The Application of High-Resolution Melting Analysis to trnL (UAA) Intron Allowed a Qualitative Identification of Apple Juice Adulterations
Abstract
1. Introduction
2. Materials and Methods
2.1. Fresh Fruits and Juices Preparation
2.2. Phylogenetic Analysis
2.3. DNA Extraction
2.4. DNA Sequencing
2.5. High Resolution Melting Analysis
3. Results and Discussion
3.1. Phylogenetic Analysis
3.2. DNA Sequencing
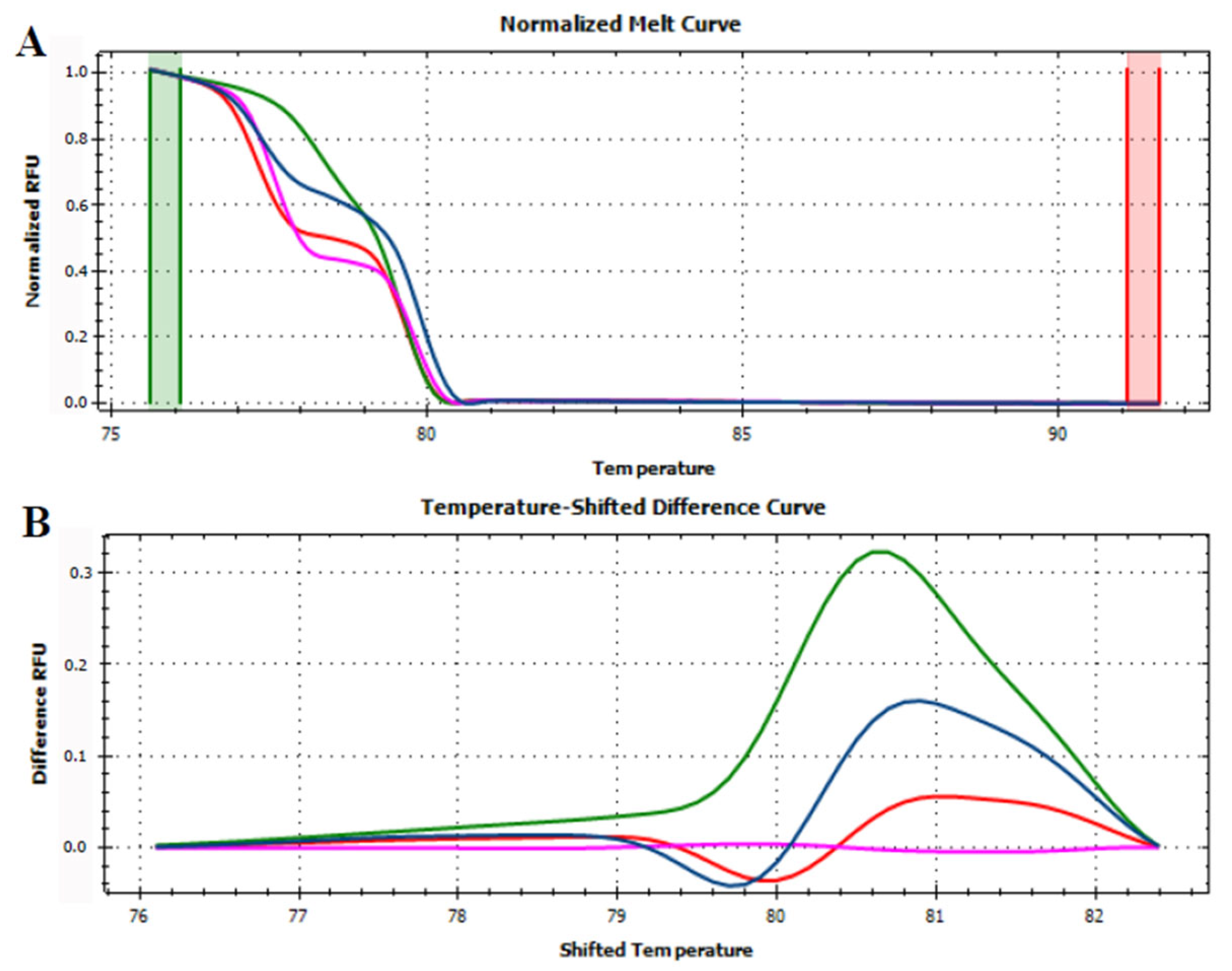
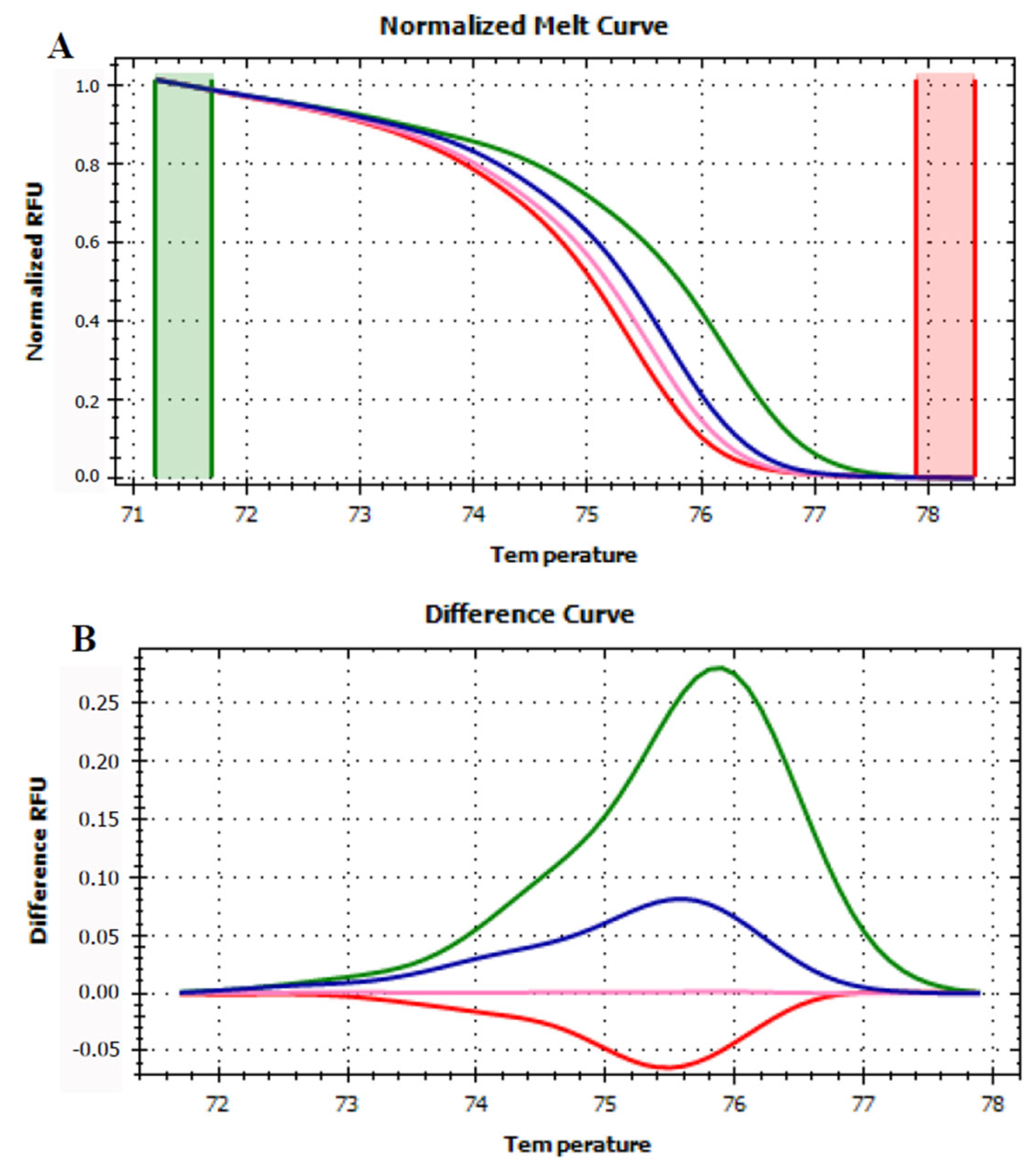
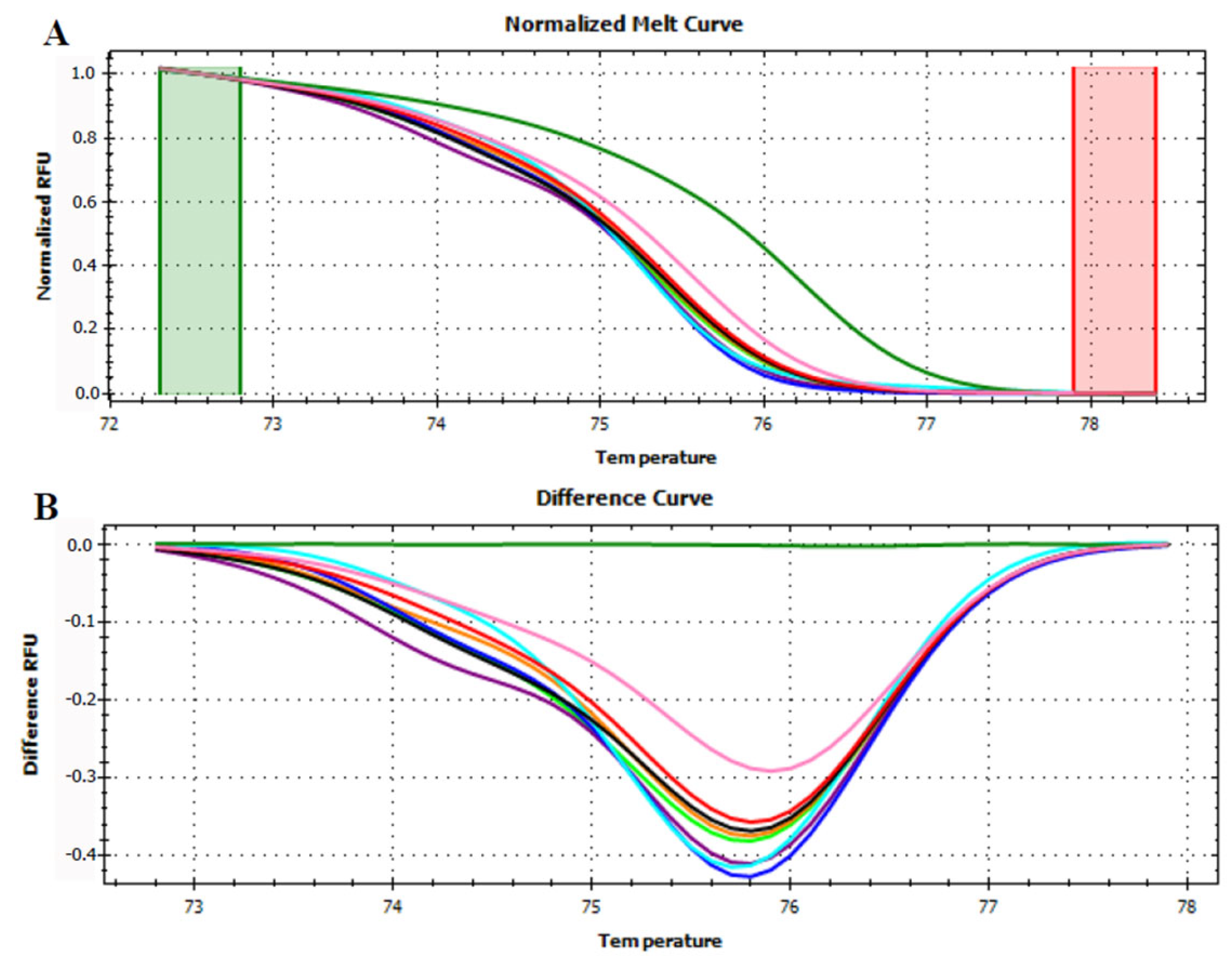
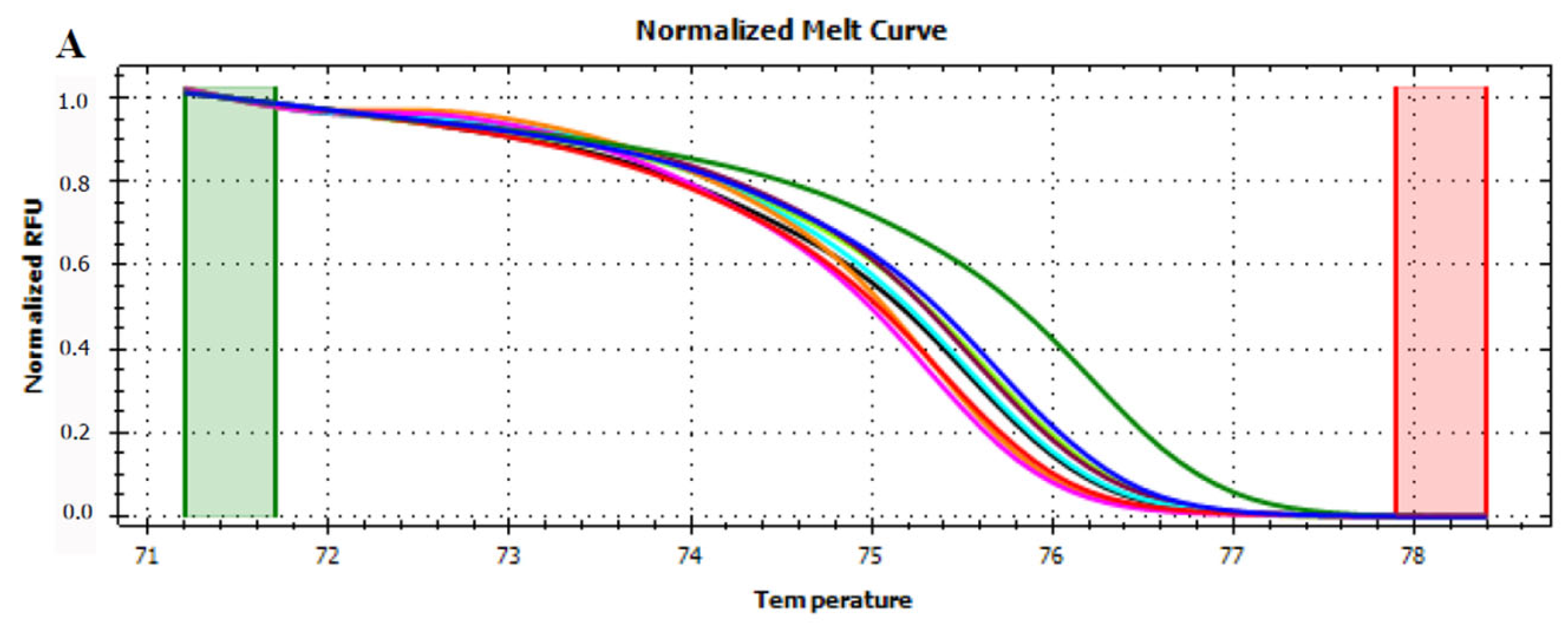
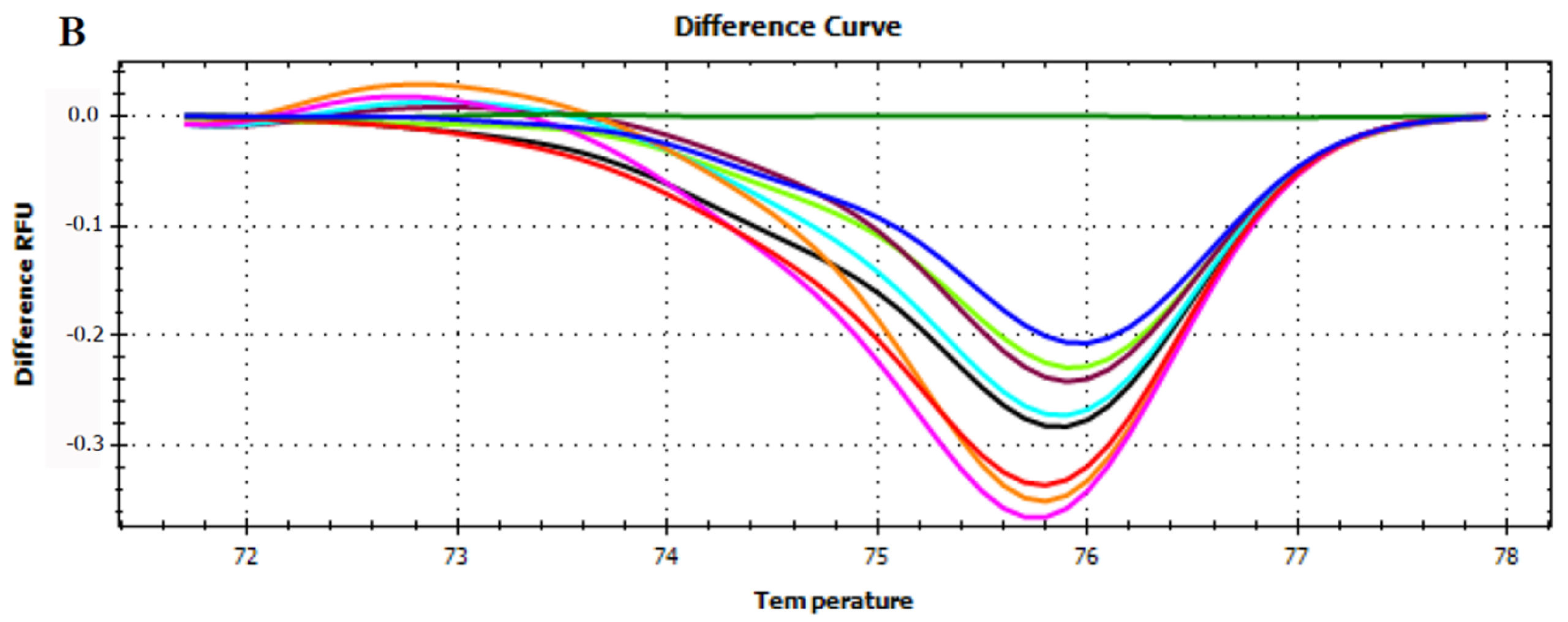
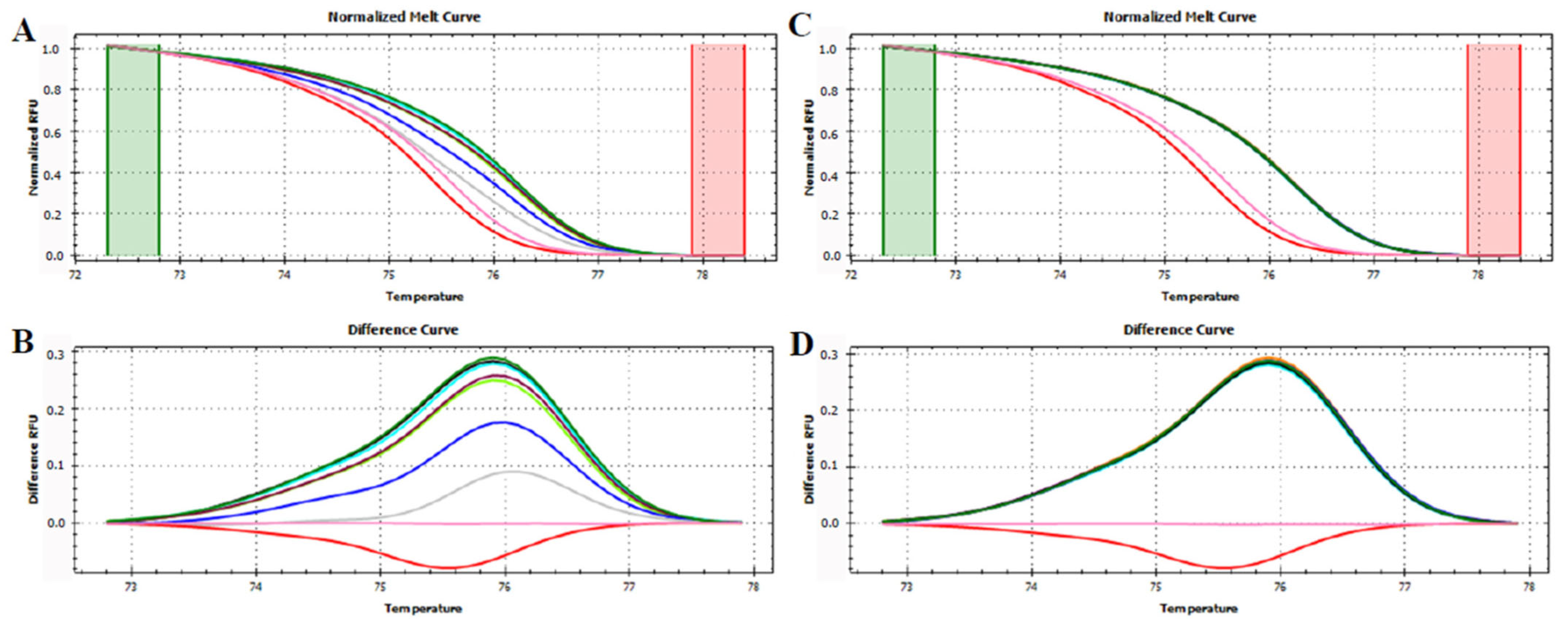
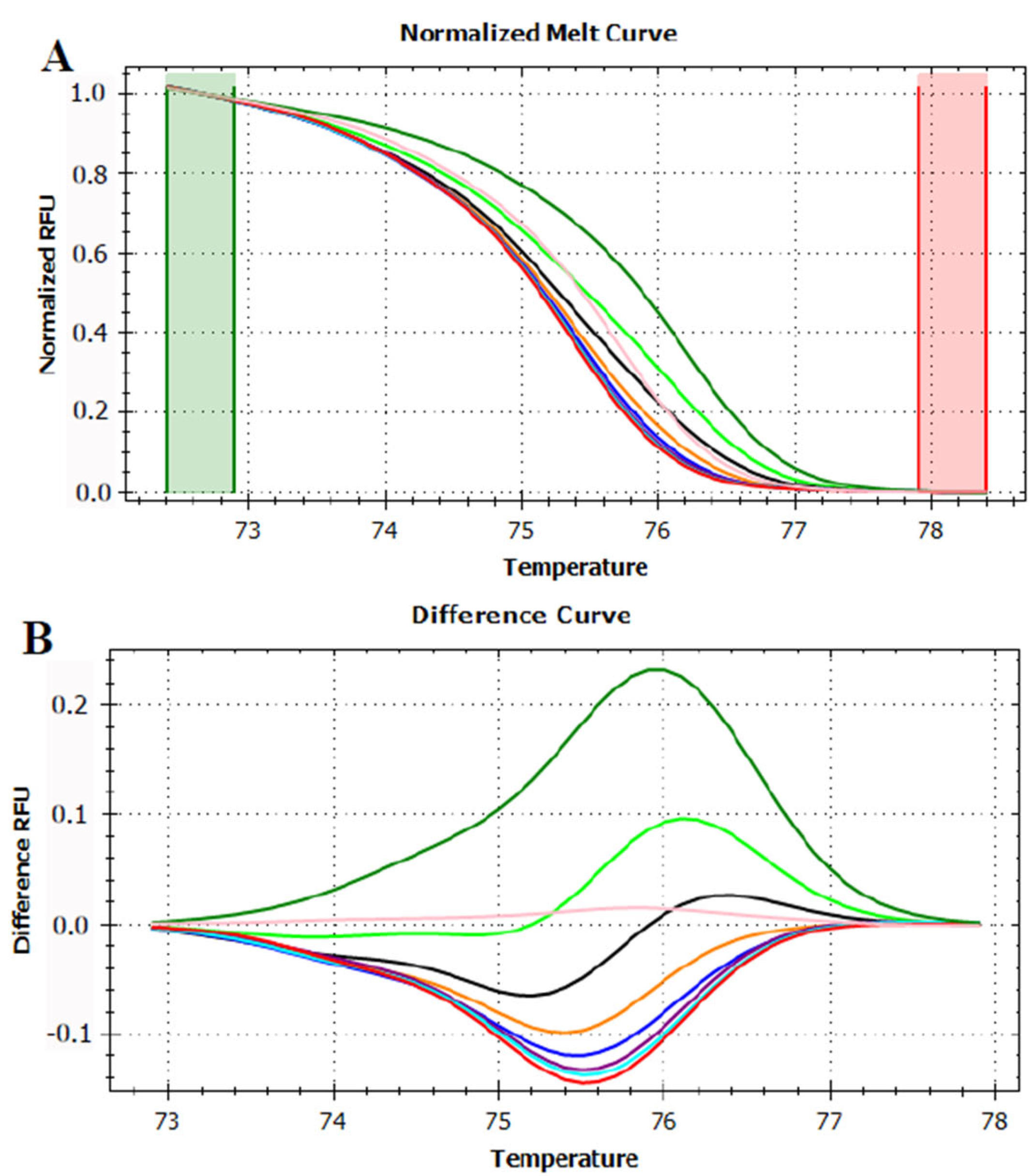
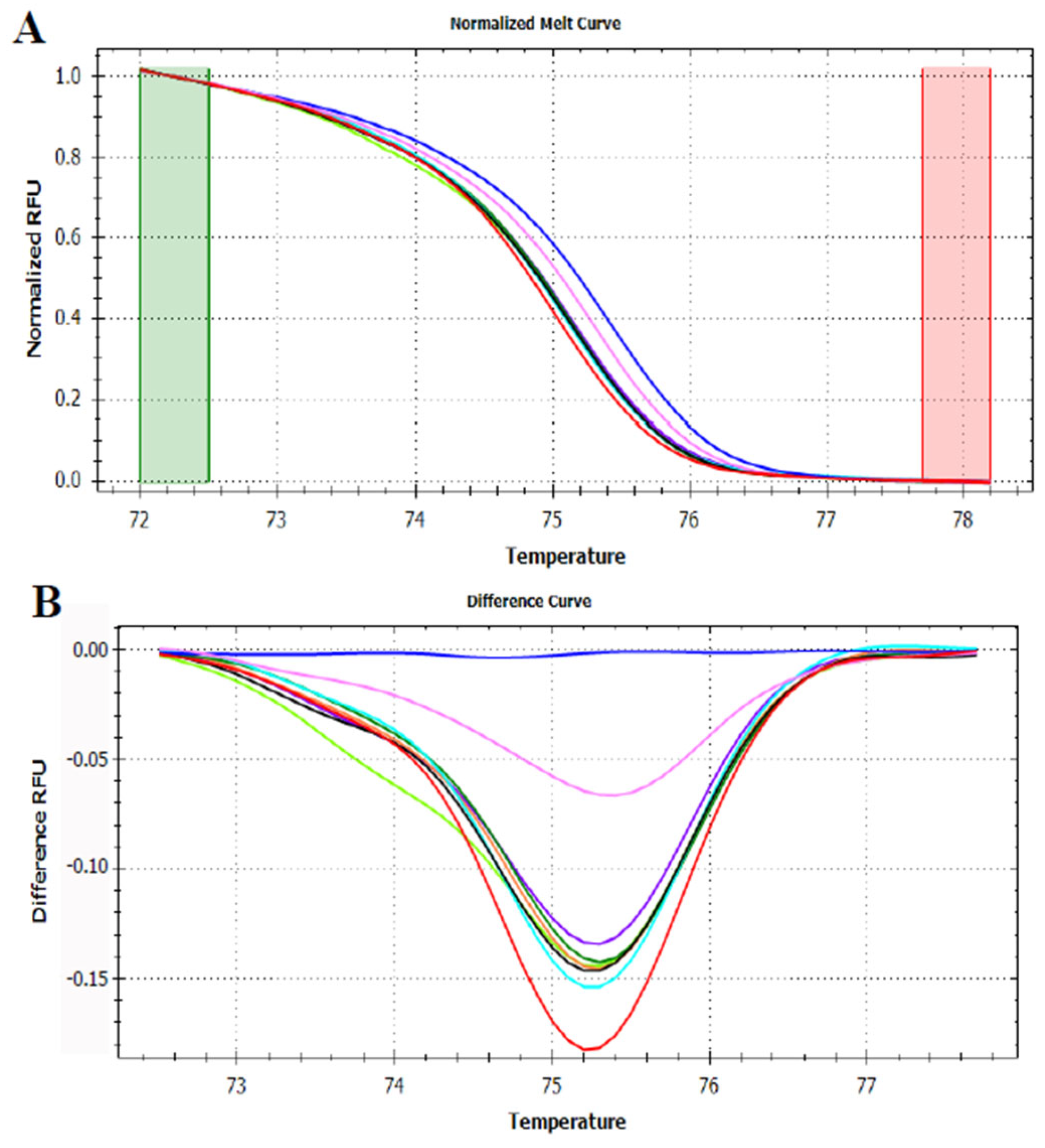
3.3. HRMA Targeting the Entire trnL (UAA) Sequence
3.4. HRMA Targeting P6 Loop Sequence
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Marconi, G.; Ferradini, N.; Russi, L.; Concezzi, L.; Veronesi, F.; Albertini, E. Genetic Characterization of the Apple Germplasm Collection in Central Italy: The Value of Local Varieties. Front. Plant Sci. 2018, 9, 1460. [Google Scholar] [CrossRef] [PubMed]
- Food and Agricultural Organization of the United Nation [FAO]. 2021. Available online: http://faostat.fao.org/beta/en/#data/qc/visualize (accessed on 14 February 2023).
- Assomela-CSO. 2017. Available online: http://www.assomela.it/index_it.html (accessed on 14 February 2023).
- British Soft Drinks Association. Market Share of Fruit Juice in the United Kingdom in 2021, by Flavor. 25 June 2022. Available online: https://www.statista.com/statistics/283986/soft-drinks-market-share-of-fruit-juice-in-the-united-kingdom-uk-by-flavour/ (accessed on 14 February 2023).
- Winsight Grocery Business. Most Appealing Fruit Juice Flavors in the United States in 2019. 1 March 2020. Available online: https://www.statista.com/statistics/1106476/fruit-juice-leading-flavors-us/ (accessed on 14 February 2023).
- El Sheikha, A.F. How to Determine the Geographical Origin of Food by Molecular Techniques. In Molecular Techniques in Food Biology; John Wiley & Sons, Ltd.: Hoboken, NJ, USA, 2018; pp. 1–26. ISBN 9781119374633. [Google Scholar]
- Wadood, S.A.; Boli, G.; Xiaowen, Z.; Hussain, I.; Yimin, W. Recent Development in the Application of Analytical Techniques for the Traceability and Authenticity of Food of Plant Origin. Microchem. J. 2020, 152, 104295. [Google Scholar] [CrossRef]
- Ruiz del Castillo, M.L.; Caja, M.M.; Blanch, G.P.; Herraiz, M. Enantiomeric Distribution of Chiral Compounds in Orange Juices According to Their Geographical Origins. J. Food Prot. 2003, 66, 1448–1454. [Google Scholar] [CrossRef] [PubMed]
- Aebersold, R.; Mann, M. Mass Spectrometry-Based Proteomics. Nature 2003, 422, 198–207. [Google Scholar] [CrossRef] [PubMed]
- Brenna, J.T.; Corso, T.N.; Tobias, H.J.; Caimi, R.J. High-Precision Continuous-Flow Isotope Ratio Mass Spectrometry. Mass Spectrom. Rev. 1997, 16, 227–258. [Google Scholar] [CrossRef]
- Longobardi, F.; Casiello, G.; Ventrella, A.; Mazzilli, V.; Nardelli, A.; Sacco, D.; Catucci, L.; Agostiano, A. Electronic Nose and Isotope Ratio Mass Spectrometry in Combination with Chemometrics for the Characterization of the Geographical Origin of Italian Sweet Cherries. Food Chem. 2015, 170, 90–96. [Google Scholar] [CrossRef]
- De Rijke, E.; Schoorl, J.C.; Cerli, C.; Vonhof, H.B.; Verdegaal, S.J.A.; Vivó-Truyols, G.; Lopatka, M.; Dekter, R.; Bakker, D.; Sjerps, M.J.; et al. The Use of δ2H and δ18O Isotopic Analyses Combined with Chemometrics as a Traceability Tool for the Geographical Origin of Bell Peppers. Food Chem. 2016, 204, 122–128. [Google Scholar] [CrossRef]
- Mimmo, T.; Camin, F.; Bontempo, L.; Capici, C.; Tagliavini, M.; Cesco, S.; Scampicchio, M. Traceability of Different Apple Varieties by Multivariate Analysis of Isotope Ratio Mass Spectrometry Data. Rapid Commun. Mass Spectrom. 2015, 29, 1984–1990. [Google Scholar] [CrossRef]
- Pii, Y.; Zamboni, A.; Santo, S.D.; Pezzotti, M.; Varanini, Z.; Pandolfini, T. Prospect on Ionomic Signatures for the Classification of Grapevine Berries According to Their Geographical Origin. Front. Plant Sci. 2017, 8, 640. [Google Scholar] [CrossRef]
- Hopfer, H.; Nelson, J.; Collins, T.S.; Heymann, H.; Ebeler, S.E. The Combined Impact of Vineyard Origin and Processing Winery on the Elemental Profile of Red Wines. Food Chem. 2015, 172, 486–496. [Google Scholar] [CrossRef]
- Zhao, H.; Yu, C.; Li, M. Effects of Geographical Origin, Variety, Season and Their Interactions on Minerals in Tea for Traceability. J. Food Compos. Anal. 2017, 63, 15–20. [Google Scholar] [CrossRef]
- Consonni, R.; Polla, D.; Cagliani, L.R. Organic and Conventional Coffee Differentiation by NMR Spectroscopy. Food Control 2018, 94, 284–288. [Google Scholar] [CrossRef]
- Consonni, R.; Cagliani, L.R.; Cogliati, C. NMR Based Geographical Characterization of Roasted Coffee. Talanta 2012, 88, 420–426. [Google Scholar] [CrossRef]
- Böhme, K.; Calo-Mata, P.; Barros-Velázquez, J.; Ortea, I. Review of Recent DNA-Based Methods for Main Food-Authentication Topics. J. Agric. Food Chem. 2019, 67, 3854–3864. [Google Scholar] [CrossRef]
- Sforza, S.E. Food Authentication Using Bioorganic Molecules; DEStech Publications, Inc.: Lancaster, PA, USA, 2013; ISBN 9781605950457. [Google Scholar]
- Barcaccia, G.; Lucchin, M.; Cassandro, M. DNA Barcoding as a Molecular Tool to Track down Mislabeling and Food Piracy. Diversity 2016, 8, 2. [Google Scholar] [CrossRef]
- Gregory, T.R. DNA Barcoding Does Not Compete with Taxonomy. Nature 2005, 434, 1067. [Google Scholar] [CrossRef]
- Miller, S.E. DNA Barcoding and the Renaissance of Taxonomy. Proc. Natl. Acad. Sci. USA 2007, 104, 4775–4776. [Google Scholar] [CrossRef]
- Reed, G.H.; Kent, J.O.; Wittwer, C.T. High-Resolution DNA Melting Analysis for Simple and Efficient Molecular Diagnostics. Pharmacogenomics 2007, 8, 597–608. [Google Scholar] [CrossRef]
- Taberlet, P.; Coissac, E.; Pompanon, F.; Gielly, L.; Miquel, C.; Valentini, A.; Vermat, T.; Corthier, G.; Brochmann, C.; Willerslev, E. Power and Limitations of the Chloroplast trnL (UAA) Intron for Plant DNA Barcoding. Nucl. Acids Res. 2007, 35, e14. [Google Scholar] [CrossRef]
- Rønning, S.B.; Rudi, K.; Berdal, K.G.; Holst-Jensen, A. Differentiation of Important and Closely Related Cereal Plant Species (Poaceae) in Food by Hybridization to an Oligonucleotide Array. J. Agric. Food Chem. 2005, 53, 8874–8880. [Google Scholar] [CrossRef]
- Ward, J.; Peakall, R.; Gilmore, S.R.; Robertson, J. A Molecular Identification System for Grasses: A Novel Technology for Forensic Botany. Forensic Sci. Int. 2005, 152, 121–131. [Google Scholar] [CrossRef] [PubMed]
- Faria, M.A.; Magalhães, A.; Nunes, M.E.; Oliveira, M.B.P.P. High Resolution Melting of TrnL Amplicons in Fruit Juices Authentication. Food Control 2013, 33, 136–141. [Google Scholar] [CrossRef]
- Chedid, E.; Rizou, M.; Kalaitzis, P. Application of High Resolution Melting Combined with DNA-Based Markers for Quantitative Analysis of Olive Oil Authenticity and Adulteration. Food Chem. X 2020, 6, 100082. [Google Scholar] [CrossRef] [PubMed]
- Taberlet, P.; Gielly, L.; Pautou, G.; Bouvet, J. Universal Primers for Amplification of Three Non-Coding Regions of Chloroplast DNA. Plant Mol. Biol. 1991, 17, 1105–1109. [Google Scholar] [CrossRef] [PubMed]
- Doyle, J.J.; Doyle, J.L. A Rapid DNA Isolation Procedure for Small Quantities of Fresh Leaf Tissue. Phytochem. Bull. 1987, 19, 11–15. [Google Scholar]
- Davies, R.W.; Waring, R.B.; Ray, J.A.; Brown, T.A.; Scazzocchio, C. Making Ends Meet: A Model for RNA Splicing in Fungal Mitochondria. Nature 1982, 300, 719–724. [Google Scholar] [CrossRef]
- Michel, F.; Jacquier, A.; Dujon, B. Comparison of Fungal Mitochondrial Introns Reveals Extensive Homologies in RNA Secondary Structure. Biochimie 1982, 64, 867–881. [Google Scholar] [CrossRef]
- Quandt, D.; Müller, K.; Stech, M.; Hilu, K.; Frey, W.; Borsch, T. Molecular Evolution of the Chloroplast TRNL-F Region in Land Plants. Monogr. Syst. Bot. Missouri Bot. Gard. 2004, 98, 13–37. [Google Scholar]
- Chen, S.; Xia, T.; Wang, Y.; Liu, J.; Chen, S. Molecular Systematics and Biogeography of Crawfurdia, Metagentiana and Tripterospermum (Gentianaceae) Based on Nuclear Ribosomal and Plastid DNA Sequences. Ann. Bot. 2005, 96, 413–424. [Google Scholar] [CrossRef]
- McDade, L.A.; Daniel, T.F.; Kiel, C.A.; Vollesen, K. Phylogenetic Relationships among Acantheae (Acanthaceae): Major Lineages Present Contrasting Patterns of Molecular Evolution and Morphological Differentiation. Syst. Bot. 2005, 30, 834–862. [Google Scholar] [CrossRef]
- Scharaschkin, T.; Doyle, J.A. Phylogeny and Historical Biogeography of Anaxagorea (Annonaceae) Using Morphology and Non-Coding Chloroplast Sequence Data. Syst. Bot. 2005, 30, 712–735. [Google Scholar] [CrossRef]
- Farrar, J.S.; Wittwer, C.T. High-Resolution Melting Curve Analysis for Molecular Diagnostics. In Molecular Diagnostics, 3rd ed.; Academic Press: Cambridge, MA, USA, 2017; pp. 79–102. [Google Scholar] [CrossRef]
- Bosmali, I.; Ganopoulos, I.; Madesis, P.; Tsaftaris, A. Microsatellite and DNA-Barcode Regions Typing Combined with High Resolution Melting (HRM) Analysis for Food Forensic Uses: A Case Study on Lentils (Lens culinaris). Food Res. Int. 2012, 46, 141–147. [Google Scholar] [CrossRef]
- Ganopoulos, I.; Argiriou, A.; Tsaftaris, A. Microsatellite High Resolution Melting (SSR-HRM) Analysis for Authenticity Testing of Protected Designation of Origin (PDO) Sweet Cherry Products. Food Control 2011, 22, 532–541. [Google Scholar] [CrossRef]
- Ganopoulos, I.; Madesis, P.; Darzentas, N.; Argiriou, A.; Tsaftaris, A. Barcode High Resolution Melting (Bar-HRM) Analysis for Detection and Quantification of PDO “Fava Santorinis” (Lathyrus clymenum) Adulterants. Food Chem. 2012, 133, 505–512. [Google Scholar] [CrossRef]
- Chaffey, N. Raven Biology of Plants. Ann. Bot. 2014, 113, vii. [Google Scholar] [CrossRef]



| Mixed Juices Composition | ||||
|---|---|---|---|---|
| Apple | Pear | Peach | Kiwi | |
| MJ1 | 99.5% | 0.5% | - | - |
| MJ2 | 99% | 1% | - | - |
| MJ3 | 95% | 5% | - | - |
| MJ4 | 90% | 10% | - | - |
| MJ5 | 75% | 25% | - | - |
| MJ6 | 50% | 50% | - | - |
| MJ7 | 99.5% | - | 0.5% | - |
| MJ8 | 99% | - | 1% | - |
| MJ9 | 95% | - | 5% | - |
| MJ10 | 90% | - | 10% | - |
| MJ11 | 75% | - | 25% | - |
| MJ12 | 50% | - | 50% | - |
| MJ13 | 99.5% | - | - | 0.5% |
| MJ14 | 99% | - | - | 1% |
| MJ15 | 95% | - | - | 5% |
| MJ16 | 90% | - | - | 10% |
| MJ17 | 75% | - | - | 25% |
| MJ18 | 50% | - | - | 50% |
| MJ19 | 25% | - | - | 75% |
| MJ20 | 10% | - | - | 90% |
| MJ21 | 5% | - | - | 95% |
| MJ22 | 1% | - | - | 99% |
| MJ23 | 0.5% | - | - | 99.5% |
| Scientific Name | P6 Loop Sequence Alignment | ||||
|---|---|---|---|---|---|
| Malus domestica | GGGCAATCCTGAGCCAAATCCTGTTTTATGAAAATAAACA | ||||
| Pyrus communis | GGGCAATCCTGAGCCAAATCCTGTTTTATGAAAATAAACA | ||||
| Prunus persica | GGGCGATCCTGAGCCAAATCCTGTTTTATTAAAACAAACA | ||||
| Actinidia chinensis | GGGCAATCCTGAGCCAAATCCTTTTTTTCGAAAACAAACA | ||||
| **** ***************** **** **** ***** | |||||
| Malus domestica | AGGGTTTCATAAACCGAAAATAAAA-AAGGATAGGTGCAG | ||||
| Pyrus communis | AGGGTTTCATAAACCGAAAATAAAA-AAGGATAGGTGCAG | ||||
| Prunus persica | AGGGTTTCATAAACCGAGAATAAAA-AAGGATAGGTGCAG | ||||
| Actinidia chinensis | AAGATT-CAGAAAGCGAAAATAAAACAAGGATAGGTGCAG | ||||
| * * ** ** *** *** ******* ************** | |||||
| Malus domestica | AGACTCAATGG | ||||
| Pyrus communis | AGACTCAATGG | ||||
| Prunus persica | AGACTCAATGG | ||||
| Actinidia chinensis | AGACTCAATGG | ||||
| *********** | |||||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Monterisi, S.; Zuluaga, M.Y.A.; Porceddu, A.; Cesco, S.; Pii, Y. The Application of High-Resolution Melting Analysis to trnL (UAA) Intron Allowed a Qualitative Identification of Apple Juice Adulterations. Foods 2023, 12, 1437. https://doi.org/10.3390/foods12071437
Monterisi S, Zuluaga MYA, Porceddu A, Cesco S, Pii Y. The Application of High-Resolution Melting Analysis to trnL (UAA) Intron Allowed a Qualitative Identification of Apple Juice Adulterations. Foods. 2023; 12(7):1437. https://doi.org/10.3390/foods12071437
Chicago/Turabian StyleMonterisi, Sonia, Monica Yorlady Alzate Zuluaga, Andrea Porceddu, Stefano Cesco, and Youry Pii. 2023. "The Application of High-Resolution Melting Analysis to trnL (UAA) Intron Allowed a Qualitative Identification of Apple Juice Adulterations" Foods 12, no. 7: 1437. https://doi.org/10.3390/foods12071437
APA StyleMonterisi, S., Zuluaga, M. Y. A., Porceddu, A., Cesco, S., & Pii, Y. (2023). The Application of High-Resolution Melting Analysis to trnL (UAA) Intron Allowed a Qualitative Identification of Apple Juice Adulterations. Foods, 12(7), 1437. https://doi.org/10.3390/foods12071437







