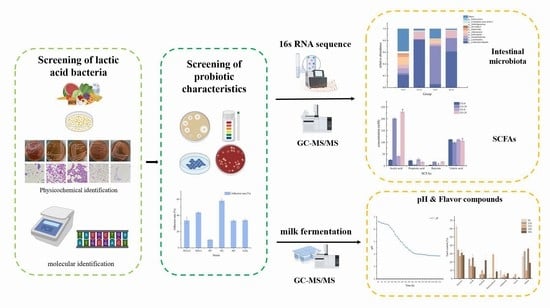
Screening, Identification and Physiological Characteristics of Lactobacillus rhamnosus M3 (1) against Intestinal Inflammation
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Collection
2.2. Reagents
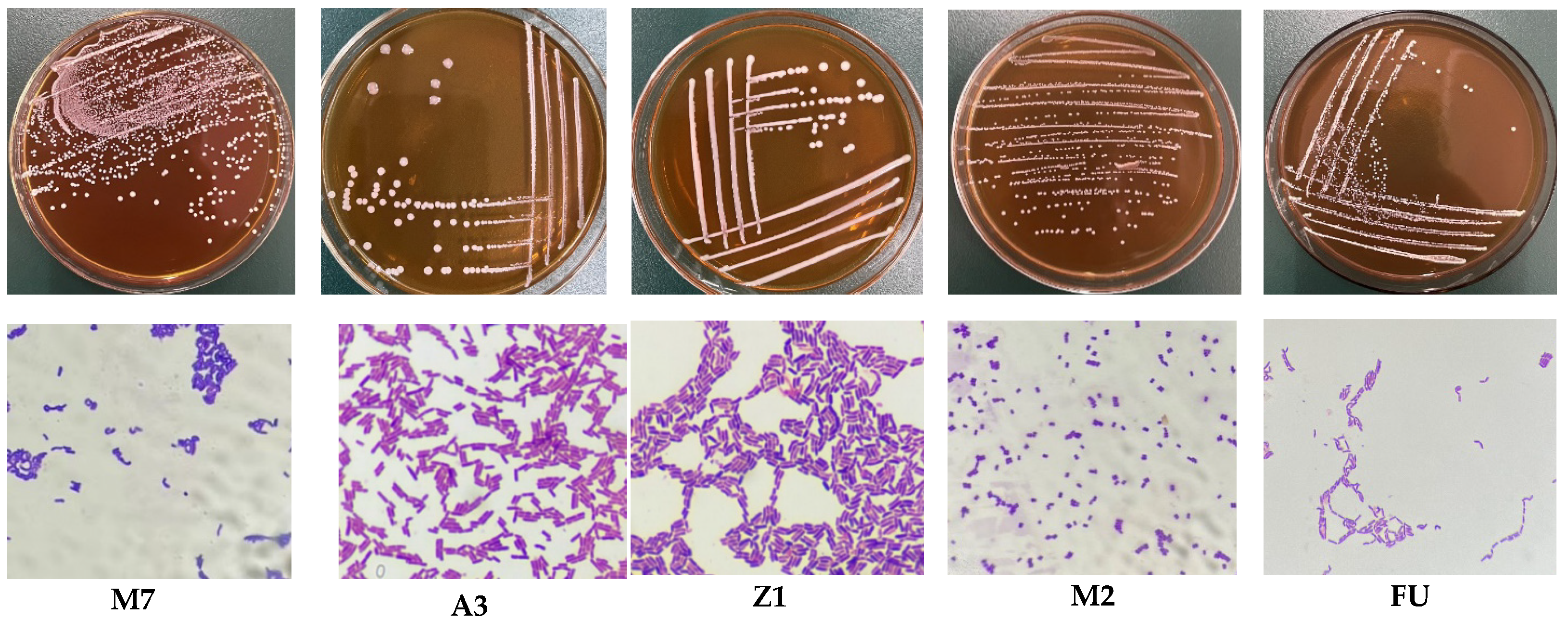
2.3. Isolation and Purification of Bacterial Strains
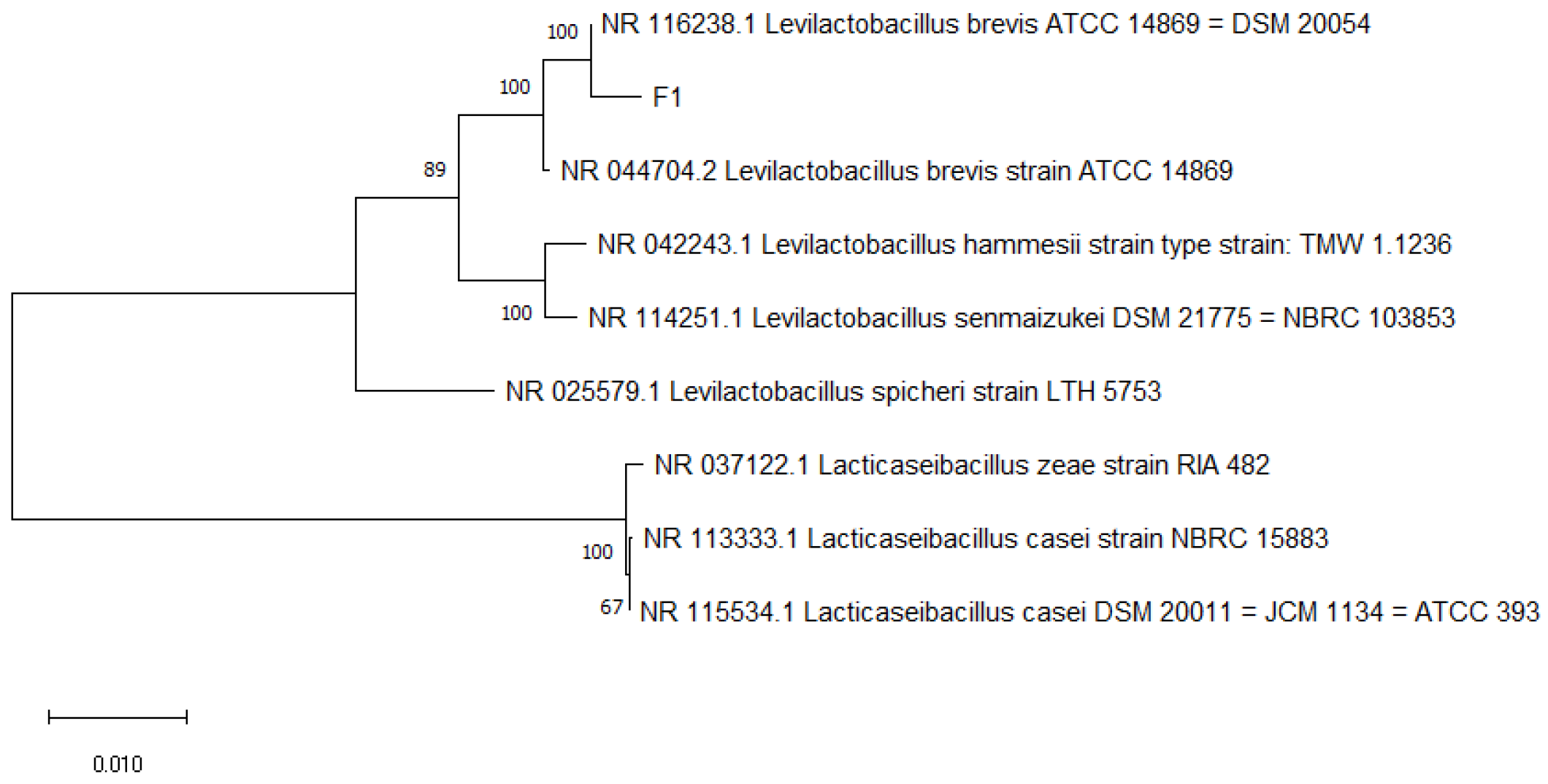
2.4. Identification of LAB
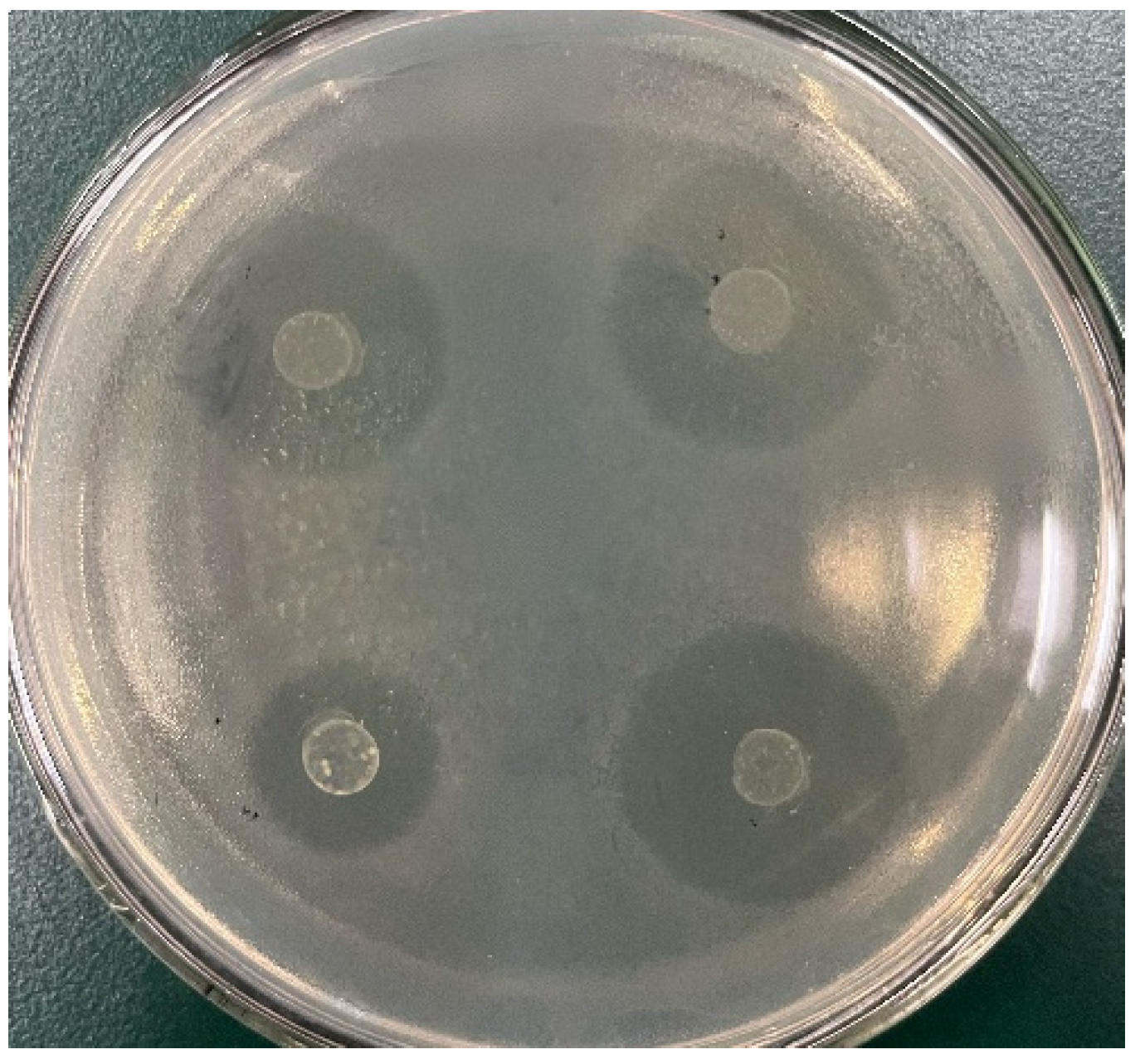
2.5. Antibacterial Experiment
2.6. Determination of LAB Tolerance
2.6.1. Acids Resistance Test
2.6.2. Bile Salt Resistance Experiment
2.6.3. Simulated Gastrointestinal Fluid Tolerance Experiment
2.7. Determination of Adhesion Ability of LAB
2.8. In Vitro Safety Experiment
2.8.1. Hemolytic Test
2.8.2. Antibiotic Sensitivity Test
2.9. In Vitro Fermentation and Detection
2.9.1. In Vitro Fermentation
- Sample collection: This study was approved by the Biomedical Research Ethics Committee of the Hunan Agricultural University (Protocol Code: Ethical Review 2022 No. 117, 10 November 2022). All participants provided written informed consent. Fecal samples were collected from 8 donors with diarrhea symptoms, all of whom had no underlying diseases and had not taken antibiotics within 3 months before collection. The collected samples were mixed with the same amount of feces and frozen in a refrigerator at −80 °C until use;
- Preparation of basal medium [19]: The medium composition (/L) was yeast extract 2.0 g, peptone 2.0 g, sodium chloride 0.1 g, potassium dihydrogen phosphate 0.04 g, sodium bicarbonate 2.0 g, calcium chloride 0.01 g, magnesium sulfate 0.01 g, hemin 50 mg, vitamin K 10 μL, bile salt 0.5 g, L-cysteine 0.5 g, and resazurin 1.0 mg. The basic medium was divided into test tubes and sterilized at 121 °C for 20 min. After sterilization, the medium was removed and placed in an anaerobic incubator for 18–24 h to ensure the container was anaerobic before the experiment. The expected color of the basic medium in the anaerobic environment was light yellow, and the medium was to turn red rapidly in the presence of oxygen;
- In vitro fermentation: The feces were taken out and mixed evenly in a sterile container, and sterile normal saline with a weight of 9 times the weight of the feces was added to mix thoroughly. The gauze was filtered to obtain a 10% fully mixed fecal suspension. Then, 10% fecal suspension was added to each test tube. The experimental group received 10% LAB suspension with a concentration of about 108 CFU/mL, and the same amount of sterile water was added to the control group. The samples were cultured in an anaerobic incubator at 37 °C for 24 h, and the samples were collected aseptically at 0 and 24 h, respectively. The samples were taken out and immediately placed in ice water to stop fermentation and then stored at −80 °C.
2.9.2. DNA Extraction and Sequencing
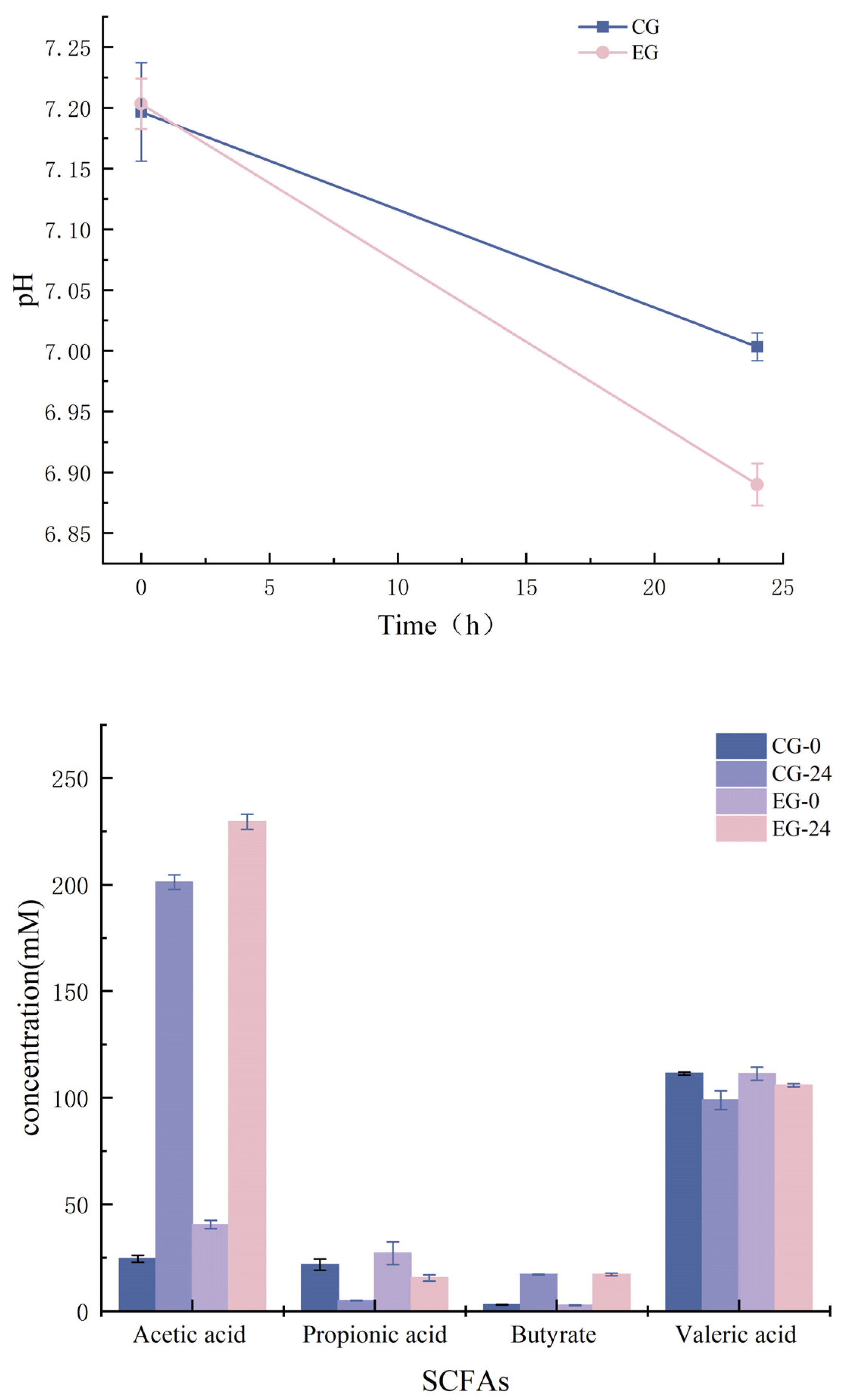
2.9.3. Detection of PH Value and Determination of Short-Chain Fatty Acids (SCFAs)
2.10. Study on the Milk Fermentation Performance
2.10.1. Preparation of Fermented Milk
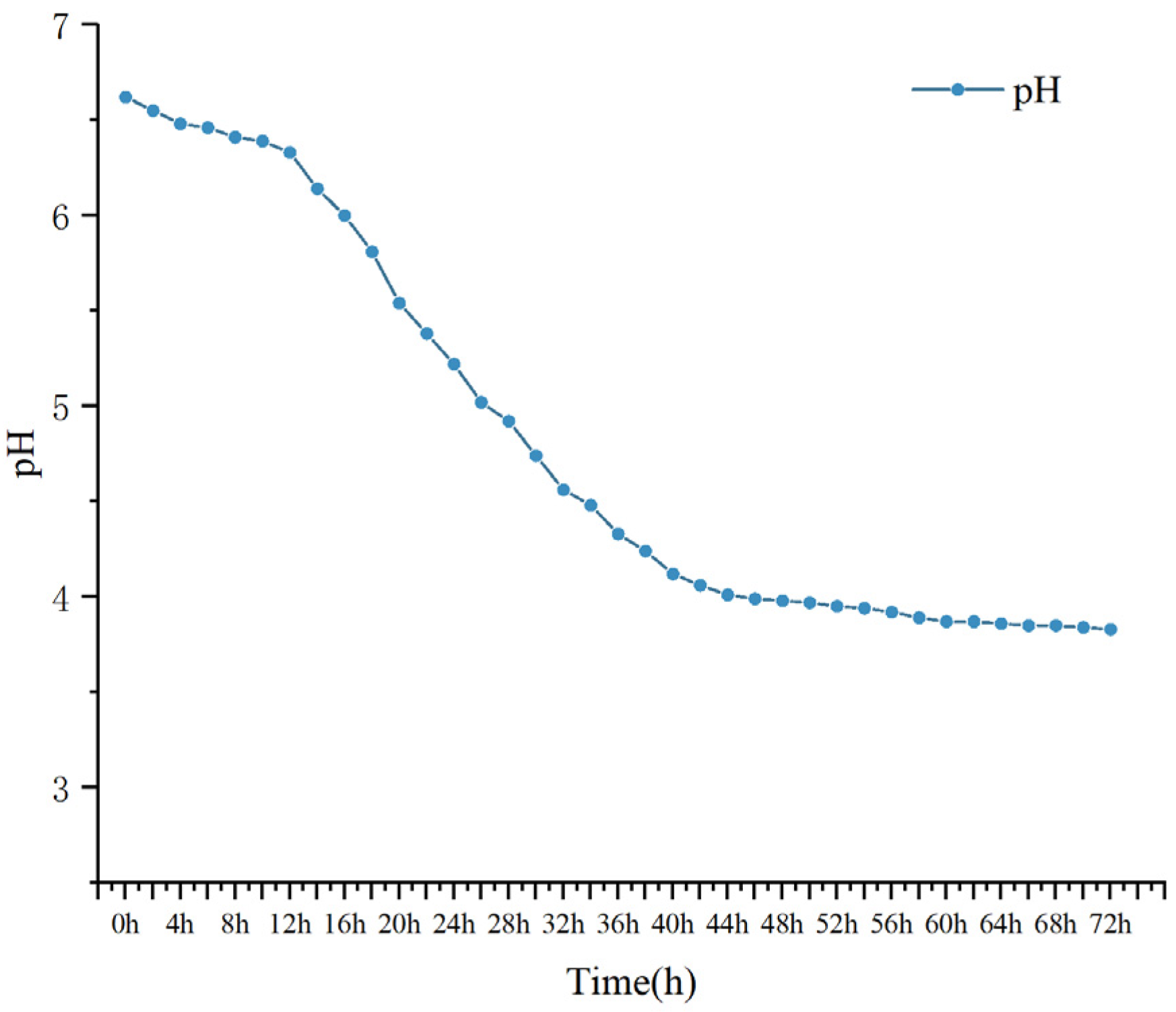
2.10.2. PH Changes
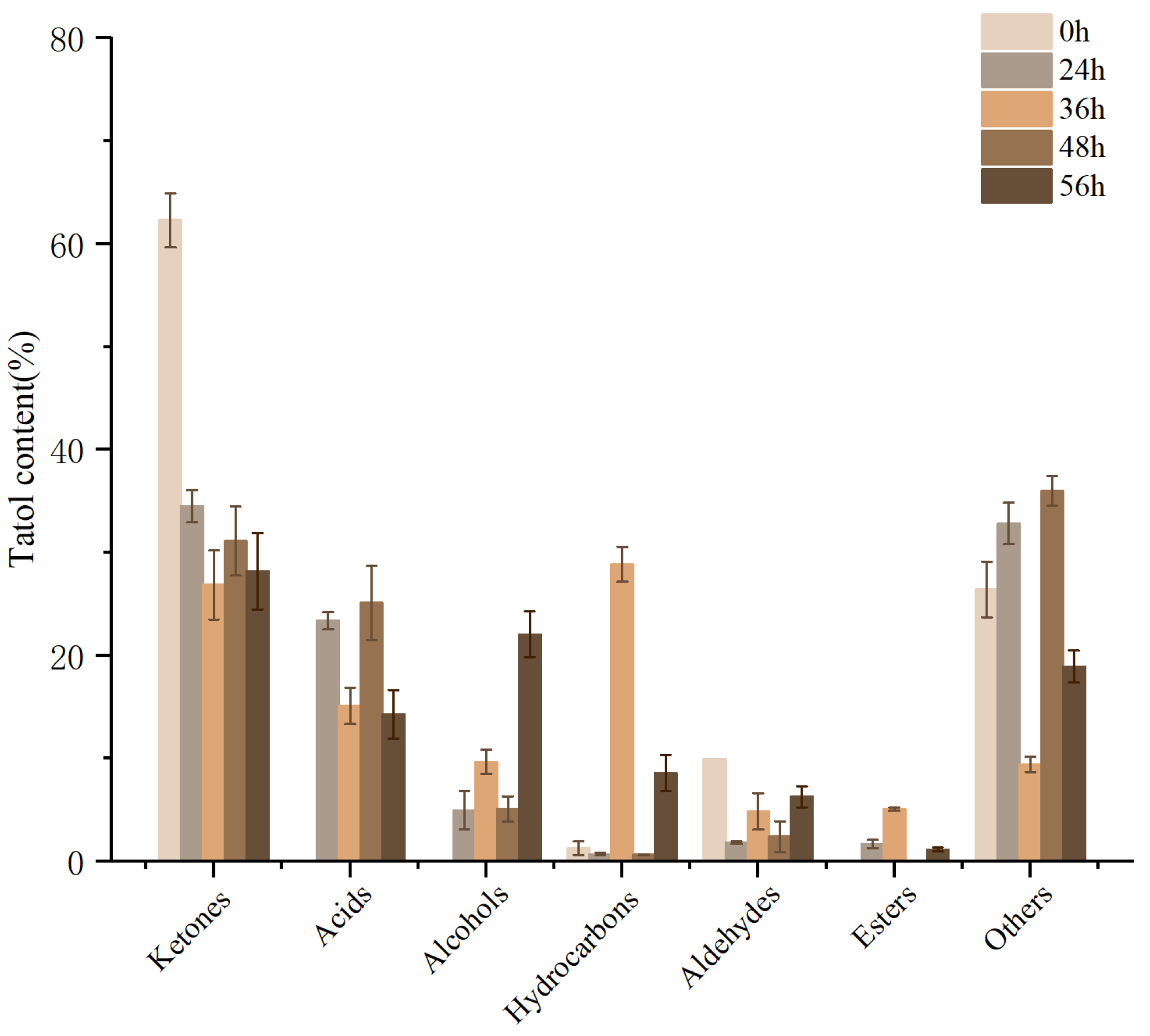
2.10.3. Determination and Analysis of Volatile Flavor Compounds
2.11. Statistical Analysis
3. Results and Discussion
3.1. Analysis of Basic Physiological Characteristics of LAB Strains
3.2. Molecular Identification Results and Analysis
3.3. Antibacterial Experiment Results and Analysis
3.4. Acid Resistance Test Results and Analysis
3.5. Bile Salt Tolerance Test Results and Analysis
3.6. Simulated Gastrointestinal Fluid Tolerance Results and Analysis
3.7. Adhesion Experiment
3.8. In Vitro Safety Experiment
3.8.1. Hemolytic Analysis
3.8.2. Drug Susceptibility Test Results and Analysis
3.9. In Vitro Fermentation Test
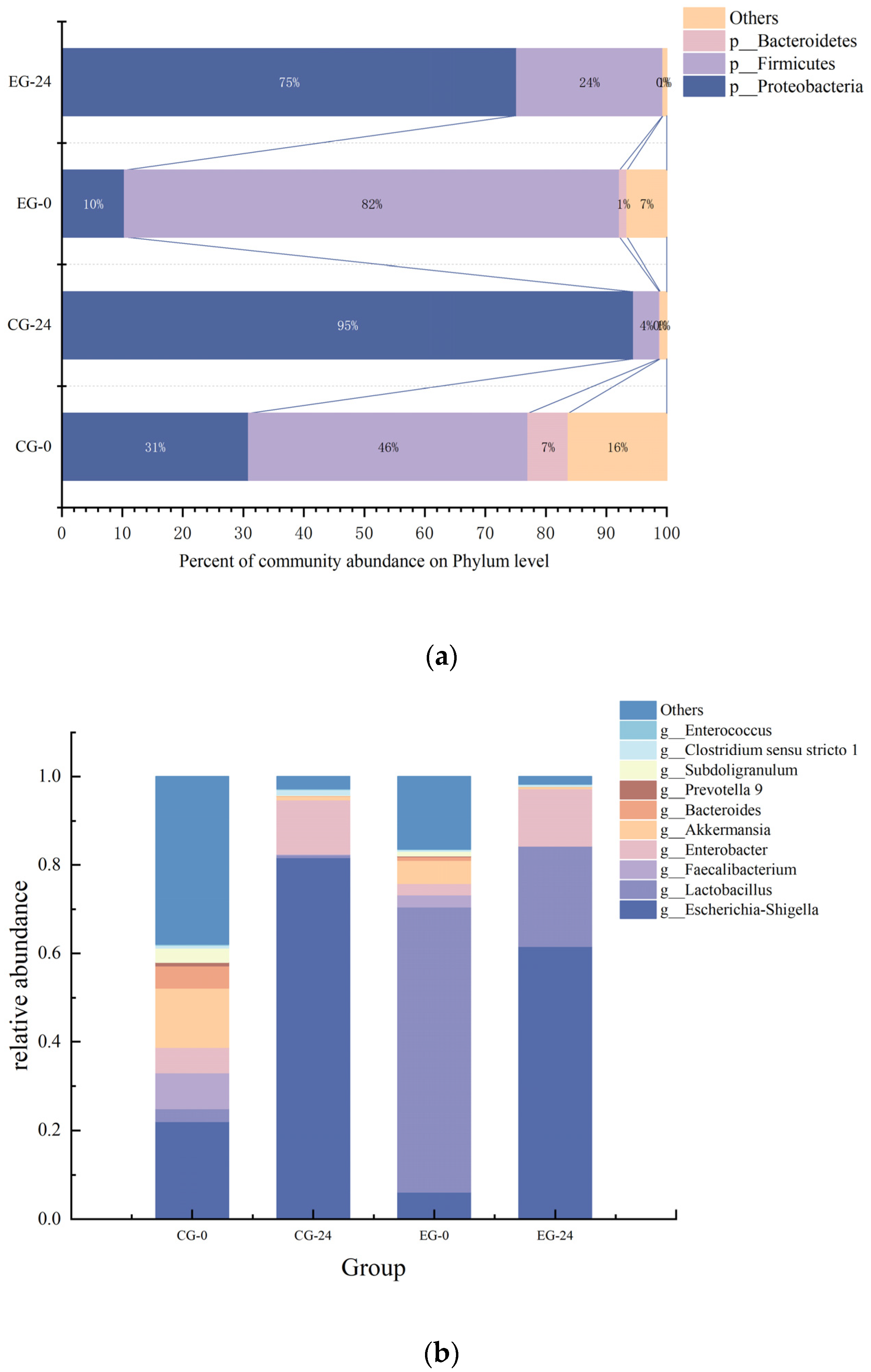
3.9.1. The Effect of M3 (1) on Intestinal Flora
3.9.2. Correlation Analysis between pH and SCFAs
3.10. Flavor Analysis
3.10.1. pH Changes
3.10.2. Analysis of Volatile Components
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Barakat, O.S.; Ibrahim, G.; Tawfik, N.; El-Kholy, W.; El-Rab, G.D. Identification and probiotic characteristics of Lactobacillus strains isolated from traditional Domiati cheese. Int. J. Microbiol. Res. 2011, 3, 59. [Google Scholar]
- Liu, H.; Hou, C.; Wang, G.; Jia, H.; Yu, H.; Zeng, X.; Thacker, P.A.; Zhang, G.; Qiao, S. Lactobacillus reuteri I5007 modulates intestinal host defense peptide expression in the model of IPEC-J2 cells and neonatal piglets. Nutrients 2017, 9, 559. [Google Scholar] [CrossRef] [PubMed]
- Edlund, C.; Nord, C.E. Effect on the human normal microflora of oral antibiotics for treatment of urinary tract infections. J. Antimicrob. Chemother. 2000, 46, 41–48. [Google Scholar] [CrossRef] [PubMed]
- Hill, C.; Guarner, F.; Reid, G.; Gibson, G.R.; Merenstein, D.J.; Pot, B.; Morelli, L.; Canani, R.B.; Flint, H.J.; Salminen, S. Expert consensus document: The International Scientific Association for Probiotics and Prebiotics consensus statement on the scope and appropriate use of the term probiotic. Nat. Rev. Gastroenterol. Hepatol. 2014, 11, 506–514. [Google Scholar] [CrossRef] [PubMed]
- Eichele, D.D.; Kharbanda, K.K. Dextran sodium sulfate colitis murine model: An indispensable tool for advancing our understanding of inflammatory bowel diseases pathogenesis. World J. Gastroenterol. 2017, 23, 6016. [Google Scholar] [CrossRef]
- Simon, E.; Călinoiu, L.F.; Mitrea, L.; Vodnar, D.C. Probiotics, prebiotics, and synbiotics: Implications and beneficial effects against irritable bowel syndrome. Nutrients 2021, 13, 2112. [Google Scholar] [CrossRef]
- Jiang, J.; Li, K.; Xiao, Y.; Zhong, A.; Tang, J.; Duan, Y.; Li, Z. Limosilactobacillus reuteri Regulating Intestinal Function: A Review. Fermentation 2023, 9, 19. [Google Scholar] [CrossRef]
- Wang, J.; Feng, J.; Liu, S.; Cai, Z.; Song, D.; Yang, L.; Nie, G. The probiotic properties of different preparations using Lactococcus lactis Z-2 on intestinal tract, blood and hepatopancreas in Cyprinus carpio. Aquaculture 2021, 543, 736911. [Google Scholar] [CrossRef]
- Kolaček, S.; Hojsak, I.; Berni Canani, R.; Guarino, A.; Indrio, F.; Orel, R.; Pot, B.; Shamir, R.; Szajewska, H.; Vandenplas, Y. ESPGHAN working Group for Probiotics and Prebiotics. Commercial probiotic products: A call for improved quality control. A position paper by the ESPGHAN working Group for Probiotics and Prebiotics. J. Pediatr. Gastroenterol. Nutr. 2017, 65, 117–124. [Google Scholar] [CrossRef]
- Dong, X.; Cai, M. Common bacterial system identification manual. Science 2001, 354–357. [Google Scholar]
- Monteiro, C.R.; do Carmo, M.S.; Melo, B.O.; Alves, M.S.; Dos Santos, C.I.; Monteiro, S.G.; Bomfim, M.R.Q.; Fernandes, E.S.; Monteiro-Neto, V. In vitro antimicrobial activity and probiotic potential of Bifidobacterium and Lactobacillus against species of Clostridium. Nutrients 2019, 11, 448. [Google Scholar] [CrossRef]
- Feng, T.; Wang, J. Oxidative stress tolerance and antioxidant capacity of lactic acid bacteria as probiotic: A systematic review. Gut Microbes 2020, 12, 1801944. [Google Scholar] [CrossRef]
- Liu, W.; Chen, M.; Duo, L.; Wang, J.; Guo, S.; Sun, H.; Menghe, B.; Zhang, H. Characterization of potentially probiotic lactic acid bacteria and bifidobacteria isolated from human colostrum. J. Dairy Sci. 2020, 103, 4013–4025. [Google Scholar] [CrossRef]
- Kuerman, M.; Bao, Y.; Guo, Y.; Guo, M. Effects of prebiotic carbohydrates on the growth promotion and cholesterol-lowering abilities of compound probiotics in vitro. LWT 2020, 118, 108703. [Google Scholar] [CrossRef]
- Sui, Y.; Liu, J.; Liu, Y.; Wang, Y.; Xiao, Y.; Gao, B.; Zhu, D. In vitro probiotic characterization of Lactobacillus strains from fermented tangerine vinegar and their cholesterol degradation activity. Food Biosci. 2021, 39, 100843. [Google Scholar] [CrossRef]
- Yasmin, I.; Saeed, M.; Khan, W.A.; Khaliq, A.; Chughtai, M.F.J.; Iqbal, R.; Tehseen, S.; Naz, S.; Liaqat, A.; Mehmood, T. In vitro probiotic potential and safety evaluation (hemolytic, cytotoxic activity) of Bifidobacterium strains isolated from raw camel milk. Microorganisms 2020, 8, 354. [Google Scholar] [CrossRef]
- Kim, H.; Kim, J.-S.; Kim, Y.; Jeong, Y.; Kim, J.-E.; Paek, N.-S.; Kang, C.-H. Antioxidant and probiotic properties of Lactobacilli and Bifidobacteria of human origins. Biotechnol. Bioprocess Eng. 2020, 25, 421–430. [Google Scholar] [CrossRef]
- Hindler, J.F.; Munro, S. Antimicrobial susceptibility testing. Clin. Microbiol. Proced. Handb. 2010. [Google Scholar] [CrossRef]
- Robinson, C.D.; Auchtung, J.M.; Collins, J.; Britton, R.A. Epidemic Clostridium difficile strains demonstrate increased competitive fitness compared to nonepidemic isolates. Infect. Immun. 2014, 82, 2815–2825. [Google Scholar] [CrossRef] [PubMed]
- Zhou, W.; Yan, Y.; Mi, J.; Zhang, H.; Lu, L.; Luo, Q.; Li, X.; Zeng, X.; Cao, Y. Simulated digestion and fermentation in vitro by human gut microbiota of polysaccharides from bee collected pollen of Chinese wolfberry. J. Agric. Food Chem. 2018, 66, 898–907. [Google Scholar] [CrossRef] [PubMed]
- Settachaimongkon, S.; Nout, M.R.; Fernandes, E.C.A.; Hettinga, K.A.; Vervoort, J.M.; van Hooijdonk, T.C.; Zwietering, M.H.; Smid, E.J.; van Valenberg, H.J. Influence of different proteolytic strains of Streptococcus thermophilus in co-culture with Lactobacillus delbrueckii subsp. bulgaricus on the metabolite profile of set-yoghurt. Int. J. Food Microbiol. 2014, 177, 29–36. [Google Scholar] [CrossRef]
- Liu, Q.; Yu, Z.; Tian, F.; Zhao, J.; Zhang, H.; Zhai, Q.; Chen, W. Surface components and metabolites of probiotics for regulation of intestinal epithelial barrier. Microb. Cell Factories 2020, 19, 23. [Google Scholar] [CrossRef] [PubMed]
- Vecchione, A.; Celandroni, F.; Mazzantini, D.; Senesi, S.; Lupetti, A.; Ghelardi, E. Compositional quality and potential gastrointestinal behavior of probiotic products commercialized in Italy. Front. Med. 2018, 5, 59. [Google Scholar] [CrossRef]
- Sharma, D.; Kapila, S.; Kapila, R. Bile salt tolerance and adhesion mechanism of probiotic bacteria. In Agricultural Science: Research and Reviews; Bhumi Publishing: Kolhapur, India, 2013; Volume II, p. 20. ISBN 978-93-91768-12-6. [Google Scholar]
- Nickzad, A.; Déziel, É. The involvement of rhamnolipids in microbial cell adhesion and biofilm development–an approach for control? Lett. Appl. Microbiol. 2014, 58, 447–453. [Google Scholar] [CrossRef]
- Ding, X.; Hu, X.; Chen, Y.; Xie, J.; Ying, M.; Wang, Y.; Yu, Q. Differentiated Caco-2 cell models in food-intestine interaction study: Current applications and future trends. Trends Food Sci. Technol. 2021, 107, 455–465. [Google Scholar] [CrossRef]
- Das, D.J.; Shankar, A.; Johnson, J.B.; Thomas, S. Critical insights into antibiotic resistance transferability in probiotic Lactobacillus. Nutrition 2020, 69, 110567. [Google Scholar] [CrossRef] [PubMed]
- Li, T.; Teng, D.; Mao, R.; Hao, Y.; Wang, X.; Wang, J. A critical review of antibiotic resistance in probiotic bacteria. Food Res. Int. 2020, 136, 109571. [Google Scholar] [CrossRef]
- Coman, M.; Verdenelli, M.; Cecchini, C.; Belà, B.; Gramenzi, A.; Orpianesi, C.; Cresci, A.; Silvi, S. Probiotic characterization of Lactobacillus isolates from canine faeces. J. Appl. Microbiol. 2019, 126, 1245–1256. [Google Scholar] [CrossRef] [PubMed]
- Martin, F.P.J.; Wang, Y.; Sprenger, N.; Yap, I.K.; Lundstedt, T.; Lek, P.; Rezzi, S.; Ramadan, Z.; Van Bladeren, P.; Fay, L.B. Probiotic modulation of symbiotic gut microbial–host metabolic interactions in a humanized microbiome mouse model. Mol. Syst. Biol. 2008, 4, 157. [Google Scholar] [CrossRef]
- Chen, L.; Xu, Y.; Chen, X.; Fang, C.; Zhao, L.; Chen, F. The maturing development of gut microbiota in commercial piglets during the weaning transition. Front. Microbiol. 2017, 8, 1688. [Google Scholar] [CrossRef] [PubMed]
- Coconnier, M.H.; Bernet, M.F.; Chauvière, G.; Servin, A.L. Adhering heat-killed human Lactobacillus acidophilus, strain LB, inhibits the process of pathogenicity of diarrhoeagenic bacteria in cultured human intestinal cells. J. Diarrhoeal Dis. Res. 1993, 11, 235–242. Available online: https://www.jstor.org/stable/23498285 (accessed on 1 April 2023).
- Wong, J.M.; De Souza, R.; Kendall, C.W.; Emam, A.; Jenkins, D.J. Colonic health: Fermentation and short chain fatty acids. J. Clin. Gastroenterol. 2006, 40, 235–243. [Google Scholar] [CrossRef] [PubMed]
- Louis, P.; Scott, K.P.; Duncan, S.H.; Flint, H.J. Understanding the effects of diet on bacterial metabolism in the large intestine. J. Appl. Microbiol. 2007, 102, 1197–1208. [Google Scholar] [CrossRef] [PubMed]
- Louis, P.; Flint, H.J. Formation of propionate and butyrate by the human colonic microbiota. Environ. Microbiol. 2017, 19, 29–41. [Google Scholar] [CrossRef]
- Venter, C.S.; Vorster, H.H.; Cummings, J.H. Effects of dietary propionate on carbohydrate and lipid metabolism in healthy volunteers. Am. J. Gastroenterol. (Springer Nat.) 1990, 85, 549–553. [Google Scholar] [PubMed]
- Hosseini, E.; Grootaert, C.; Verstraete, W.; Van de Wiele, T. Propionate as a health-promoting microbial metabolite in the human gut. Nutr. Rev. 2011, 69, 245–258. [Google Scholar] [CrossRef]
- Willing, B.P.; Dicksved, J.; Halfvarson, J.; Andersson, A.F.; Lucio, M.; Zheng, Z.; Järnerot, G.; Tysk, C.; Jansson, J.K.; Engstrand, L. A pyrosequencing study in twins shows that gastrointestinal microbial profiles vary with inflammatory bowel disease phenotypes. Gastroenterology 2010, 139, 1844–1854.e1841. [Google Scholar] [CrossRef] [PubMed]
- Donohoe, D.R.; Holley, D.; Collins, L.B.; Montgomery, S.A.; Whitmore, A.C.; Hillhouse, A.; Curry, K.P.; Renner, S.W.; Greenwalt, A.; Ryan, E.P. A Gnotobiotic Mouse Model Demonstrates That Dietary Fiber Protects against Colorectal Tumorigenesis in a Microbiota-and Butyrate-Dependent MannerFiber–Microbiota–Butyrate Axis in Tumor Suppression. Cancer Discov. 2014, 4, 1387–1397. [Google Scholar] [CrossRef] [PubMed]
- Onyszkiewicz, M.; Gawrys-Kopczynska, M.; Sałagaj, M.; Aleksandrowicz, M.; Sawicka, A.; Koźniewska, E.; Samborowska, E.; Ufnal, M. Valeric acid lowers arterial blood pressure in rats. Eur. J. Pharmacol. 2020, 877, 173086. [Google Scholar] [CrossRef] [PubMed]
- Shiratsuchi, H.; Shimoda, M.; Imayoshi, K.; Noda, K.; Osajima, Y. Volatile flavor compounds in spray-dried skim milk powder. J. Agric. Food Chem. 1994, 42, 984–988. [Google Scholar] [CrossRef]
- Chen, C.; Zhao, S.; Hao, G.; Yu, H.; Tian, H.; Zhao, G. Role of lactic acid bacteria on the yogurt flavour: A review. Int. J. Food Prop. 2017, 20, S316–S330. [Google Scholar] [CrossRef]
- Vazquez-Landaverde, P.A.; Velazquez, G.; Torres, J.; Qian, M. Quantitative determination of thermally derived off-flavor compounds in milk using solid-phase microextraction and gas chromatography. J. Dairy Sci. 2005, 88, 3764–3772. [Google Scholar] [CrossRef] [PubMed]
- Ott, A.; Germond, J.-E.; Baumgartner, M.; Chaintreau, A. Aroma comparisons of traditional and mild yogurts: Headspace gas chromatography quantification of volatiles and origin of α-diketones. J. Agric. Food Chem. 1999, 47, 2379–2385. [Google Scholar] [CrossRef]
- Coolbear, T.; Weimer, B.C.; Wilkinson, M. Lactic acid bacteria: Lactic acid bacteria in flavor development. In Encyclopedia of Dairy Sciences: Second Edition; Elsevier Inc.: Amsterdam, The Netherlands, 2011; pp. 160–165. [Google Scholar] [CrossRef]
- Delgado, F.J.; González-Crespo, J.; Cava, R.; García-Parra, J.; Ramírez, R. Characterisation by SPME–GC–MS of the volatile profile of a Spanish soft cheese PDO Torta del Casar during ripening. Food Chem. 2010, 118, 182–189. [Google Scholar] [CrossRef]
- Axelsson, L. Lactic acid bacteria: Classification and physiology. Food Sci. Technol. N. Y. Marcel Dekker 2004, 139, 1–66. [Google Scholar]
- Tsai, Y.K.; Lin, T.H. Sequence, organization, transcription and regulation of lactose and galactose operons in Lactobacillus rhamnosus TCELL-1. J. Appl. Microbiol. 2006, 100, 446–459. [Google Scholar] [CrossRef] [PubMed]
- Dalié, D.; Deschamps, A.; Richard-Forget, F. Lactic acid bacteria–Potential for control of mould growth and mycotoxins: A review. Food Control 2010, 21, 370–380. [Google Scholar] [CrossRef]
- Cheng, H. Volatile flavor compounds in yogurt: A review. Crit. Rev. Food Sci. Nutr. 2010, 50, 938–950. [Google Scholar] [CrossRef]
- Smit, G.; Smit, B.A.; Engels, W.J. Flavour formation by lactic acid bacteria and biochemical flavour profiling of cheese products. FEMS Microbiol. Rev. 2005, 29, 591–610. [Google Scholar] [CrossRef]
- Curioni, P.; Bosset, J. Key odorants in various cheese types as determined by gas chromatography-olfactometry. Int. Dairy J. 2002, 12, 959–984. [Google Scholar] [CrossRef]
- Acree, T.; Barnard, J.; Cunningham, D. A procedure for the sensory analysis of gas chromatographic effluents. Food Chem. 1984, 14, 273–286. [Google Scholar] [CrossRef]
- Aryana, K.J.; Olson, D.W. A 100-Year Review: Yogurt and other cultured dairy products. J. Dairy Sci. 2017, 100, 9987–10013. [Google Scholar] [CrossRef]


| Name of Material or Reagent | Source or Manufacturer |
|---|---|
| Staphylococcus aureus (S. aureus) ATCC 6538 | Center for Microbial Species Preservation, Chinese Academy of Sciences (Beijing, China) |
| Listeria monocytogenes (L. monocytogenes) ATCC 19115 | |
| Escherichia coli (E. coli) CGMCC 9181 | |
| Pseudomonas aeruginosa | Laboratory of Food Science and Technology College of Hunan Agricultural University (Changsha, China) |
| Bacillus subtilis | |
| Soybean peptone, tryptone | Thermo Fisher Oxoid (Shanghai, China) |
| MRS agar medium, MRS broth, technical agar powder | Guangdong Huankai Microbial Technology Co., Ltd. (Guangzhou, China) |
| Blood agar plate, Gram staining kit | |
| Pig bile salt | |
| Pepsin | Beijing Soleibao Technology Co., Ltd. (Beijing, China) |
| Sensitivity paper | Hangzhou Microbial Reagent Co., Ltd. (Hangzhou, China) |
| DNA Bacterial Extraction Kit | Hangzhou Beiwo Medical Technology Co., Ltd. (Hangzhou, China) |
| Trypsin | Sinopharm Chemical Reagent Co., Ltd. (Shanghai, China) |
| Fetal calf serum | Biological Industries Co., Ltd. (Guangzhou, China) |
| Dulbecco’s modified eagle medium high glucose medium | |
| Trypsin-EDTA | |
| Short-chain fatty acid standard | Shanghai Macklin Biochemical Co., Ltd. (Shanghai, China) |
| Item | Drug Category | Drug Content on Paper (μg/Piece) | Evaluation Criteria (mm) | ||
|---|---|---|---|---|---|
| S | I | R | |||
| Clindamycin | Lincomycin class | 2.0 | ≥21.00 | 15.00–20.00 | ≤14.00 |
| Streptomycin | Aminoglycosides | 10.0 | ≥15.00 | 12.00–14.00 | ≤11.00 |
| Gentamicin | Aminoglycosides | 10.0 | ≥15.00 | 13.00–14.00 | <12.00 |
| Kanamycin | Aminoglycosides | 30.0 | ≥18.00 | 14.00–17.00 | ≤13.00 |
| Vancomycin | Polypeptide Class | 30.0 | >17.00 | 14.00–17.00 | <14.00 |
| Tetracycline | Tetracycline class | 30.0 | ≥19.00 | 15.00–18.00 | ≤14.00 |
| Chloramphenicol | β-Lactams | 30.0 | ≥18.00 | 13.00–17.00 | ≤12.00 |
| Ampicillin | Penicillin class | 10.0 | ≥17.00 | 14.00–16.00 | ≤13.00 |
| Metronidazole | Nitroimidazole class | 5.0 | ≥16.00 | 11.00–15.00 | ≤10.00 |
| Bacitracin | Polypeptide Class | 0.4 | ≥10.00 | - | <10.00 |
| Cotrimoxazole | Sulfonamides | 25.0 | ≥16.00 | 10.00–16.00 | <10.00 |
| Neomycin | Coumarin Class | 30.0 | ≥17.00 | 13.00–16.00 | <12.00 |
| Ofloxacin | Quinolones | 5.0 | ≥16.00 | 13.00–15.00 | ≤12.00 |
| Strain | Source | Strain Name | Strain Number | Login Number | Similarity% |
|---|---|---|---|---|---|
| E5 | Fermented bamboo shoots | L. plantarum | L. plantarum strain NRRL B-14768 | NR_042394.1 | 99.66% |
| F9 | Fermented bamboo shoots | L. plantarum | L. plantarum strain 3335 | MT613628.1 | 99.86% |
| C2 | Fermented bamboo shoots | L. plantarum | L. plantarum strain BCH-2 | KX388383.1 | 99.86% |
| A1 | Fermented bamboo shoots | L. plantarum | L. plantarum strain JCM 1149 | NR_115605.1 | 99.79% |
| G3 | Fermented bamboo shoots | L. brevis | L. brevis strain 23 | MH681601.1 | 99.86% |
| C4 | Fermented bamboo shoots | L. brevis | L. Brevis strain ATCC 14869 | NR_044704.2 | 99.52% |
| D10 | Fermented vegetables | L. brevis | L. brevis strain ATCC 14869 | NR_044704.2 | 99.45% |
| F10 | Fermented vegetables | L. brevis | L. brevis strain NWAFU1541 | MG551215.1 | 99.86% |
| D6 | Fermented cowpeas | L. plantarum | L. plantarum strain 3784 | MT538636.1 | 99.86% |
| D11 | Fermented cowpeas | L. plantarum | L. plantarum strain thankcomeLP1 | MZ045749.1 | 99.86% |
| G5 | Fermented bamboo shoots | L. plantarum | L. plantarum strain TMPC 33321 | OM265411.1 | 100.00% |
| G7 | Fermented bamboo shoots | L. plantarum | L. plantarum strain LAB-12 | MW928452.1 | 99.93% |
| F11(2) | Fermented vegetables | L. brevis | L. brevis strain 1997 | MT597799.1 | 99.80% |
| B11 | Fermented vegetables | L. plantarum | L. plantarum strain Sourdough_B11 | MG754629.1 | 99.86% |
| C7 | Pickled Chili | L. plantarum | L. plantarum strain MLG4-3-1 | MT473372.1 | 100.00% |
| C1 | Fermented bamboo shoots | L. brevis | L. brevis strain 1997 | MT597799.1 | 99.86% |
| Fa16 | Infant feces | L. paracasei | L. paracasei strain HBUAS52231 | MH472956.1 | 92.55% |
| Fb5 | Feces of obese people | Enterococcus avium (E. avium) | E. avium strain MG4610 | ON631294.1 | 96.73% |
| Fa17 | Infant feces | Bifidobacterium breve (B. breve) | B. breve strain 1176 | MT573641.1 | 92.27% |
| Fa1 | Infant feces | L. paracasei | L. paracasei strain 5601 | MT510433.1 | 97.93% |
| F2 | Feces of obese people | L. paracasei | L. paracasei strain 6582 | MT463826.1 | 98.24% |
| F5 | Feces of obese people | B. crudilactis | B. crudilactis strain C4/12B | MG639889.1 | 96.30% |
| F3 | Feces of obese people | S. salivarius | S. salivarius strain 1740 | MT597596.1 | 96.28% |
| F9 | Feces of obese people | E. faecium | E. faecium strain CDCP37 | MT814619.1 | 95.54% |
| F6 | Feces of obese people | Weissella cibaria (W. cibaria) | W. cibaria strain M.D.W.YAN2-10 | JF690894.1 | 97.42% |
| F8 | Feces of obese people | E. faecium | E. faecium isolate NSY | EU047802.1 | 96.17% |
| M7 | Raw milk | L. rhamnosus | L. rhamnosus strain 2949 | MT611888.1 | 96.08% |
| M3(1) | Raw milk | L. rhamnosus | L. rhamnosus strain YIT 0105 (=ATCC 7469) | AB008211.1 | 97.06% |
| M2 | Raw milk | P.pentosaceus | P.pentosaceus s strain F8S2 | KF245570.1 | 97.62% |
| M19 | Raw milk | P.pentosaceus | P.pentosaceus strain 6359 | MT463768.1 | 97.04% |
| M8(1) | Raw milk | L. parabuchneri | L. parabuchneri strain B3BL14G | MW700852.1 | 97.64% |
| M14 | Raw milk | B. crudilactis | B. crudilactis strain C4/12B strain C4/12B | MG639889.1 | 97.17% |
| M9 | Raw milk | Bifidobacterium psychraerophilum (B. psychraerophilum) | B. psychraerophilum strain DA-50B | KJ128206.1 | 96.26% |
| M6 | Raw milk | L. paracasei | L. paracasei strain 6493 | MT515921.1 | 97.91% |
| M5 | Raw milk | L. paracasei | L. paracasei strain 4888 | MT505631.1 | 97.53% |
| M3(2) | Raw milk | L. paracasei | L. paracasei strain 6522 | MT515950.1 | 97.70% |
| M2(2) | Raw milk | B. crudilactis | B. crudilactis strain C4/12B | MG639889.1 | 96.70% |
| M8(2) | Raw milk | L. parabuchneri | L. parabuchneri strain 2012 | MT604605.1 | 98.49% |
| Strain | S. aureus (mm) | L. monocytogenes (mm) | E. coli (mm) | Bacillus subtilis (mm) | Pseudomonas aeruginosa (mm) |
|---|---|---|---|---|---|
| M16 | 19.51 ± 0.49 | 10.97 ± 0.49 | - | 17.44 ± 0.58 | 17.69 ± 1.60 |
| M6 | 19.45 ± 0.51 | - | 10.32 ± 0.00 | 10.44 ± 0.14 | 18.17 ± 0.47 |
| FU | 17.23 ± 1.55 | 10.16 ± 0.00 | 10.37 ± 0.21 | 12,70 ± 0.00 | 18.80 ± 0.06 |
| F2 | 17.57 ± 0.27 | 11.36 ± 0.00 | - | 9.70 ± 0.16 | 14.79 ± 0.89 |
| M11 (2) | 16.99 ± 0.33 | 11.02 ± 0.34 | 9.23 ± 0.41 | 12.55 ± 3.77 | 17.46 ± 0.24 |
| M7 | 17.02 ± 0.00 | 13.79 ± 0.13 | 8.02 ± 0.00 | - | 15.37 ± 0.27 |
| M5 | 16.13 ± 0.07 | 11.37 ± 0.05 | 14.52 ± 0.26 | 12.16 ± 1.89 | 18.59 ± 0.49 |
| M3 (2) | 16.02 ± 0.00 | 18.05 ± 0.23 | 8.93 ± 0.11 | - | 18.68 ± 0.48 |
| M19 | 15.56 ± 0.46 | 14.25 ± 0.33 | - | - | 21.07 ± 0.19 |
| M12 | 14.83 ± 0.37 | 15.42 ± 0.10 | - | 16.21 ± 1.31 | 16.54 ± 0.06 |
| M21 (2) | 15.10 ± 0.04 | - | - | 15.12 ± 1.14 | 20.45 ± 2.61 |
| M3 (1) | 15.04 ± 0.06 | 14.48 ± 0.18 | 9.60 ± 0.20 | 13.41 ± 1.39 | 20.09 ± 0.21 |
| M12 (1) | 14.87 ± 0.37 | 15.75 ± 0.27 | - | 11.77 ± 0.21 | 14.99 ± 0.49 |
| Z2 | 13.11 ± 0.61 | 11.63 ± 0.27 | - | 15.53 ± 0.55 | 14.33 ± 1.35 |
| D2 | 13.57 ± 0.23 | 11.75 ± 0.29 | - | - | 12.05 ± 0.71 |
| Z1 | 12.84 ± 0.18 | 9.79 ± 0.29 | - | 16.05 ± 0.89 | 18.80 ± 0.10 |
| Strain | pH = 2 | PH = 3 | ||
|---|---|---|---|---|
| 0 h (l g CFU/mL) | 3 h (l g CFU/mL) | 0 h (l g CFU/mL) | 3 h (l g CFU/mL) | |
| M3 (1) | 6.53 ± 0.090 a | 5.56 ± 0.100 c | 6.26 ± 0.055 a | 6.28 ± 0.078 b |
| M5 | 7.23 ± 0.416 cd | 7.11 ± 0.090 f | 7.28 ± 0.072 c | 7.27 ± 0.046 h |
| M3 (2) | 7.25 ± 0.092 cd | ND a | 7.12 ± 0.115 c | 6.80 ± 0.118 f |
| M7 | 6.75 ± 0.075 b | 4.20 ± 0.020 b | 7.15 ± 0.070 c | 6.37 ± 0.076 c |
| M16 | 7.18 ± 0.072 cd | ND a | 7.24 ± 0.049 c | 6.63 ± 0.058 e |
| M6 | 7.79 ± 0.011 e | ND a | 7.19 ± 0.061 c | 6.87 ± 0.012 f |
| M11 (2) | 6.43 ± 0.117 a | ND a | 6.64 ± 0.036 b | 6.23 ± 0.006 bc |
| M2 | 7.31 ± 0.021 d | 6.28 ± 0.062 d | 7.35 ± 0.105 c | 7.02 ± 0.015 g |
| M19 | 6.81 ± 0.067 b | ND a | 6.64 ± 0.288 b | 6.19 ± 0.055 b |
| M12 (1) | 6.41 ± 0.031 a | ND a | 6.35 ± 0.104 a | 6.23 ± 0.055 bc |
| M21 (2) | 7.12 ± 0.093 c | 6.68 ± 0.387 e | 7.23 ± 0.110 c | 6.21 ± 0.025 g |
| Z1 | 7.13 ± 0.105 c | ND a | 7.29 ± 0.077 c | 6.20 ± 0.025 b |
| Z2 | 7.26 ± 0.164 cd | ND a | 7.34 ± 0.115 c | 6.21 ± 0.058 bc |
| D2 | 7.13 ± 0.095 a | ND a | 6.37 ± 0.066 a | 6.21 ± 0.081 b |
| F2 | 6.84 ± 0.095 c | ND a | 6.77 ± 0.110 b | 5.63 ± 0.031 a |
| FU | 6.98 ± 0.053 b | ND a | 6.61 ± 0.061 b | 6.41 ± 0.031 d |
| Strain | 0.20% | 0.30% | ||
|---|---|---|---|---|
| 0 h (l g CFU/mL) | 3 h (l g CFU/mL) | 0 h (l g CFU/mL) | 3 h (l g CFU/mL) | |
| M3 (1) | 6.32 ± 0.023 a | 6.04 ± 0.012 a | 6.33 ± 0.036 a | 6.32 ± 0.080 a |
| M2 | 7.30 ± 0.067 d | 7.28 ± 0.056 d | 6.61 ± 0.034 b | 6.34 ± 0.024 b |
| M7 | 6.49 ± 0.015 b | 6.49 ± 0.004 b | 7.49 ± 0.010 c | 6.84 ± 0.034 b |
| M5 | 7.10 ± 0.007 c | 7.08 ± 0.003 c | 7.11 ± 0.031 d | 7.06 ± 0.027 c |
| M21 (2) | 7.17 ± 0.052 c | 7.11 ± 0.000 c | 7.06 ± 0.554 e | 7.48 ± 0.005 d |
| Strain | Simulated Gastric Juice (pH = 2) | Simulated Intestinal Fluid (pH = 6.8) | ||
|---|---|---|---|---|
| 0 h (l g CFU/mL) | 3 h (l g CFU/mL) | 0 h (l g CFU/mL) | 3 h (l g CFU/mL) | |
| M5 | 8.65 ± 0.006 a | 6.46 ± 0.012 c | 5.23 ± 0.035 c | 5.12 ± 0.017 c |
| M3 (1) | 8.79 ± 0.014 d | 6.33 ± 0.010 b | 5.11 ± 0.024 d | 5.04 ± 0.113 d |
| M21 (2) | 9.80 ± 0.018 c | 7.23 ± 0.018 d | 5.01 ± 0.017 b | 4.94 ± 0.017 c |
| M2 | 8.07 ± 0.032 a | 4.27 ± 0.002 a | 3.51 ± 0.001 a | 2.87 ± 0.130 b |
| M7 | 8.09 ± 0.042 a | ND e | 3.48 ± 0.011 a | 2.65 ± 0.171 a |
| Item | Size of Inhibition Zone of M3 (1) (mm) | Sensitive Level | Size of Inhibition Zone of M21 (2) (mm) | Sensitive Level | Size of Inhibition Zone of M5 (mm) | Sensitive Level |
|---|---|---|---|---|---|---|
| Clindamycin | 31.53 ± 0.34 | S | 29.06 ± 0.58 | S | 18.35 ± 0.97 | I |
| Streptomycin | 9.47 ± 0.52 | R | 7.00 ± 0.00 | R | 7.94 ± 0.58 | R |
| Gentamicin | 11.29 ± 0.43 | R | 9.967 ± 0.613 | R | 10.29 ± 0.99 | R |
| Kanamycin | 9.95 ± 0.62 | R | 10.69 ± 0.27 | R | 7.00 ± 0.00 | R |
| Vancomycin | 7.00 ± 0.00 | R | 7.00 ± 0.00 | R | 7.00 ± 0.00 | R |
| Tetracycline | 34.73 ± 0.87 | S | 21.90 ± 0.62 | S | 17.91 ± 0.33 | S |
| Chloramphenicol | 33.74 ± 0.23 | S | 31.43 ± 0.02 | S | 23.76 ± 1.56 | S |
| Ampicillin | 22.70 ± 0.50 | S | 16.92 ± 0.16 | I | 27.88 ± 0.86 | S |
| Metronidazole | 7.00 ± 0.00 | R | 7.00 ± 0.00 | R | 8.90 ± 0.78 | R |
| Bacitracin | 7.00 ± 0.00 | R | 7.00 ± 0.00 | R | 7.00 ± 0.00 | R |
| Cotrimoxazole | 7.00 ± 0.00 | R | 7.00 ± 0.00 | R | 13.31 ± 0.43 | I |
| Neomycin | 32.34 ± 0.16 | S | 16.89 ± 0.14 | I | 23.47 ± 2.37 | S |
| Ofloxacin | 17.50 ± 0.02 | S | 8.16 ± 0.27 | R | 7.00 ± 0.00 | R |
| NO | Name | Formula | CAS | Relative Amount (%) | ||||
|---|---|---|---|---|---|---|---|---|
| 0 h | 24 h | 36 h | 48 h | 56 h | ||||
| 1 | 2-Nonanone | C9H18O | 821-55-6 | 8.928 ± 0.833 ab | 10.495 ± 0.265 b | 8.897 ± 0.881 ab | 8.3663 ± 0.969 a | 10.528 ± 1.221 b |
| 2 | 2-Butanone, 3-methyl- | C5H10O | 563-80-4 | 1.674 ± 1.176 b | ND | ND | 0.472 ± 0.041 a | ND |
| 3 | Pyrolo[3,2-d]pyrimidin-2,4(1H,3H)-dione | C6H5N3O2 | 65996-50-1 | ND | ND | ND | 0.477 ± 0.049 b | ND |
| 4 | 2-Undecanone | C11H22O | 112-12-9 | ND | ND | 2.087 ± 0.363 c | ND | 1.433 ± 0.492 b |
| 5 | Isophorone | C9H14O | 78-59-1 | ND | ND | 3.003 ± 0.444 b | ND | 4.67 ± 0.459 c |
| 6 | 1-Pentanone, 1-(4-methylphenyl)- | C12H16O | 1671-77-8 | ND | ND | 0.433 ± 0.335 b | ND | ND |
| 7 | Acetoin | C4H8O2 | 513-86-0 | ND | ND | ND | 0.480 ± 0.052 b | 0.367 ± 0.169 b |
| 8 | 2-Heptanone | C7H14O | 110-43-0 | 51.697 ± 0.624 d | 24.010 ± 1.297 c | 12.437 ± 1.325 a | 21.320 ± 2.236 b | 11.193 ± 1.376 a |
| 9 | Acetic acid | C2H4O2 | 64-19-7 | ND | 23.368 ± 0.830 c | 9.790 ± 0.840 b | 25.107 ± 3.583 c | 14.278 ± 12.378 bc |
| 10 | Hexanoic acid | C6H12O2 | 142-62-1 | ND | ND | 2.868 ± 0.636 b | ND | ND |
| 11 | Octanoic acid | C8H16O2 | 124-07-2 | ND | ND | 2.457 ± 0.289 b | ND | ND |
| 12 | 2-Heptanol | C7H16O | 543-49-7 | ND | 4.980 ± 4.888 b | 9.680 ± 9.183 c | 5.100 ± 6.227 b | 20.243 ± 16.089 d |
| 13 | 2-Nonanol | C9H20O | 628-99-9 | ND | ND | ND | ND | 1.827 ± 0.129b |
| 14 | Longifolene | C15H24 | 475-20-7 | 1.310 ± 0.156 c | 0.723 ± 0.136 b | ND | 0.677 ± 0.038 b | 0.813 ± 0.095 b |
| 15 | Benzene, 1-methyl-3-(1-methylethyl)- | C10H14 | 535-77-3 | ND | ND | 0.897 ± 0.055 c | ND | 0.423 ± 0.146 b |
| 16 | Benzene, 1,2,4,5-tetramethyl- | C10H14 | 95-93-2 | ND | ND | 9.323 ± 0.280 b | ND | ND |
| 17 | Benzene, 1-ethyl-2,4-dimethyl- | C10H14 | 874-41-9 | ND | ND | 3.700 ± 0.252 b | ND | 4.137 ± 0.623 b |
| 18 | Benzene, pentamethyl- | C11H16 | 700-12-9 | ND | ND | 3.547 ± 0.407 c | ND | 1.130 ± 0.308 b |
| 19 | Benzene, 1,2,3,5-tetramethyl- | C10H14 | 527-53-7 | ND | ND | 8.500 ± 0.440 b | ND | ND |
| 20 | Benzene, 2-ethyl-1,4-dimethyl- | C10H14 | 1758-88-9 | ND | ND | 2.430 ± 0.234 b | ND | 0.577 ± 0.096 c |
| 21 | Benzene, 1-methyl-4-propyl- | C10H14 | 1074-55-1 | ND | ND | 0.457 ± 0.012 b | ND | ND |
| 22 | 2,4-Di-tert-butylphenol | C14H22O | 96-76-4 | ND | ND | ND | ND | 0.590 ± 0.020 b |
| 23 | Benzene, 1-ethyl-3,5-dimethyl- | C10H14 | 934-74-7 | ND | ND | ND | ND | 0.920 ± 0.485 b |
| 24 | 2-Propenal | C3H4O | 107-02-8 | ND | ND | ND | 0.600 ± 0.044 | ND |
| 25 | Benzaldehyde, 3,4-dimethyl- | C9H10O | 5973-71-7 | ND | ND | 2.957 ± 2.561 b | ND | 5.510 ± 0.907 c |
| 26 | Decanal | C10H20O | 112-31-2 | ND | ND | 0.783 ± 0.035 b | ND | ND |
| 27 | Nonanal | C9H18O | 124-19-6 | 9.957 ± 3.669 b | 1.857 ± 0.136 a | 1.133 ± 0.162 a | 1.817 ± 1.436 a | 0.793 ± 0.121 a |
| 28 | Hydrogen isocyanate | CHNO | 75-13-8 | ND | 0.531 ± 0.036 b | 5.0827 ± 0.173 c | ND | ND |
| 29 | Formic acid, octyl ester | C9H18O2 | 112-32-3 | ND | 1.170 ± 0.363 b | ND | ND | ND |
| 30 | Methyl formate | C2H4O2 | 107-31-3 | ND | ND | ND | 0.050 ± ND a | 1.173 ± 0.197 b |
| 31 | Oxime-, methoxy-phenyl-_ | C8H9NO2 | 1000222-86-6 | 26.417 ± 2.709 c | 31.740 ± 1.834 d | 8.430 ± 0.559 a | 34.863 ± 0.953 d | 18.093 ± 4.510 b |
| 32 | Dodecane, 4,6-dimethyl- | C14H30 | 61141-72-8 | ND | 0.203 ± 0.077 b | ND | 0.271 ± 0.061 b | ND |
| 33 | Nonane, 4,5-dimethyl- | C11H24 | 17302-23-7 | ND | 0.107 ± 0.035 b | ND | ND | 0.153 ± 0.021 c |
| 34 | Naphthalene, 1,2,3,5,6,7,8,8a-octahydro-1,8a-dimethyl-7-(1-methylethenyl)-, [1R-(1.alpha.,7.beta.,8a.alpha.)]- | C15H24 | 4630-7-3 | ND | 0.773 ± 0.068 b | ND | ND | ND |
| 35 | Benzene, 1-ethyl-4-(1-methylethyl)- | C11H16 | 4218-48-8 | ND | ND | 0.623 ± 0.055 b | ND | ND |
| 36 | Benzene, 1-ethyl-2,4,5-trimethyl- | C11H16 | 17851-27-3 | ND | ND | ND | ND | 0.587 ± 0.032 b |
| 37 | 2-Oxo-4-phenyl-6-(4-chlorophenyl)-1,2-dihydropyrimidine | C16H11ClN2O | 24030-13-5 | ND | ND | ND | 0.363 ± 0.015 b | ND |
| 38 | 3,5-Dimethylbenzylhexylamine | C15H25N | 1000491-60-9 | ND | ND | 0.090 ± 0.082 b | ND | ND |
| 39 | Cyclobutanecarbohydrazide | C5H10N2O | 1000489-21-7 | ND | ND | 0.140 ± 0.030 b | ND | ND |
| 40 | 1,2-Benzenediol, O-(4-ethylbenzoyl)-O’-propargyloxycarbonyl- | C19H16O5 | 1000329-75-1 | ND | ND | 0.153 ± 0.023 b | ND | ND |
| 41 | 2-Butanol, 3-methyl- | C5H12O | 598-75-4 | ND | ND | ND | 0.473 ± 0.410 b | ND |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jiang, J.; Li, K.; Wang, Y.; Wu, Z.; Ma, H.; Zheng, S.; Li, Z. Screening, Identification and Physiological Characteristics of Lactobacillus rhamnosus M3 (1) against Intestinal Inflammation. Foods 2023, 12, 1628. https://doi.org/10.3390/foods12081628
Jiang J, Li K, Wang Y, Wu Z, Ma H, Zheng S, Li Z. Screening, Identification and Physiological Characteristics of Lactobacillus rhamnosus M3 (1) against Intestinal Inflammation. Foods. 2023; 12(8):1628. https://doi.org/10.3390/foods12081628
Chicago/Turabian StyleJiang, Jiayan, Ke Li, Yuanliang Wang, Zhongqin Wu, Huiqin Ma, Shilin Zheng, and Zongjun Li. 2023. "Screening, Identification and Physiological Characteristics of Lactobacillus rhamnosus M3 (1) against Intestinal Inflammation" Foods 12, no. 8: 1628. https://doi.org/10.3390/foods12081628
APA StyleJiang, J., Li, K., Wang, Y., Wu, Z., Ma, H., Zheng, S., & Li, Z. (2023). Screening, Identification and Physiological Characteristics of Lactobacillus rhamnosus M3 (1) against Intestinal Inflammation. Foods, 12(8), 1628. https://doi.org/10.3390/foods12081628