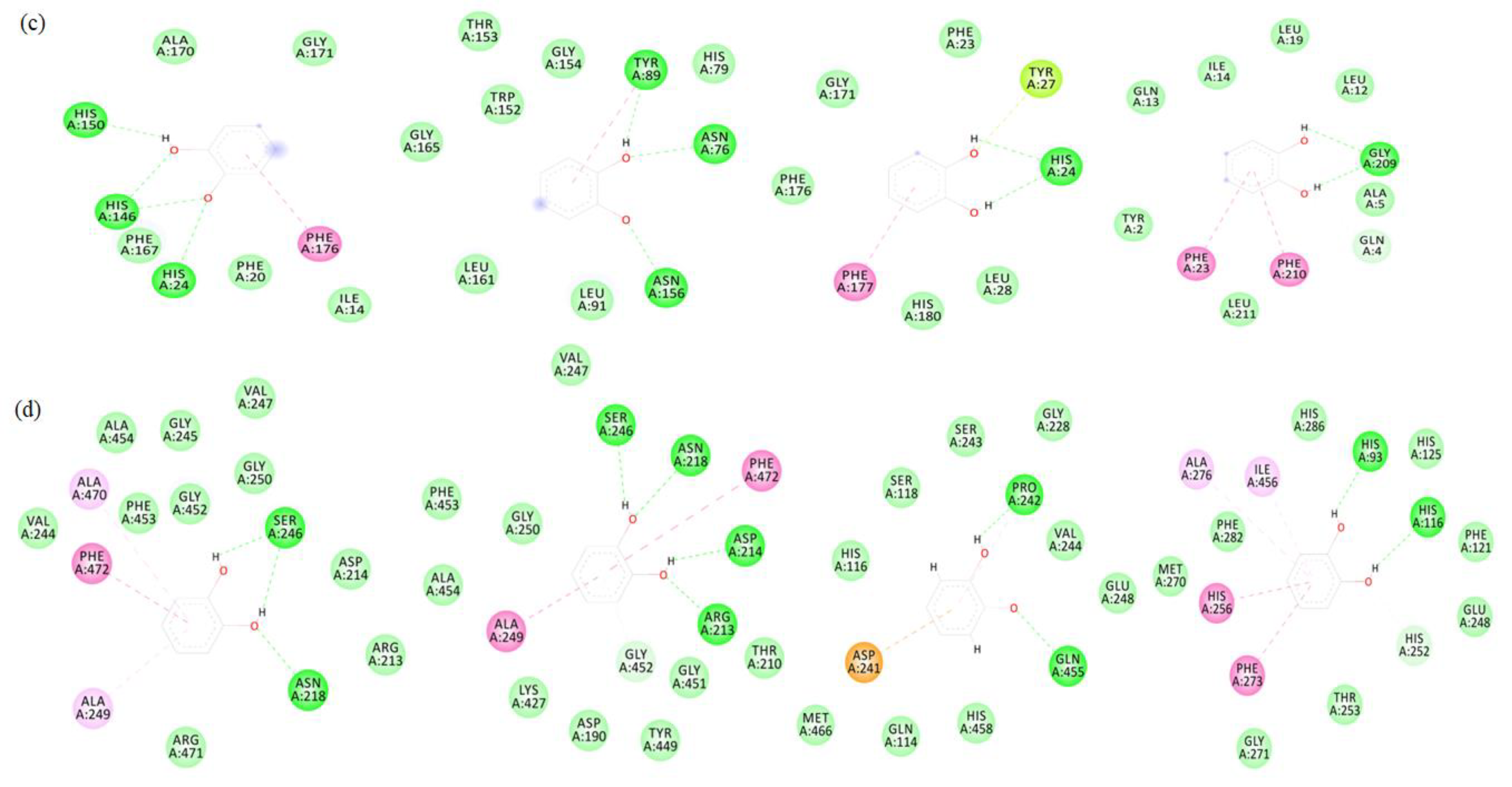
3.5. Molecular Docking
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Mayer, A.M. Polyphenol oxidases in plants and fungi: Going places? A review. Phytochemistry 2006, 67, 2318–2331. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.J.; Uyama, H. Tyrosinase inhibitors from natural and synthetic sources: Structure, inhibition mechanism and perspective for the future. Cell Mol. Life 2005, 62, 1707–1723. [Google Scholar] [CrossRef] [PubMed]
- Tinello, F.; Lante, A. Recent advances in controlling polyphenol oxidase activity of fruit and vegetable products. Innov. Food Sci. Emerg. Technol. 2018, 50, 73–83. [Google Scholar] [CrossRef]
- Yoruk, R.; Marshall, M.M.R. Physicochemical properties and function of plant polyphenol oxidase: A review. J. Food Biochem. 2003, 27, 361–422. [Google Scholar] [CrossRef]
- Derardja, A.E.; Pretzler, M.; Kampatsikas, I.; Barkat, M.; Rompel, A. Purification and Characterization of Latent Polyphenol Oxidase from Apricot (Prunus armeniaca L.). J. Agric. Food Chem. 2017, 65, 8203–8212. [Google Scholar] [CrossRef]
- Jia, S.; Jiang, S.; Chen, Y.; Wei, Y.; Shao, X. Comparison of Inhibitory Effects of Cinnamic Acid, β-Cyclodextrin, L-Cysteine, and Ascorbic Acid on Soluble and Membrane-Bound Polyphenol Oxidase in Peach Fruit. Foods 2023, 12, 167. [Google Scholar] [CrossRef]
- Liu, F.; Zhao, J.; Wen, X.; Ni, Y. Purification and structural analysis of membrane-bound polyphenol oxidase from Fuji apple. Food Chem. 2015, 183, 72–77. [Google Scholar] [CrossRef]
- Zaini, N.A.M.; Osman, A.; Hamid, A.; Ebrahimpour, N.; Saari, N. Purification and characterization of membrane-bound polyphenoloxidase (mPPO) from Snake fruit [Salacca zalacca (Gaertn.) Voss]. Food Chem. 2013, 136, 407–414. [Google Scholar] [CrossRef] [PubMed]
- Han, Q.; Liu, F.; Li, M.; Wang, K.; Ni, Y. Comparison of biochemical properties of membrane-bound and soluble polyphenol oxidase from Granny Smith apple (Malus × domestlca Borkh.). Food Chem. 2019, 289, 657–663. [Google Scholar] [CrossRef]
- Wang, F.; Zhou, H.; Cheng, F.; Niu, H.; Yuan, L.; Yi, J.; Zhou, L. Comparison of the characterization and the temperature/pressure stability of soluble and membrane-bound polyphenol oxidase from ‘Lijiang’ snow peach. LWT 2021, 146, 111401. [Google Scholar] [CrossRef]
- Ma, Y.; Chen, Y.Y.; Liu, P.; Meng, A.L.; Deng, L.; Xue, W.; Chen, F.; Che, Z.M. Comparative study of the biochemical properties of membrane-bound and soluble polyphenol oxidase from Prunus mume. LWT 2022, 171, 114156. [Google Scholar] [CrossRef]
- Hong, X.; Luo, X.; Wang, L.; Gong, D.; Zhang, G. New Insights into the Inhibition of Hesperetin on Polyphenol Oxidase: Inhibitory Kinetics, Binding Characteristics, Conformational Change and Computational Simulation. Foods 2023, 12, 905. [Google Scholar] [CrossRef] [PubMed]
- Bleoancă, I.; Saje, K.; Mihalcea, L.; Oniciuc, E.A.; Smole-Mozina, S.; Nicolau, A.I.; Borda, D. Contribution of high pressure and thyme extract to control Listeria monocytogenes in fresh cheese-A hurdle approach. Innov. Food Sci. Emerg. Technol. 2016, 38, 7–14. [Google Scholar] [CrossRef]
- Li, W.; He, X.; Chen, Y.; Lei, L.; Li, F.; Zhao, J.; Zeng, K.; Ming, J. Improving antioxidant activity and modifying Tartary buckwheat bran by steam explosion treatment. LWT 2022, 170, 114106. [Google Scholar] [CrossRef]
- Makroo, H.A.; Srivastava, B.; Jabeen, A. Influence of mild electric field (MEF) on polyphenol oxidase and quality attributes of pineapple juice during ohmic heating. LWT 2022, 156, 113021. [Google Scholar] [CrossRef]
- Basak, S.; Chakraborty, S. The potential of nonthermal techniques to achieve enzyme inactivation in fruit products. Trends Food Sci. Technol. 2022, 123, 114–129. [Google Scholar] [CrossRef]
- Gomes, M.R.A.; Ledward, D.A. Effect of high-pressure treatment on the activity of some polyphenoloxidases. Food Chem. 1996, 56, 1–5. [Google Scholar] [CrossRef]
- Sulaiman, A.; Soo, M.J.; Yoon, M.M.; Farid, M.; Silva, F.V. Modeling the polyphenoloxidase inactivation kinetics in pear, apple and strawberry purees after high pressure processing. J. Food Eng. 2015, 147, 89–94. [Google Scholar] [CrossRef]
- Tsikrika, K.; O’Brien, N.; Rai, D.K. The effect of high pressure processing on polyphenol oxidase activity, phytochemicals and proximate composition of irish potato cultivars. Foods 2019, 8, 517. [Google Scholar] [CrossRef] [PubMed]
- Wu, S.; Yang, R. Effect of high-pressure processing on polyphenol oxidase, melanosis and quality in ready-to-eat (RTE) crabs during storage. LWT 2023, 178, 114607. [Google Scholar] [CrossRef]
- Tiberio, A.L.; Leite, T.S.; Cristianini, M. High isostatic pressure and thermal processing of gal fruit (Euterpe oleracea Martius): Effect on pulp color and inactivation of peroxidase and polyphenol oxidase. Food Res. Int. 2018, 105, 853–862. [Google Scholar]
- Zhou, H.; Wang, F.; Niu, H.; Yuan, L.; Tian, J.; Cai, S.; Bi, X.; Zhou, L. Structural studies and molecular dynamic simulations of polyphenol oxidase treated by high pressure processing. Food Chem. 2022, 372, 131243. [Google Scholar] [CrossRef] [PubMed]
- Hata, H.; Nishiyama, M.; Kitao, A. Molecular dynamics simulation of proteins under high pressure: Structure, function and thermodynamics. BBA-Gen Subj. 2020, 1864, 129395. [Google Scholar] [CrossRef]
- Yi, J.; Jiang, B.; Zhang, Z.; Liao, X.; Zhang, Y.; Hu, X. Effect of ultrahigh hydrostatic pressure on the activity and structure of mushroom (Agaricus bisporus) polyphenoloxidase. J. Agric. Food Chem. 2012, 60, 593–599. [Google Scholar] [CrossRef] [PubMed]
- Zhou, L.; Liu, W.; Terefe, N.S. The inactivation kinetics of soluble and membrane-bound polyphenol oxidase in pear during thermal and high-pressure processing. Food Bioprocess Technol. 2018, 11, 1039–1049. [Google Scholar] [CrossRef]
- Zhou, H.; Bie, S.; Li, J.; Yuan, L.; Zhou, L. Comparison on inhibitory effect and mechanism of inhibitors on sPPO and mPPO purified from ‘Lijiang snow’ peach by combining multispectroscopic analysis, molecular docking and molecular dynamics simulation. Food Chem. 2023, 400, 134048. [Google Scholar] [CrossRef]
- Laemmli, U.K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 1970, 227, 680–685. [Google Scholar] [CrossRef]
- Terefe, N.S.; Delon, A.; Buckow, R.; Versteeg, C. Blueberry polyphenol oxidase: Characterization and the kinetics of thermal and high pressure activation and inactivation. Food Chem. 2015, 188, 193–200. [Google Scholar] [CrossRef]
- Chen, G.; Miao, M.; Jiang, B.; Jin, J.; Campanella, O.H.; Feng, B. Effects of high hydrostatic pressure on lipase from Rhizopus chinensis: I. Conformational changes. Innov. Food Sci. Emerg. Technol. 2017, 41, 267–276. [Google Scholar] [CrossRef]
- Pronk, S.; Páll, S.; Schulz, R.; Larsson, P. GROMACS 4.5: A high-throughput and highly parallel open source molecular simulation toolkit. Bioinformatics 2013, 29, 845–854. [Google Scholar] [CrossRef]
- Liu, F.; Zhao, J.; Gan, Z.; Ni, Y. Comparison of membrane-bound and soluble polyphenol oxidase in Fuji apple (Malus domestica Borkh. cv. Red Fuji). Food Chem. 2015, 173, 86–91. [Google Scholar] [CrossRef] [PubMed]
- Li, F. Purification, kinetic parameters, and isoforms of polyphenol oxidase from “Xushu 22” sweet potato skin. J. Food Biochem. 2020, 44, e123452. [Google Scholar] [CrossRef] [PubMed]
- Hunt, M.D.; Eannetta, N.T.; Yu, H. cDNA cloning and expression of potato polyphenol oxidase. Plant Mol. Biol. 1993, 21, 59–68. [Google Scholar] [CrossRef] [PubMed]
- Krogh, A.; Larsson, B.; Von-Heijne, G.; Sonnhammer, E.L. Predicting transmembrane protein topology with a hidden Markov model: Application to complete genomes. J. Mol. Biol. 2001, 305, 567–580. [Google Scholar] [CrossRef]
- Darnell, S.J.; Riese, M.; Edlunde, E.G.; Blattner, F.R. Enhancing B-cell epitope predictions by integrating protein sequence and structural bioinformatics. Biophys. J. 2014, 106, 207a. [Google Scholar] [CrossRef]
- Mune Mune, M.A.; Stănciuc, N.; Grigore-Gurgu, L.; Aprodu, I.; Borda, D. Structural changes induced by high pressure processing in Bambara bean proteins at different pH. LWT 2020, 124, 109187. [Google Scholar] [CrossRef]
- Hou, Z.; Zhao, L.; Wang, Y.; Liao, X. Effects of high pressure on activities and properties of superoxide dismutase from chestnut rose. Food Chem. 2019, 294, 557–564. [Google Scholar] [CrossRef]
- Hu, W.; Zhang, Y.; Wang, Y.; Zhou, L.; Leng, X.; Liao, X.; Hu, X. Aggregation and homogenization, surface charge and structural change, and inactivation of mushroom tyrosinase in an aqueous system by subcritical/supercritical carbon dioxide. Langmuir 2011, 27, 909–916. [Google Scholar] [CrossRef] [PubMed]
- Gaigalas, A.K.; Li, L.; Henderson, O.; Vogt, R.; Barr, J.; Marti, G.; Weaver, J.; Schwartz, A. The development of fluorescence intensity standards. J. Res. Natl. Inst. Stan. 2001, 106, 381–389. [Google Scholar] [CrossRef]
- Grigera, J.R.; McCarthy, A.N. The behavior of the hydrophobic effect under pressure and protein denaturation. Biophys. J. 2010, 98, 1626–1631. [Google Scholar] [CrossRef]
- Baildya, N.; Ghosh, N.N.; Chattopadhyay, A.P. Inhibitory activity of hydroxychloroquine on COVID-19 main protease: An insight from MD-simulation studies. J. Mol. Struct. 2020, 1219, 128595. [Google Scholar] [CrossRef] [PubMed]
- Munishkina, L.A.; Fink, A.L. Fluorescence as a method to reveal structures and membrane-interactions of amyloidogenic proteins. BBA-Biomembranes 2007, 1768, 1862–1885. [Google Scholar] [CrossRef]
- Li, Y.; Miao, M.; Liu, M.; Jiang, B.; Zhang, T.; Chen, X. Sorbitol counteracts high hydrostatic pressure-induced denaturation of inulin fructotransferase. Int. J. Biol. Macromol. 2014, 70, 251–256. [Google Scholar] [CrossRef] [PubMed]
- Gaykema, W.P.J.; Hol, W.G.J.; Vereijken, J.M.; Soeter, N.M.; Bak, H.J.; Beintema, J.J. 3.2 Å structure of the copper-containing, oxygen-carrying protein Panulirus interruptus haemocyanin. Nature 1984, 309, 23–29. [Google Scholar] [CrossRef]
- Si, Y.; Yin, S.; Park, D.; Chung, H.; Yan, L.; Lu, Z.; Zhou, H.M.; Yang, J.M.; Qian, G.Y.; Park, Y.D. Tyrosinase inhibition by isophthalic acid: Kinetics and computational simulation. Int. J. Biol. Macromol. 2011, 48, 700–704. [Google Scholar] [CrossRef]
- Ismaya, W.T.; Rozeboom, H.J.; Weijn, A.; Mes, J.J.; Fusetti, F.; Wichers, H.J.; Dijkstra, B.W. Crystal structure of Agaricus bisporus mushroom tyrosinase: Identity of the tetramer subunits and interaction with tropolone. Biochemistry 2011, 50, 5477–5486. [Google Scholar] [PubMed]
- Gao, X.; Yin, Y.; Yan, J.; Zhang, J.; Ma, H.; Zhou, C. Separation, biochemical characterization and salt-tolerant mechanisms of alkaline protease from Aspergillus oryzae. J. Sci. Food Agric. 2019, 99, 3359–3366. [Google Scholar] [CrossRef]
- Rabbani, G.; Ahmad, E.; Khan, M.V.; Ashraf, M.T.; Bhat, R.; Khan, R.H. Impact of structural stability of cold adapted candida antarctica lipase b (calb): In relation to pH, chemical and thermal denaturation. RSC Adv. 2015, 5, 20115–20131. [Google Scholar]
- McDonald, I.K.; Thornton, J.M. Satisfying hydrogen bonding potential in proteins. J. Mol. Biol. 1994, 238, 777–793. [Google Scholar]
- Derreumaux, P.; Schlick, T. The loop opening/closing motion of the enzyme triosephosphate isomerase. Biophys. J. 1998, 74, 72–81. [Google Scholar] [CrossRef]
- Gerstein, M.; Lesk, A.M.; Chothia, C. Structural mechanisms for domain movements in proteins. Biochemistry 1994, 33, 6739–6749. [Google Scholar] [CrossRef] [PubMed]
- Brown, E.; Hooper, L.; Ho, T.; Gresham, H. Integrin-associated protein: A 50-kD plasma membrane antigen physically and functionally associated with integrins. J. Cell Biol. 1990, 111, 2785–2794. [Google Scholar] [CrossRef] [PubMed]
- Schumacher, M.A.; Tonthat, N.K.; Lee, J.; Rodriguez-Castañeda, F.A.; Chinnam, N.B.; Kalliomaa-Sanford, A.K.; Ng, I.W.; Barge, M.T.; Shaw, P.L.R.; Barillà, D. Structures of archaeal DNA segregation machinery reveal bacterial and eukaryotic linkages. Science 2015, 349, 1120–1124. [Google Scholar] [CrossRef]
- Zhang, C.; Ma, Y.; Gao, F.; Zhao, Y.; Cai, S.; Pang, M. The free, esterified, and insoluble-bound phenolic profiles of Rhus chinensis Mill. fruits and their pancreatic lipase inhibitory activities with molecular docking analysis. J. Funct. Foods 2018, 40, 729–735. [Google Scholar] [CrossRef]
- Huang, Y.; Zhang, X.; Suo, H. Interaction between β-lactoglobulin and EGCG under high-pressure by molecular dynamics simulation. PLoS ONE 2021, 16, e0255866. [Google Scholar] [CrossRef] [PubMed]



 : α-helix;
: α-helix;  : coil;
: coil;  : β-sheet;
: β-sheet;  : 3-10 helix;
: 3-10 helix;  : isolated beta bridge).
: isolated beta bridge).
 : α-helix;
: α-helix;  : coil;
: coil;  : β-sheet;
: β-sheet;  : 3-10 helix;
: 3-10 helix;  : isolated beta bridge).
: isolated beta bridge).
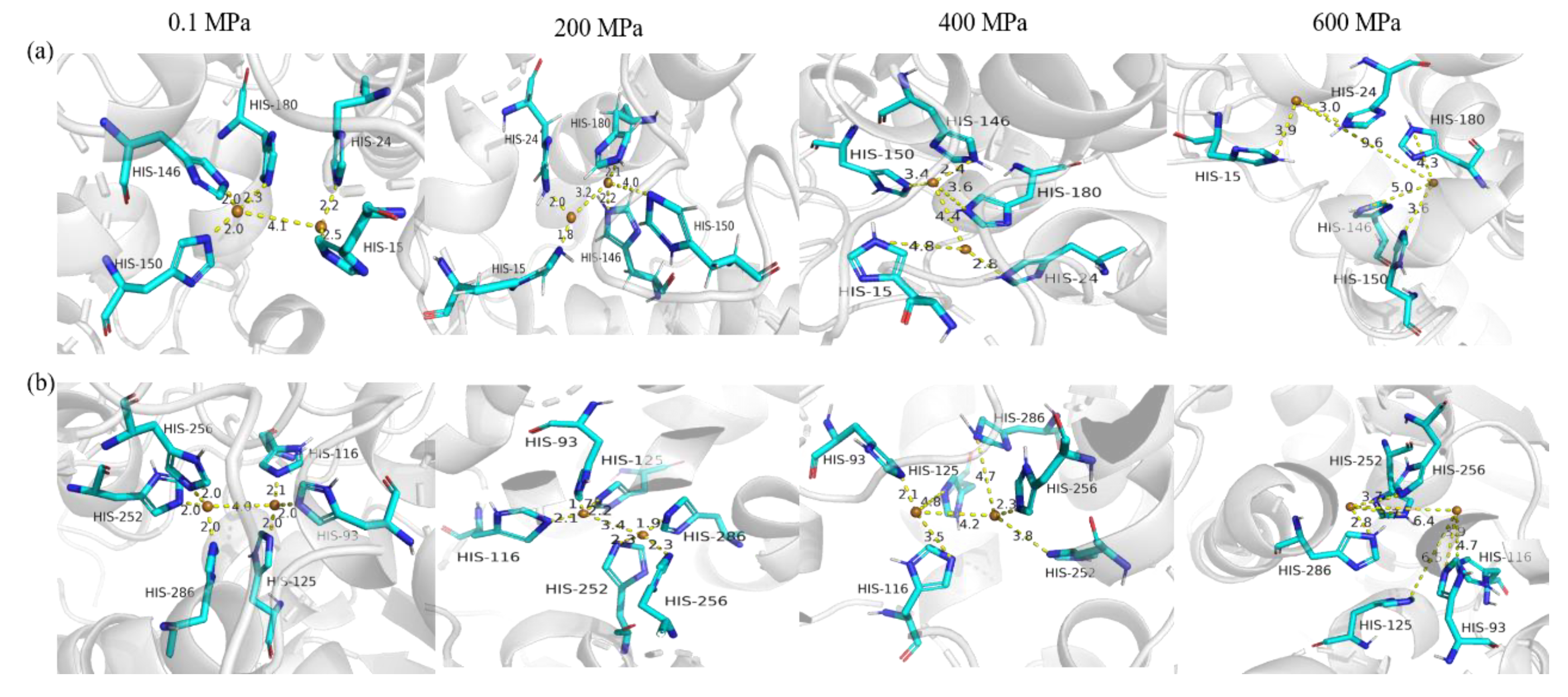

 ) represents van der Waals force; dark green (
) represents van der Waals force; dark green ( ) represents hydrogen bond; other colors represent pi conjugation.
) represents hydrogen bond; other colors represent pi conjugation.
 ) represents van der Waals force; dark green (
) represents van der Waals force; dark green ( ) represents hydrogen bond; other colors represent pi conjugation.
) represents hydrogen bond; other colors represent pi conjugation.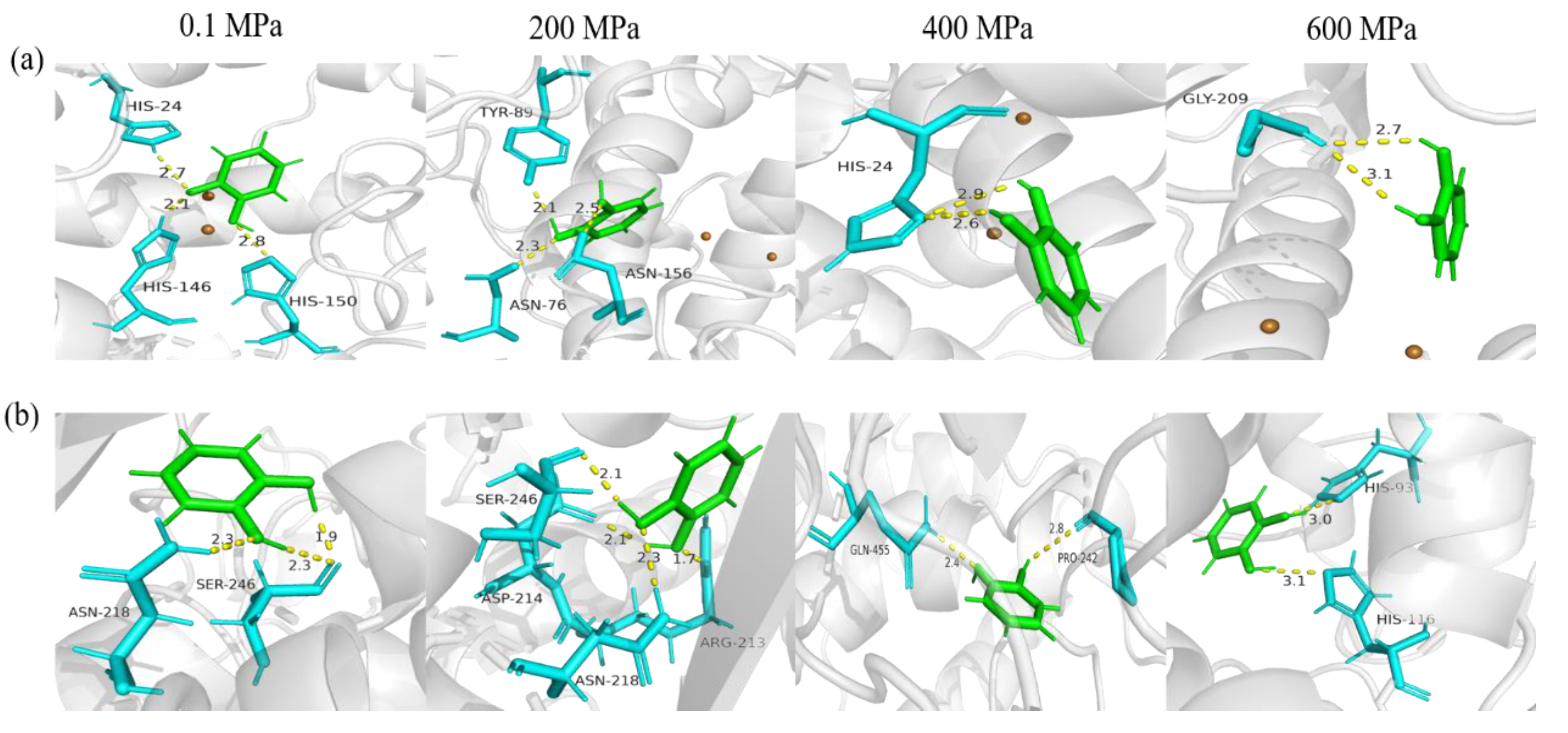
| Purification Stages | Total Activity (U) | Total Protein (mg) | Specific Activity (U/mg) | Yield (%) | Purification Fold | |
|---|---|---|---|---|---|---|
| sPPO | Crude extract | 106,600 | 41.82 | 2549.02 | 100 | 1 |
| 40–80% (NH4)2SO4 precipitation | 49,650 | 12.05 | 4120.33 | 28.81 | 1.62 | |
| DEAE Sepharose fast flow | 18,350 | 0.9812 | 18,701.59 | 2.35 | 7.34 | |
| mPPO | Crude extract | 185,240 | 53.45 | 3465.67 | 100 | 1 |
| 40–80% (NH4)2SO4 precipitation | 106,340 | 19.06 | 5579.22 | 35.66 | 1.61 | |
| DEAE Sepharose fast flow | 86,360 | 2.188 | 39,469.83 | 4.10 | 11.39 | |
| Sephacryl S-200 HR 16/60 | 53,150 | 0.553 | 96,112.16 | 1.04 | 27.73 | |
| Pressure (MPa) | Volume (nm3) | Rg (nm) | SASA (nm2) | Hydrogen Bonds | |
|---|---|---|---|---|---|
| sPPO | 0.1 | 372.242 | 2.311 | 12,113.271 | 185.995 |
| 200 | 364.894 | 2.295 | 11,972.615 | 186.592 | |
| 400 | 361.495 | 2.262 | 11,603.713 | 180.403 | |
| 600 | 348.742 | 2.162 | 12,109.229 | 171.095 | |
| mPPO | 0.1 | 682.572 | 2.264 | 16,219.570 | 406.263 |
| 200 | 678.115 | 2.258 | 16,358.267 | 411.625 | |
| 400 | 670.246 | 2.247 | 16,714.924 | 409.374 | |
| 600 | 665.864 | 2.232 | 16,993.183 | 402.333 | |
| Docking Indexes | 0.1 MPa | 200 MPa | 400 MPa | 600 MPa | |
|---|---|---|---|---|---|
| sPPO | T-Score | 4.065 | 4.696 | 4.016 | 3.503 |
| Number of hydrogen bonds | 3 | 3 | 2 | 2 | |
| Amino acid residues involved in hydrogen bonds | His24, His146, His150 | Asn76, Trp89, Asn156 | His24 | Gly209 | |
| Average distance | 2.53 Å | 2.3 Å | 2.75 Å | 2.90 Å | |
| mPPO | T-Score | 4.196 | 4.319 | 3.93 | 3.866 |
| Number of hydrogen bonds | 3 | 4 | 2 | 2 | |
| Amino acid residues involved in hydrogen bonds | Asn218, Ser246 | Arg213, Asp214, Asn218, Ser246 | Pro242, Gln455 | His93, His116 | |
| Average distance | 2.17 Å | 2.05 Å | 2.60 Å | 3.05 Å |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhou, H.; Bie, S.; Li, Z.; Zhou, L. Comparing the Effect of HPP on the Structure and Stability of Soluble and Membrane-Bound Polyphenol Oxidase from ‘Lijiang Snow’ Peach: Multispectroscopic and Molecular Dynamics Simulation. Foods 2023, 12, 1820. https://doi.org/10.3390/foods12091820
Zhou H, Bie S, Li Z, Zhou L. Comparing the Effect of HPP on the Structure and Stability of Soluble and Membrane-Bound Polyphenol Oxidase from ‘Lijiang Snow’ Peach: Multispectroscopic and Molecular Dynamics Simulation. Foods. 2023; 12(9):1820. https://doi.org/10.3390/foods12091820
Chicago/Turabian StyleZhou, Hengle, Shenke Bie, Zi Li, and Linyan Zhou. 2023. "Comparing the Effect of HPP on the Structure and Stability of Soluble and Membrane-Bound Polyphenol Oxidase from ‘Lijiang Snow’ Peach: Multispectroscopic and Molecular Dynamics Simulation" Foods 12, no. 9: 1820. https://doi.org/10.3390/foods12091820
APA StyleZhou, H., Bie, S., Li, Z., & Zhou, L. (2023). Comparing the Effect of HPP on the Structure and Stability of Soluble and Membrane-Bound Polyphenol Oxidase from ‘Lijiang Snow’ Peach: Multispectroscopic and Molecular Dynamics Simulation. Foods, 12(9), 1820. https://doi.org/10.3390/foods12091820








