Microbial and Biochemical Profile of Different Types of Greek Table Olives
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Description
2.2. Library Construction and Sequencing
2.3. Bioinformatics and Data Analysis
2.4. Biochemical Analysis
2.4.1. Reagents and Chemicals
2.4.2. Preparation of Samples and Extracts
2.4.3. pH Measurement
2.4.4. Total Polyphenol Content
2.4.5. Antioxidant Activity
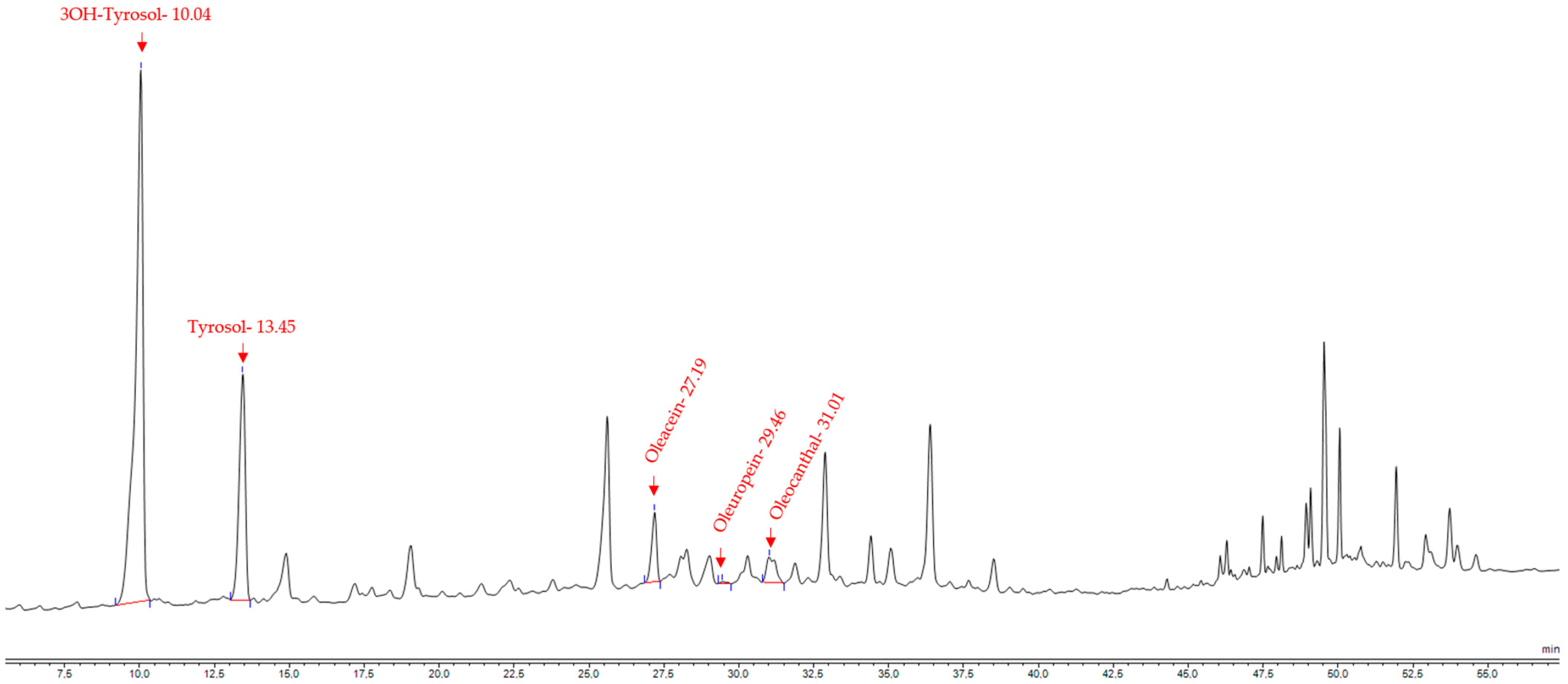
2.4.6. Quantification of Oleuropein, Oleacein, Oleocanthal and 3-Hydroxytyrosol with HPLC
2.4.7. Statistical Analysis
3. Results
3.1. Amplicon Metabarcoding Analysis
3.1.1. 16S rRNA Bacterial Assessment
3.1.2. 18S rRNA Eukaryotic Assessment
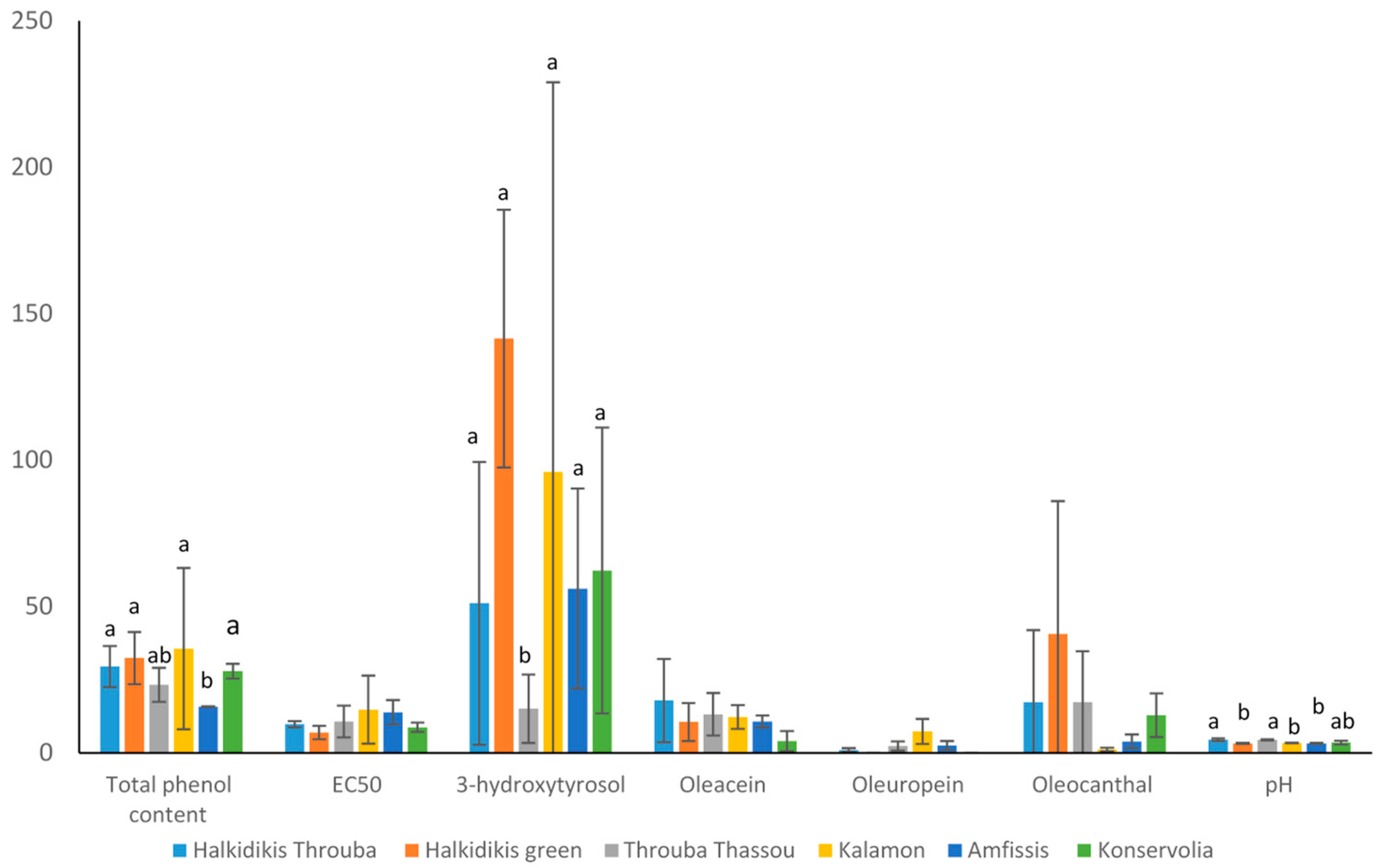
3.2. Biochemical Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
References
- Loumou, A.; Giourga, C. Olive Groves: “The Life and Identity of the Mediterranean”. Agric. Hum. Values 2003, 20, 87–95. [Google Scholar] [CrossRef]
- International Olive Council Table Olives Consumption. Available online: https://www.internationaloliveoil.org/wp-content/uploads/2020/12/OT-W901-23-11-2020-C.pdf (accessed on 15 April 2021).
- Tassou, C.C.; Panagou, E.Z.; Katsaboxakis, K.Z. Microbiological and Physicochemical Changes of Naturally Black Olives Fermented at Different Temperatures and NaCl Levels in the Brines. Food Microbiol. 2002, 19, 605–615. [Google Scholar] [CrossRef]
- Cillidag, S.I. Table Olive Processing Technologies. In Present and Future of the Mediterranean Olive Sector; Arcas, N., Arroyo López, F.N., Caballero, J., D’Andria, R., Fernández, M., Fernandez, E.R., Garrido, A., López-Miranda, J., Msallem, M., Parras, M., et al., Eds.; CIHEAM/IOC: Zaragoza, Spain, 2013; pp. 64–67. [Google Scholar]
- Perpetuini, G.; Prete, R.; Garcia-Gonzalez, N.; Khairul Alam, M.; Corsetti, A. Table Olives More than a Fermented Food. Foods 2020, 9, 178. [Google Scholar] [CrossRef]
- Bonatsou, S.; Tassou, C.; Panagou, E.; Nychas, G.J. Table Olive Fermentation Using Starter Cultures with Multifunctional Potential. Microorganisms 2017, 5, 30. [Google Scholar] [CrossRef]
- Corsetti, A.; Perpetuini, G.; Schirone, M.; Tofalo, R.; Suzzi, G. Application of Starter Cultures to Table Olive Fermentation: An Overview on the Experimental Studies. Front. Microbiol. 2012, 3, 248. [Google Scholar] [CrossRef] [PubMed]
- Heperkan, D. Microbiota of Table Olive Fermentations and Criteria of Selection for Their Use as Starters. Front. Microbiol. 2013, 4, 143. [Google Scholar] [CrossRef] [PubMed]
- Argyri, K.; Doulgeraki, A.I.; Manthou, E.; Grounta, A.; Argyri, A.A.; Nychas, G.J.; Tassou, C.C. Microbial Diversity of Fermented Greek Table Olives of Halkidiki and Konservolia Varieties from Different Regions as Revealed by Metagenomic Analysis. Microorganisms 2020, 8, 1241. [Google Scholar] [CrossRef]
- Tzamourani, A.P.; Di Napoli, E.; Paramithiotis, S.; Oikonomou-Petrovits, G.; Panagiotidis, S.; Panagou, E.Z. Microbiological and Physicochemical Characterization of Green Table Olives of Halkidiki and Conservolea Varieties Processed by the Spanish Method on Industrial Scale. Int. J. Food Sci. Technol. 2021, 56, 3845–3857. [Google Scholar] [CrossRef]
- Panagou, E.Z.; Katsaboxakis, C.Z. Effect of Different Brining Treatments on the Fermentation of Cv. Conservolea Green Olives Processed by the Spanish-Method. Food Microbiol. 2006, 23, 199–204. [Google Scholar] [CrossRef] [PubMed]
- Lanza, B. Abnormal Fermentations in Table-Olive Processing: Microbial Origin and Sensory Evaluation. Front. Microbiol. 2013, 4, 91. [Google Scholar] [CrossRef]
- Bleve, G.; Tufariello, M.; Durante, M.; Grieco, F.; Ramires, F.A.; Mita, G.; Tasioula-Margari, M.; Logrieco, A.F. Physico-Chemical Characterization of Natural Fermentation Process of Conservolea and Kalamàta Table Olives and Developement of a Protocol for the Pre-Selection of Fermentation Starters. Food Microbiol. 2015, 46, 368–382. [Google Scholar] [CrossRef]
- Tsapatsaris, S.; Kotzekidou, P. Application of Central Composite Design and Response Surface Methodology to the Fermentation of Olive Juice by Lactobacillus plantarum and Debaryomyces hansenii. Int. J. Food Microbiol. 2004, 95, 157–168. [Google Scholar] [CrossRef] [PubMed]
- Ettayebi, K.; Errachidi, F.; Jamai, L.; Tahri-Jouti, M.A.; Sendide, K.; Ettayebi, M. Biodegradation of Polyphenols with Immobilized Candida Tropicalis under Metabolic Induction. FEMS Microbiol. Lett. 2003, 223, 215–219. [Google Scholar] [CrossRef]
- Hernández, A.; Martín, A.; Aranda, E.; Pérez-Nevado, F.; Córdoba, M.G. Identification and Characterization of Yeast Isolated from the Elaboration of Seasoned Green Table Olives. Food Microbiol. 2007, 24, 346–351. [Google Scholar] [CrossRef] [PubMed]
- Mantzouridou, F.; Tsimidou, M.Z. Microbiological Quality and Biophenol Content of Hot Air-Dried Thassos Cv. Table Olives upon Storage. Eur. J. Lipid Sci. Technol. 2011, 113, 786–795. [Google Scholar] [CrossRef]
- Roubos, K.; Moustakas, M.; Aravanopoulos, F. Molecular Identification of Greek Olive (Olea Europaea) Cultivars Based on Microsatellite Loci. Genet. Mol. Res. 2010, 9, 1865–1876. [Google Scholar] [CrossRef] [PubMed]
- International Olive Oil Council Table Olives Production. Available online: https://www.internationaloliveoil.org/wp-content/uploads/2020/12/OT-CE-901-23-11-2020-P.pdf (accessed on 15 April 2021).
- DOEPEL The Table Olive Sector in Numbers. Available online: https://doepel.gr/?page_id=15483&lang=en_US (accessed on 8 September 2021).
- Zoidou, E.; Melliou, E.; Gikas, E.; Tsarbopoulos, A.; Magiatis, P.; Skaltsounis, A.L. Identification of Throuba Thassos, a Traditional Greek Table Olive Variety, as a Nutritional Rich Source of Oleuropein. J. Agric. Food Chem. 2010, 58, 46–50. [Google Scholar] [CrossRef] [PubMed]
- Vekiari, S.A.; Oreopoulou, V.; Kourkoutas, Y.; Kamoun, N.; Msallem, M.; Psimouli, V.; Arapoglou, D. Characterization and Seasonal Variation of the Quality of Virgin Olive Oil of the Throumbolia and Koroneiki Varieties from Southern Greece. Grasa Aceites 2010, 61, 221–231. [Google Scholar] [CrossRef]
- Kalogereas, S.A. Table Olives; Hermes Publications: Athens, Greece, 1932. [Google Scholar]
- Panagou, E.Z. Greek Dry-Salted Olives: Monitoring the Dry-Salting Process and Subsequent Physico-Chemical and Microbiological Profile during Storage under Different Packing Conditions at 4 and 20 °C. LWT—Food Sci. Technol. 2006, 39, 323–330. [Google Scholar] [CrossRef]
- Koudounas, K.; Banilas, G.; Michaelidis, C.; Demoliou, C.; Rigas, S.; Hatzopoulos, P. A Defence-Related Olea Europaea β-Glucosidase Hydrolyses and Activates Oleuropein into a Potent Protein Cross-Linking Agent. J. Exp. Bot. 2015, 66, 2093–2106. [Google Scholar] [CrossRef]
- Hugenholtz, P.; Goebel, B.M.; Pace, N.R. Impact of Culture-Independent Studies on the Emerging Phylogenetic View of Bacterial Diversity. J. Bacteriol. 1998, 180, 4765–4774. [Google Scholar] [CrossRef]
- Ercolini, D. High-Throughput Sequencing and Metagenomics: Moving Forward in the Culture-Independent Analysis of Food Microbial Ecology. Appl. Environ. Microbiol. 2013, 79, 3148–3155. [Google Scholar] [CrossRef] [PubMed]
- Panagou, E.Z.; Schillinger, U.; Franz, C.M.A.P.; Nychas, G.J. Microbiological and Biochemical Profile of Cv. Conservolea Naturally Black Olives during Controlled Fermentation with Selected Strains of Lactic Acid Bacteria. Food Microbiol. 2008, 25, 348–358. [Google Scholar] [CrossRef] [PubMed]
- Bonatsou, S.; Paramithiotis, S.; Panagou, E.Z. Evolution of Yeast Consortia during the Fermentation of Kalamata Natural Black Olives upon Two Initial Acidification Treatments. Front. Microbiol. 2018, 8, 2673. [Google Scholar] [CrossRef] [PubMed]
- Kazou, M.; Tzamourani, A.; Panagou, E.Z.; Tsakalidou, E. Unraveling the Microbiota of Natural Black Cv. Kalamata Fermented Olives through 16S and ITS Metataxonomic Analysis. Microorganisms 2020, 8, 672. [Google Scholar] [CrossRef] [PubMed]
- Doulgeraki, A.I.; Hondrodimou, O.; Iliopoulos, V.; Panagou, E.Z. Lactic Acid Bacteria and Yeast Heterogeneity during Aerobic and Modified Atmosphere Packaging Storage of Natural Black Conservolea Olives in Polyethylene Pouches. Food Control 2012, 26, 49–57. [Google Scholar] [CrossRef]
- Michailidou, S.; Trikka, F.; Pasentsis, K.; Petrovits, G.E.; Kyritsi, M.; Argiriou, A. Insights into the Evolution of Greek Style Table Olives Microbiome Stored under Modified Atmosphere: Biochemical Implications on the Product Quality. Food Control 2021, 130, 108286. [Google Scholar] [CrossRef]
- Visioli, F.; Galli, C. Olive Oil Phenols and Their Potential Effects on Human Health. J. Agric. Food Chem. 1998, 46, 4292–4296. [Google Scholar] [CrossRef]
- Kouka, P.; Priftis, A.; Stagos, D.; Angelis, A.; Stathopoulos, P.; Xinos, N.; Skaltsounis, A.; Mamoulakis, C.; Tsatsakis, A.; Spandidos, D.; et al. Assessment of the Antioxidant Activity of an Olive Oil Total Polyphenolic Fraction and Hydroxytyrosol from a Greek Olea Europaea Variety in Endothelial Cells and Myoblasts. Int. J. Mol. Med. 2017, 40, 703–712. [Google Scholar] [CrossRef]
- Pelucchi, C.; Bosetti, C.; Negri, E.; Lipworth, L.; La Vecchia, C. Olive Oil and Cancer Risk: An Update of Epidemiological Findings through 2010. Curr. Pharm. Des. 2011, 17, 805–812. [Google Scholar] [CrossRef]
- Soler-Rivas, C.; Espiń, J.C.; Wichers, H.J. Oleuropein and Related Compounds. J. Sci. Food Agric. 2000, 80, 1013–1023. [Google Scholar] [CrossRef]
- Alagna, F.; Mariotti, R.; Panara, F.; Caporali, S.; Urban, S.; Veneziani, G.; Esposto, S.; Taticchi, A.; Rosati, A.; Rao, R.; et al. Olive Phenolic Compounds: Metabolic and Transcriptional Profiling during Fruit Development. BMC Plant Biol. 2012, 12, 162. [Google Scholar] [CrossRef] [PubMed]
- Charoenprasert, S.; Mitchell, A. Factors Influencing Phenolic Compounds in Table Olives (Olea europaea). J. Agric. Food Chem. 2012, 60, 7081–7095. [Google Scholar] [CrossRef]
- Pang, K.L.; Chin, K.Y. The Biological Activities of Oleocanthal from a Molecular Perspective. Nutrients 2018, 10, 570. [Google Scholar] [CrossRef]
- Lombardo, G.E.; Lepore, S.M.; Morittu, V.M.; Arcidiacono, B.; Colica, C.; Procopio, A.; Maggisano, V.; Bulotta, S.; Costa, N.; Mignogna, C.; et al. Effects of Oleacein on High-Fat Diet-Dependent Steatosis, Weight Gain, and Insulin Resistance in Mice. Front. Endocrinol. 2018, 9, 116. [Google Scholar] [CrossRef]
- Michailidou, S.; Petrovits, G.E.; Kyritsi, M.; Argiriou, A. Amplicon Metabarcoding Data of Prokaryotes and Eukaryotes Present in ‘Kalamata’ Table Olives Packaged under Modified Atmosphere. Data Brief 2021, 38, 107314. [Google Scholar] [CrossRef] [PubMed]
- Klindworth, A.; Pruesse, E.; Schweer, T.; Peplies, J.; Quast, C.; Horn, M.; Glöckner, F.O. Evaluation of General 16S Ribosomal RNA Gene PCR Primers for Classical and Next-Generation Sequencing-Based Diversity Studies. Nucleic Acids Res. 2013, 41, e1. [Google Scholar] [CrossRef]
- Chemidlin Prévost-Bouré, N.; Christen, R.; Dequiedt, S.; Mougel, C.; Lelièvre, M.; Jolivet, C.; Shahbazkia, H.R.; Guillou, L.; Arrouays, D.; Ranjard, L. Validation and Application of a PCR Primer Set to Quantify Fungal Communities in the Soil Environment by Real-Time Quantitative PCR. PLoS ONE 2011, 6, e24166. [Google Scholar] [CrossRef]
- Bolyen, E.; Rideout, J.R.; Dillon, M.R.; Bokulich, N.A.; Abnet, C.C.; Al-Ghalith, G.A.; Alexander, H.; Alm, E.J.; Arumugam, M.; Asnicar, F.; et al. Reproducible, Interactive, Scalable and Extensible Microbiome Data Science Using QIIME 2. Nat. Biotechnol. 2019, 37, 852–857. [Google Scholar] [CrossRef] [PubMed]
- Martin, M. Cutadapt Removes Adapter Sequences from High-Throughput Sequencing Reads. EMBnet. J. 2011, 17, 10–12. [Google Scholar] [CrossRef]
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.A.; Holmes, S.P. DADA2: High-Resolution Sample Inference from Illumina Amplicon Data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glöckner, F.O. The SILVA Ribosomal RNA Gene Database Project: Improved Data Processing and Web-Based Tools. Nucleic Acids Res. 2013, 41, D590–D596. [Google Scholar] [CrossRef]
- RStudio Team. RStudio: Integrated Development for R. Rstudio; PBC: Boston, MA, USA, 2020. [Google Scholar]
- McMurdie, P.J.; Holmes, S. Phyloseq: A Bioconductor Package for Handling and Analysis of High-Throughput Phylogenetic Sequence Data. Pac. Symp. Biocomput. 2012, 2012, 235–246. [Google Scholar] [CrossRef]
- Albertsen, M.; Karst, S.M.; Ziegler, A.S.; Kirkegaard, R.H.; Nielsen, P.H. Back to Basics—The Influence of DNA Extraction and Primer Choice on Phylogenetic Analysis of Activated Sludge Communities. PLoS ONE 2015, 10, e0132783. [Google Scholar] [CrossRef]
- Wickham, H. Ggplot2, 3rd ed.; Gentleman, R., Hornik, K., Parmigiani, G., Eds.; Springer: New York, NY, USA, 2016. [Google Scholar]
- Boskou, G.; Salta, F.N.; Chrysostomou, S.; Mylona, A.; Chiou, A.; Andrikopoulos, N.K. Antioxidant Capacity and Phenolic Profile of Table Olives from the Greek Market. Food Chem. 2006, 94, 558–564. [Google Scholar] [CrossRef]
- Lanza, B.; Di Serio, M.G.; Iannucci, E.; Russi, F.; Marfisi, P. Nutritional, Textural and Sensorial Characterisation of Italian Table Olives (Olea europaea L. Cv. ‘Intosso d’Abruzzo’). Int. J. Food Sci. Technol. 2010, 45, 67–74. [Google Scholar] [CrossRef]
- Brand-Williams, W.; Cuvelier, M.E.; Berset, C. Use of a Free Radical Method to Evaluate Antioxidant Activity. LWT—Food Sci. Technol. 1995, 28, 25–30. [Google Scholar] [CrossRef]
- Capuzzo, C.; Firrao, G.; Mazzon, L.; Squartini, A.; Girolami, V. “Candidatus Erwinia Dacicola”, a Coevolved Symbiotic Bacterium of the Olive Fly Bactrocera Oleae (Gmelin). Int. J. Syst. Evol. Microbiol. 2005, 55, 1641–1647. [Google Scholar] [CrossRef]
- Bintsis, T. Foodborne Pathogens. AIMS Microbiol. 2017, 3, 529–563. [Google Scholar] [CrossRef]
- Arroyo-López, F.N.; Medina, E.; Ruiz-Bellido, M.Á.; Romero-Gil, V.; Montes-Borrego, M.; Landa, B.B. Enhancement of the Knowledge on Fungal Communities in Directly Brined Aloreña de Málaga Green Olive Fermentations by Metabarcoding Analysis. PLoS ONE 2016, 11, e0163135. [Google Scholar] [CrossRef] [PubMed]
- Doulgeraki, A.I.; Pramateftaki, P.; Argyri, A.A.; Nychas, G.J.; Tassou, C.C.; Panagou, E.Z. Molecular Characterization of Lactic Acid Bacteria Isolated from Industrially Fermented Greek Table Olives. LWT—Food Sci. Technol. 2013, 50, 353–356. [Google Scholar] [CrossRef]
- Tzamourani, A.; Oikonomou-Petrovits, G.; Panagiotidis, S.; Nychas, G.J.; Panagou, E. Effect of Packaging on Microbial Survival and Physicochemical Characteristics of Non-Thermally Preserved Green Spanish-Style Olives. Proceedings 2020, 70, 61. [Google Scholar] [CrossRef]
- Lucena-Padrós, H.; Caballero-Guerrero, B.; Maldonado-Barragán, A.; Ruiz-Barba, J.L. Microbial Diversity and Dynamics of Spanish-Style Green Table-Olive Fermentations in Large Manufacturing Companies through Culture-Dependent Techniques. Food Microbiol. 2014, 42, 154–165. [Google Scholar] [CrossRef] [PubMed]
- Kavroulakis, N.; Ntougias, S. Bacterial and β-Proteobacterial Diversity in Olea Europaea Var. Mastoidis- and O. Europaea Var. Koroneiki-Generated Olive Mill Wastewaters: Influence of Cultivation and Harvesting Practice on Bacterial Community Structure. World J. Microbiol. Biotechnol. 2011, 27, 57–66. [Google Scholar] [CrossRef]
- Campos, C.; Gomes, L.; Rei, F.T.; Nobre, T. Olive Fruit Fly Symbiont Population: Impact of Metamorphosis. Front. Microbiol. 2022, 13, 868458. [Google Scholar] [CrossRef] [PubMed]
- Prohic, A.; Jovovic Sadikovic, T.; Krupalija-Fazlic, M.; Kuskunovic-Vlahovljak, S. Malassezia Species in Healthy Skin and in Dermatological Conditions. Int. J. Dermatol. 2016, 55, 494–504. [Google Scholar] [CrossRef]
- Costa, D.; Fernandes, T.; Martins, F.; Pereira, J.A.; Tavares, R.M.; Santos, P.M.; Baptista, P.; Lino-Neto, T. Illuminating Olea europaea L. Endophyte Fungal Community. Microbiol. Res. 2021, 245, 126693. [Google Scholar] [CrossRef]
- Benítez-Cabello, A.; Ramiro-García, J.; Romero-Gil, V.; Medina, E.; Arroyo-López, F.N. Fungal Biodiversity in Commercial Table Olive Packages. Food Microbiol. 2022, 107, 104082. [Google Scholar] [CrossRef]
- Benítez-Cabello, A.; Romero-Gil, V.; Medina-Pradas, E.; Garrido-Fernández, A.; Arroyo-López, F.N. Exploring Bacteria Diversity in Commercialized Table Olive Biofilms by Metataxonomic and Compositional Data Analysis. Sci. Rep. 2020, 10, 11381. [Google Scholar] [CrossRef]
- Arroyo-López, F.N.; Romero-Gil, V.; Bautista-Gallego, J.; Rodríguez-Gómez, F.; Jiménez-Díaz, R.; García-García, P.; Querol, A.; Garrido-Fernández, A. Yeasts in Table Olive Processing: Desirable or Spoilage Microorganisms? Int. J. Food Microbiol. 2012, 160, 42–49. [Google Scholar] [CrossRef]
- Hajjaj, H.; Blanc, P.; Groussac, E.; Uribelarrea, J.L.; Goma, G.; Loubiere, P. Kinetic Analysis of Red Pigment and Citrinin Production by Monascus Ruber as a Function of Organic Acid Accumulation. Enzyme Microb. Technol. 2000, 27, 619–625. [Google Scholar] [CrossRef]
- Panagou, E.Z.; Katsaboxakis, C.Z.; Nychas, G.J. Heat Resistance of Monascus Ruber Ascospores Isolated from Thermally Processed Green Olives of the Conservolea Variety. Int. J. Food Microbiol. 2002, 76, 11–18. [Google Scholar] [CrossRef]
- Panagou, E.Z.; Skandamis, P.N.; Nychas, G.J. Modelling the Combined Effect of Temperature, PH and Aw on the Growth Rate of Monascus Ruber, a Heat-Resistant Fungus Isolated from Green Table Olives. J. Appl. Microbiol. 2003, 94, 146–156. [Google Scholar] [CrossRef]
- Ramírez, E.; García-García, P.; De Castro, A.; Romero, C.; Brenes, M. Debittering of Black Dry-Salted Olives. Eur. J. Lipid Sci. Technol. 2013, 115, 1319–1324. [Google Scholar] [CrossRef]
- Sozbilen, G.S.; Baysal, A.H. Microbial Profile and Bacterial Characterisation of Naturally Debittered Hurma Olives Compared to Non-debittered Erkence Variety during Ripening Period. Int. J. Food Sci. Technol. 2016, 51, 2099–2105. [Google Scholar] [CrossRef]
- Panagou, E.Z.; Tassou, C.C.; Katsaboxakis, K.Z. Microbiological, Physicochemical and Organoleptic Changes in Dry-salted Olives of Thassos Variety Stored under Different Modified Atmospheres at 4 and 20 °C. Int. J. Food Sci. Technol. 2002, 37, 635–641. [Google Scholar] [CrossRef]
- Patriarca, A. Alternaria in Food Products. Curr. Opin. Food Sci. 2016, 11, 1–9. [Google Scholar] [CrossRef]
- Visconti, A.; Logrieco, A.; Bottalico, A. Natural Occurrence of Alternaria Mycotoxins in Olives—Their Production and Possible Transfer into the Oil. Food Addit. Contam. 1986, 3, 323–330. [Google Scholar] [CrossRef]
- Woods, D.F.; Kozak, I.M.; O’Gara, F. Microbiome and Functional Analysis of a Traditional Food Process: Isolation of a Novel Species (Vibrio Hibernica) with Industrial Potential. Front. Microbiol. 2020, 11, 647. [Google Scholar] [CrossRef]
- Breidt, F.; Medina, E.; Wafa, D.; Pérez-Díaz, I.; Franco, W.; Huang, H.Y.; Johanningsmeier, S.D.; Kim, J.H. Characterization of Cucumber Fermentation Spoilage Bacteria by Enrichment Culture and 16S RDNA Cloning. J. Food Sci. 2013, 78, 470–476. [Google Scholar] [CrossRef]
- Raimondi, S.; Luciani, R.; Sirangelo, T.M.; Amaretti, A.; Leonardi, A.; Ulrici, A.; Foca, G.; D’Auria, G.; Moya, A.; Zuliani, V.; et al. Microbiota of Sliced Cooked Ham Packaged in Modified Atmosphere throughout the Shelf Life: Microbiota of Sliced Cooked Ham in MAP. Int. J. Food Microbiol. 2019, 289, 200–208. [Google Scholar] [CrossRef]
- Tao, Z.; Wu, X.; Liu, W.; Takahashi, H.; Xie, S.; Ohshima, C.; He, Q. Prevalence of Histamine-Forming Bacteria in Two Kinds of Salted Fish at Town Markets of Guangdong Province of South China. J. Food Prot. 2022, 85, 956–960. [Google Scholar] [CrossRef]
- Illikoud, N.; Rossero, A.; Chauvet, R.; Courcoux, P.; Pilet, M.F.; Charrier, T.; Jaffrès, E.; Zagorec, M. Genotypic and Phenotypic Characterization of the Food Spoilage Bacterium Brochothrix Thermosphacta. Food Microbiol. 2019, 81, 22–31. [Google Scholar] [CrossRef]
- Ferrocino, I.; Rantsiou, K.; Cocolin, L. Microbiome and -Omics Application in Food Industry. Int. J. Food Microbiol. 2022, 377, 109781. [Google Scholar] [CrossRef] [PubMed]
- Blekas, G.; Vassilakis, C.; Harizanis, C.; Tsimidou, M.; Boskou, D.G. Biophenols in Table Olives. J. Agric. Food Chem. 2002, 50, 3688–3692. [Google Scholar] [CrossRef] [PubMed]
- Marsilio, V.; Seghetti, L.; Iannucci, E.; Russi, F.; Lanza, B.; Felicioni, M. Use of a Lactic Acid Bacteria Starter Culture during Green Olive (Olea europaea L. Cv Ascolana Tenera) Processing. J. Sci. Food Agric. 2005, 85, 1084–1090. [Google Scholar] [CrossRef]
- Johnson, R.; Melliou, E.; Zweigenbaum, J.; Mitchell, A.E. Quantitation of Oleuropein and Related Phenolics in Cured Spanish-Style Green, California-Style Black Ripe, and Greek-Style Natural Fermentation Olives. J. Agric. Food Chem. 2018, 66, 2121–2128. [Google Scholar] [CrossRef]
- Mougiou, N.; Trikka, F.; Trantas, E.; Ververidis, F.; Makris, A.; Argiriou, A.; Vlachonasios, K.E. Expression of Hydroxytyrosol and Oleuropein Biosynthetic Genes Are Correlated with Metabolite Accumulation during Fruit Development in Olive, Olea Europaea, Cv. Koroneiki. Plant Physiol. Biochem. 2018, 128, 41–49. [Google Scholar] [CrossRef]
- Johnson, R.L.; Mitchell, A.E. Reducing Phenolics Related to Bitterness in Table Olives. J. Food Qual. 2018, 2018, 3193185. [Google Scholar] [CrossRef]
- García-Serrano, P.; Brenes-Álvarez, M.; Romero, C.; Medina, E.; García-García, P.; Brenes, M. Physicochemical and Microbiological Assessment of Commercial Dehydrated Black Olives. Food Control 2023, 145, 109417. [Google Scholar] [CrossRef]
- López-García, E.; Benítez-Cabello, A.; Rodríguez-Gómez, F.; Romero-Gil, V.; Garrido-Fernández, A.; Jiménez-Díaz, R.; Arroyo-López, F.N. Bacterial Metataxonomic Analysis of Industrial Spanish-Style Green Table Olive Fermentations. Food Control 2022, 137, 108969. [Google Scholar] [CrossRef]
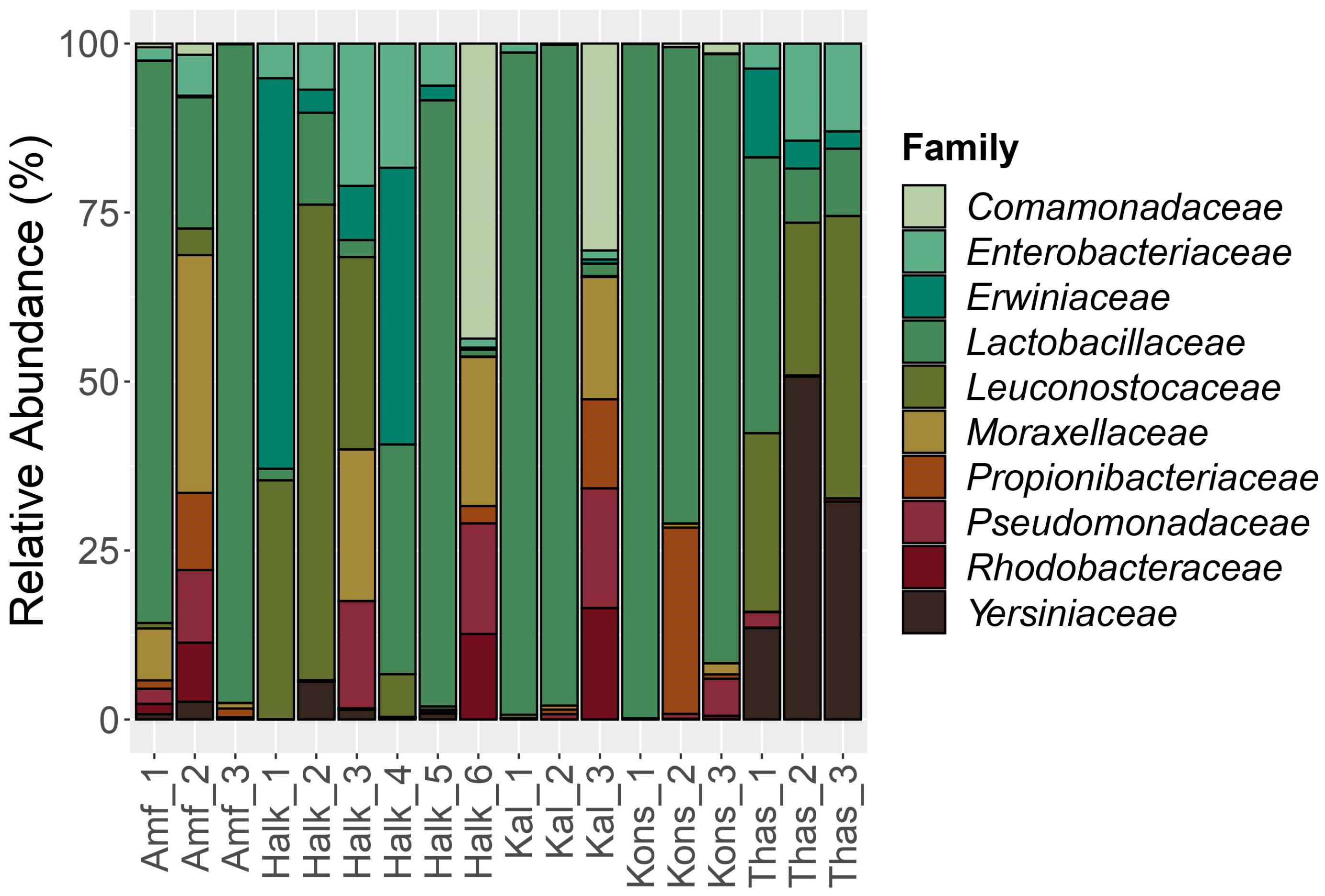
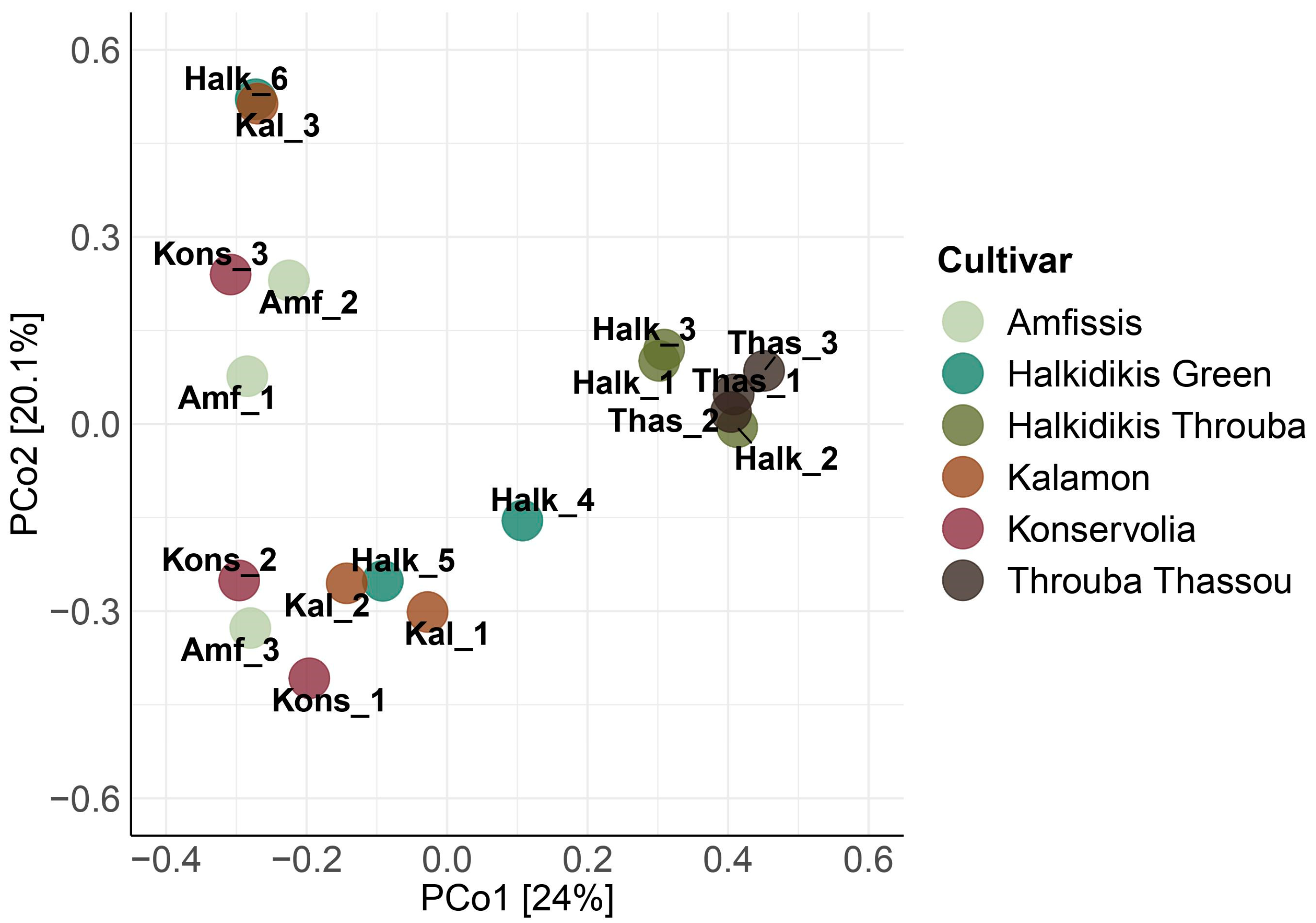

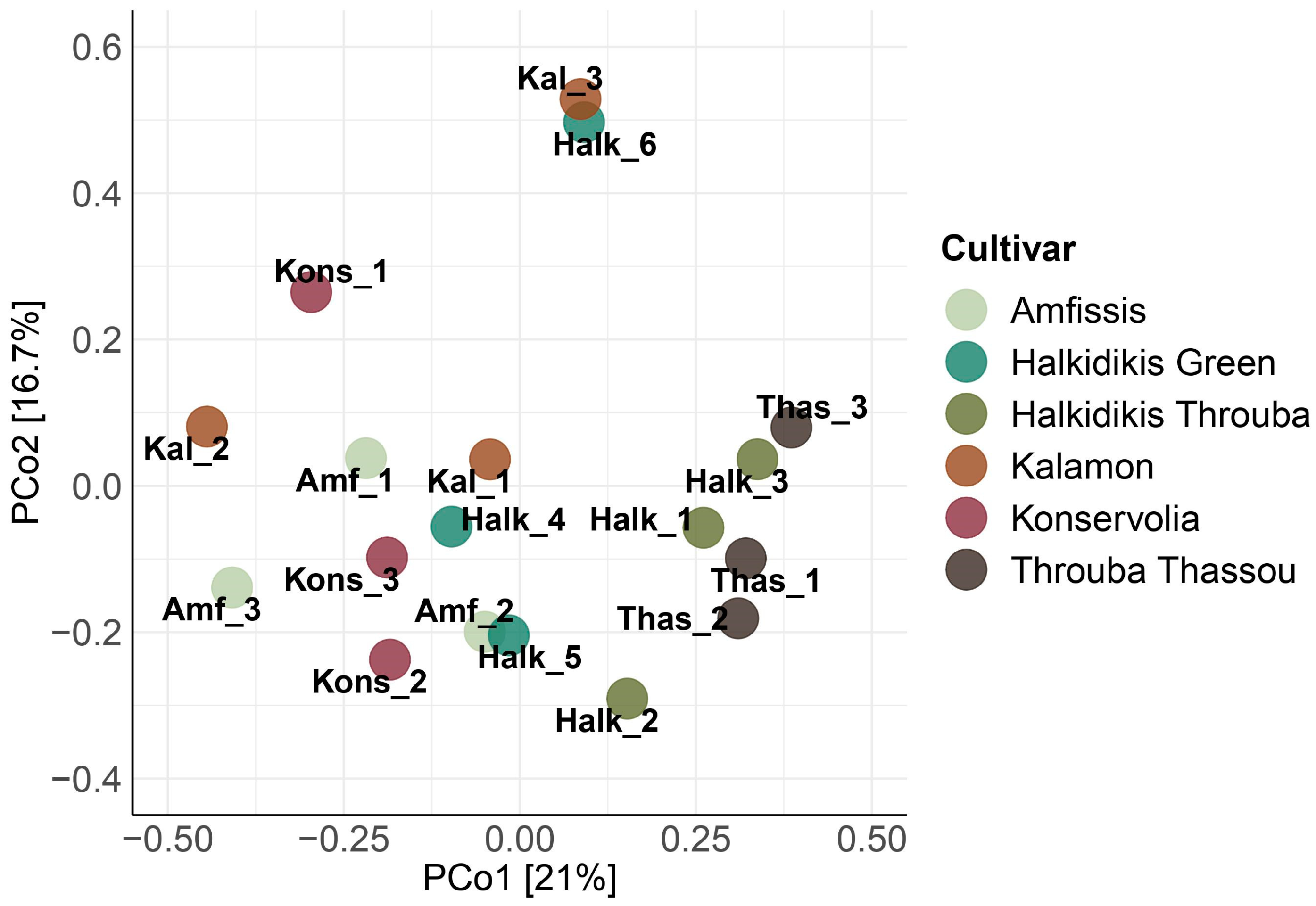
| Sample Name | Cultivar | Region | Olive Color | Fermentation Type |
|---|---|---|---|---|
| Halk_1 | Halkidikis (unpacked) | Halkidiki | Black | Salt-dried |
| Halk_2 | Halkidikis (unpacked) | Halkidiki | Black | Salt-dried |
| Halk_3 | Halkidikis (unpacked) | Halkidiki | Black | Salt-dried |
| Halk_4 | Halkidikis (unpacked) | Halkidiki | Green | Spanish-style |
| Halk_5 | Halkidikis (unpacked) | Halkidiki | Green | Spanish-style |
| Halk_6 | Halkidikis (packed in brine, jar) | Halkidiki | Green | Spanish-style |
| Thas_1 | Throuba Thassou (unpacked) | Thassos | Black | Salt-dried |
| Thas_2 | Throuba Thassou (packed in MA, pouch) | Thassos | Black | Salt-dried |
| Thas_3 | Throuba Thassou (unpacked) | Thassos | Black | Salt-dried |
| Kal_1 | Kalamon (packed in MA, pouch) | Messinia | Black | Greek-style |
| Kal_2 | Kalamon (packed in MA, pouch) | Messinia | Black | Greek-style |
| Kal_3 | Kalamon (packed in brine, jar) | Messinia | Black | Greek-style |
| Amf_1 | Amfissis (packed in MA, pouch) | Central Greece | Black | Greek-style |
| Amf_2 | Amfissis (packed in MA, pouch) | Central Greece | Black | Greek-style |
| Amf_3 | Amfissis (packed in MA, pouch) | Central Greece | Black | Greek-style |
| Kons_1 | Konservolia (packed in MA, pouch) | Central Greece | Green | Spanish-style |
| Kons_2 | Konservolia (packed in MA, pouch) | Central Greece | Green | Spanish-style |
| Kons_3 | Konservolia (packed in brine under MA, plastic container) | Central Greece | Green | Spanish-style |
| Sample Name | Observed ASVs | Shannon | Simpson | Inverse Simpson |
|---|---|---|---|---|
| Halk_1 | 34 | 2.32 | 0.84 | 6.11 |
| Halk_2 | 44 | 1.80 | 0.62 | 2.61 |
| Halk_3 | 95 | 3.53 | 0.94 | 17.27 |
| Halk_4 | 49 | 2.57 | 0.82 | 5.47 |
| Halk_5 | 104 | 3.39 | 0.94 | 16.01 |
| Halk_6 | 361 | 4.01 | 0.95 | 21.67 |
| Thas_1 | 57 | 2.17 | 0.78 | 4.50 |
| Thas_2 | 44 | 2.45 | 0.85 | 6.78 |
| Thas_3 | 39 | 2.48 | 0.84 | 6.44 |
| Kal_1 | 26 | 1.67 | 0.68 | 3.10 |
| Kal_2 | 36 | 2.28 | 0.83 | 5.77 |
| Kal_3 | 469 | 4.48 | 0.97 | 34.50 |
| Amf_1 | 261 | 3.56 | 0.88 | 8.53 |
| Amf_2 | 456 | 5.10 | 0.98 | 56.15 |
| Amf_3 | 82 | 3.20 | 0.93 | 15.11 |
| Kons_1 | 47 | 2.27 | 0.78 | 4.51 |
| Kons_2 | 78 | 2.95 | 0.91 | 10.58 |
| Kons_3 | 131 | 2.18 | 0.72 | 3.52 |
| Sample Name | Observed ASVs | Shannon | Simpson | Inverse Simpson |
|---|---|---|---|---|
| Halk_1 | 29 | 1.22 | 0.59 | 2.44 |
| Halk_2 | 24 | 0.45 | 0.17 | 1.21 |
| Halk_3 | 52 | 2.38 | 0.82 | 5.69 |
| Halk_4 | 20 | 1.63 | 0.68 | 3.17 |
| Halk_5 | 29 | 2.15 | 0.83 | 5.98 |
| Halk_6 | 54 | 2.58 | 0.82 | 5.41 |
| Thas_1 | 35 | 2.10 | 0.77 | 4.44 |
| Thas_2 | 22 | 1.42 | 0.58 | 2.37 |
| Thas_3 | 41 | 1.73 | 0.66 | 2.92 |
| Kal_1 | 25 | 1.54 | 0.67 | 3.05 |
| Kal_2 | 16 | 0.92 | 0.38 | 1.62 |
| Kal_3 | 99 | 2.86 | 0.88 | 8.08 |
| Amf_1 | 12 | 0.10 | 0.03 | 1.03 |
| Amf_2 | 26 | 1.08 | 0.43 | 1.77 |
| Amf_3 | 20 | 1.35 | 0.57 | 2.35 |
| Kons_1 | 10 | 1.48 | 0.68 | 3.08 |
| Kons_2 | 27 | 0.95 | 0.46 | 1.84 |
| Kons_3 | 22 | 0.34 | 0.11 | 1.12 |
| Sample Name | Total Phenol Content | EC50 | 3-Hydroxytyrosol | Oleacein | Oleuropein | Oleocanthal |
|---|---|---|---|---|---|---|
| Halk_1 | 256.57 ± 18.21 | 9.92 ± 1.48 | 106.66 ± 0.85 | 11.63 ± 0.03 | N.D. | 1.66 ± 0.14 |
| Halk_2 | 375.53 ± 7.73 | 8.74 ± 0.72 | 27.12 ± 0.66 | 34.19 ± 0.69 | 1.15 ± 0.04 | 45.74 ± 0.64 |
| Halk_3 | 253.42 ± 10.13 | 10.83 ± 1.27 | 19.47 ± 0.43 | 7.86 ± 0.56 | 0.99 ± 0.13 | 4.71 ± 0.23 |
| Halk_4 | 297.60 ± 0.94 | 5.07 ± 0.40 | 143.68 ± 3.75 | 6.94 ± 0.09 | N.D. | 79.43 ± 1.12 |
| Halk_5 | 250.75 ± 4.88 | 9.50 ± 0.31 | 96.46 ± 1.38 | 6.75 ± 0.17 | N.D. | 1.90 ± 0.14 |
| Halk_6 | 422.79 ± 6.33 | 6.39 ± 0.16 | 184.31 ± 6.09 | 18.05 ± 0.93 | N.D. | N.D. |
| Thas_1 | 278.43 ± 13.84 | 8.69 ± 1.10 | 10.72 ± 0.06 | 8.70 ± 0.23 | N.D. | 3.41 ± 0.53 |
| Thas_2 | 166.99 ± 0.27 | 6.63 ± 0.45 | 6.31 ± 0.72 | 21.53 ± 0.04 | 1.59 ± 0.16 | 11.50 ± 0.35 |
| Thas_3 | 250.27 ± 0.96 | 16.84 ± 0.58 | 28.32 ± 1.38 | 9.37 ± 0.94 | 3.20 ± 0.38 | 36.93 ± 0.30 |
| Kal_1 | 125.79 ± 0.04 | 26.46 ± 0.41 | 11.45 ± 0.18 | 9.65 ± 0.72 | N.D. | 1.14 ± 0.05 |
| Kal_2 | 282.09 ± 8.14 | 14.68 ± 0.89 | 27.27 ± 0.80 | 10.31 ± 0.57 | 7.36 ± 0.23 | N.D. |
| Kal_3 | 660.61 ± 1.51 | 3.29 ± 0.47 | 249.33 ± 20.58 | 16.97 ± 1.59 | N.D. | N.D. |
| Amf_1 | 167.47 ± 0.28 | 9.10 ± 0.37 | 91.93 ± 1.90 | 8.81 ± 0.73 | N.D. | 3.98 ± 0.17 |
| Amf_2 | 149.42 ± 5.46 | 16.90 ± 0.33 | 52.38 ± 2.56 | 10.69 ± 0.35 | 2.59 ± 0.43 | N.D. |
| Amf_3 | 157.03 ± 3.34 | 15.68 ± 1.11 | 24.04 ± 0.15 | 12.88 ± 0.82 | N.D. | N.D. |
| Kons_1 | 267.26 ± 7.88 | 10.48 ± 0.33 | 14.17 ± 0.14 | 6.49 ± 0.15 | N.D. | 12.86 ± 0.33 |
| Kons_2 | 261.25 ± 2.09 | 8.38 ± 0.44 | 111.70 ± 0.38 | 1.65 ± 0.04 | N.D. | N.D. |
| Kons_3 | 308.19 ± 8.11 | 7.48 ± 0.15 | 60.95 ± 3.63 | N.D. | N.D. | N.D. |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mougiou, N.; Tsoureki, A.; Didos, S.; Bouzouka, I.; Michailidou, S.; Argiriou, A. Microbial and Biochemical Profile of Different Types of Greek Table Olives. Foods 2023, 12, 1527. https://doi.org/10.3390/foods12071527
Mougiou N, Tsoureki A, Didos S, Bouzouka I, Michailidou S, Argiriou A. Microbial and Biochemical Profile of Different Types of Greek Table Olives. Foods. 2023; 12(7):1527. https://doi.org/10.3390/foods12071527
Chicago/Turabian StyleMougiou, Niki, Antiopi Tsoureki, Spyros Didos, Ioanna Bouzouka, Sofia Michailidou, and Anagnostis Argiriou. 2023. "Microbial and Biochemical Profile of Different Types of Greek Table Olives" Foods 12, no. 7: 1527. https://doi.org/10.3390/foods12071527
APA StyleMougiou, N., Tsoureki, A., Didos, S., Bouzouka, I., Michailidou, S., & Argiriou, A. (2023). Microbial and Biochemical Profile of Different Types of Greek Table Olives. Foods, 12(7), 1527. https://doi.org/10.3390/foods12071527










