Storage Fungi and Mycotoxins Associated with Rice Samples Commercialized in Thailand
Abstract
1. Introduction
2. Materials and Methods
2.1. Rice Collection and Fungal Isolation
2.2. Morphological Study
2.3. DNA Extraction, Polymerase Chain Reaction Amplification, and Sequence-Based Identification
2.4. Multiple Sequence Alignments and Phylogenetic Analyses
2.5. Fungal Extraction for Mycotoxin Detection
2.6. QuEChERS Sample Extraction and Clean-Up
2.7. LC-(ESI)-MS/MS Conditions
2.8. Method Validation Procedure and Matrix Effects Study
3. Results
3.1. Total Seed-Borne Fungal Infections Using Blotting Method
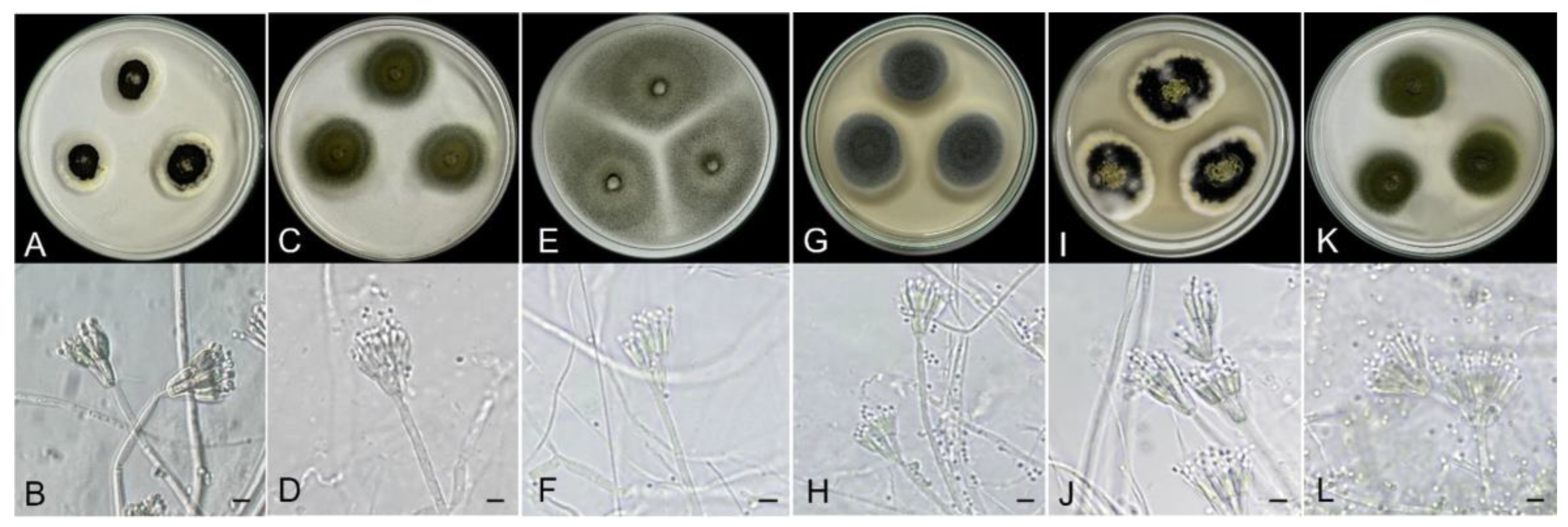
3.2. Preliminary Fungal Identification Using Agar Plate Method
3.3. DNA Sequence-Based Data of Fungal Isolates Using BLAST NCBI Database Tool
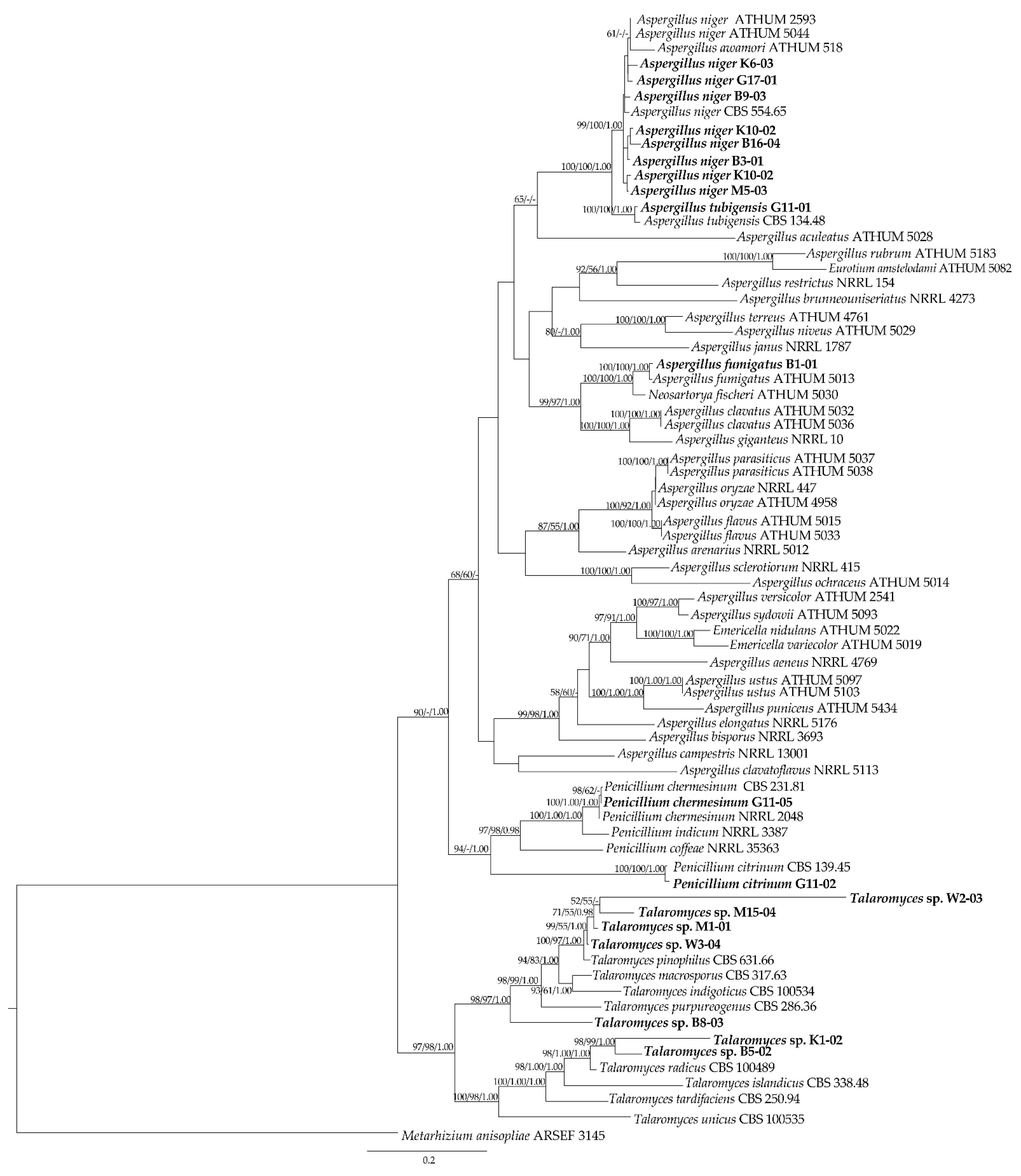
3.4. Molecular Data of Fungal Isolates Based on Multi-Locus Phylogenetic Analysis
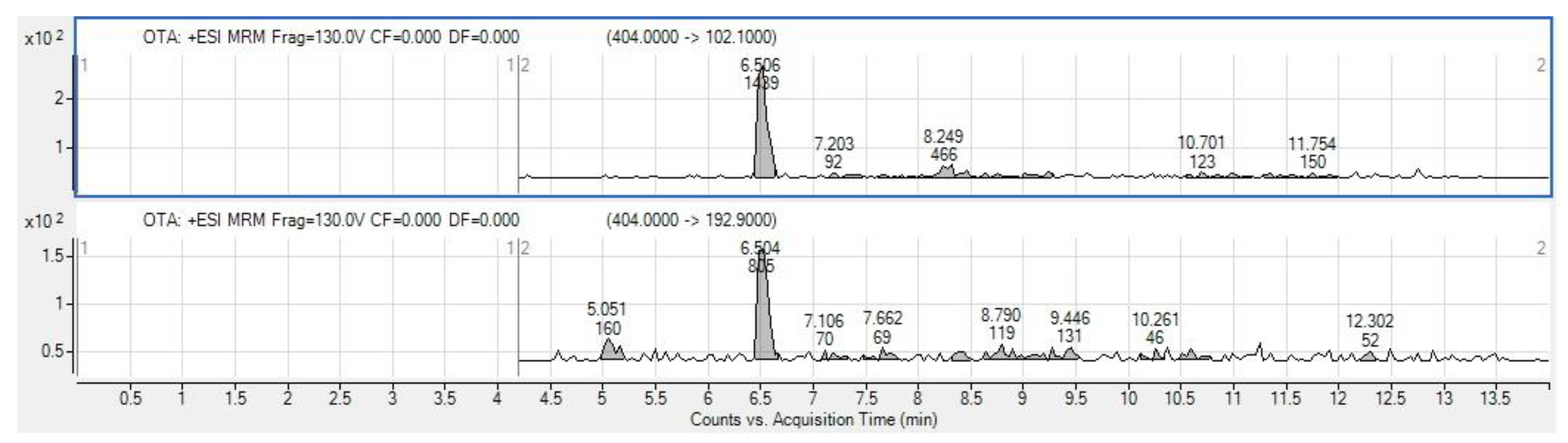
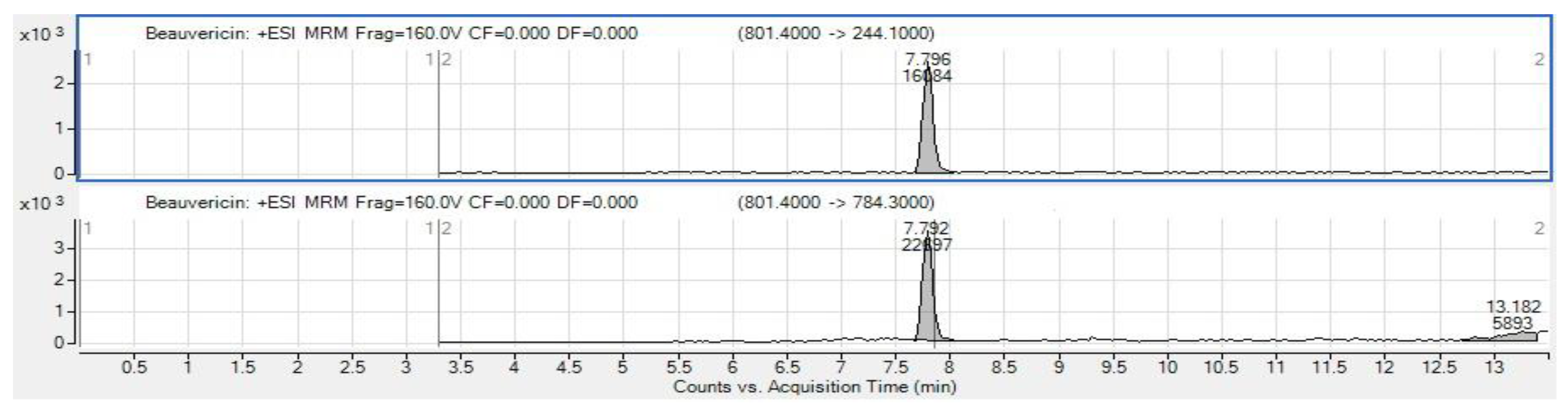
3.5. Mycotoxin Analysis
4. Discussion and Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Muthayya, S.; Sugimoto, J.D.; Montgomery, S.; Maberly, G.F. An overview of global rice production, supply, trade, and consumption. Ann. N. Y. Acad. Sci. 2014, 1324, 7–14. [Google Scholar] [CrossRef] [PubMed]
- Lu, B.-R. Taxonomy of the genus Oryza (Poaceae): Historical perspective and current status. Int. Rice Res. Notes 1999, 24, 4–8. [Google Scholar]
- Suebpongsang, P.; Ekasingh, B.; Cramb, R. Commercialisation of rice farming in northeast Thailand. In White Gold: The Commercialisation of Rice Farming in the Lower Mekong Basin; Palgrave Macmillan: Singapore, 2020; pp. 39–68. [Google Scholar]
- Groopman, J.; Kensler, T.; Wu, F. Food Safety: Mycotoxins-Occurrence and Toxic Effects. In Encyclopedia of Human Nutrition; Elsevier Inc.: Amsterdam, The Netherlands, 2012; pp. 337–341. [Google Scholar]
- Majeed, S.; De Boevre, M.; De Saeger, S.; Rauf, W.; Tawab, A.; Rahman, M.; Iqbal, M. Multiple mycotoxins in rice: Occurrence and health risk assessment in children and adults of Punjab, Pakistan. Toxins 2018, 10, 77. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Wang, Q.; Huang, J.; Chen, Z.; Liu, S.; Wang, X.; Wang, F. Mycotoxin contamination and presence of mycobiota in rice sold for human consumption in China. Food Control 2019, 98, 19–23. [Google Scholar] [CrossRef]
- World Health Organization. Mycotoxins; WHO: Geneva, Switzerland, 2018; Available online: https://www.who.int/news-room/fact-sheets/detail/mycotoxins (accessed on 21 September 2022).
- Omotayo, O.P.; Omotayo, A.O.; Babalola, O.O.; Mwanza, M. Comparative study of aflatoxin contamination of winter and summer ginger from the North West Province of South Africa. Toxicol. Rep. 2019, 6, 489–495. [Google Scholar] [CrossRef]
- Phoku, J.; Barnard, T.; Potgieter, N.; Dutton, M. Mycotoxigenic potentials of the genera: Aspergillus, Fusarium and Penicillium isolated from houseflies (Musca domestica L.). Acta Trop. 2017, 168, 29–36. [Google Scholar] [CrossRef]
- Reddy, K.; Reddy, C.; Abbas, H.; Abel, C.; Muralidharan, K. Mycotoxigenic fungi, mycotoxins, and management of rice grains. Food Microbiol. 2008, 27, 287–317. [Google Scholar] [CrossRef]
- Some Naturally Occurring Substances: Food Items and Constituents, Heterocyclic Aromatic Amines and Mycotoxins. In IARC Monographs on the Evaluation of the Carcinogenic Risk of Chemicals to Humans; IARC Working Group on the Evaluation of Carcinogenic Risks to Humans: Lyon, France, 1993; p. 56.
- EFSA. International frameworks dealing with human risk assessment of combined exposure to multiple chemicals. EFSA J. 2013, 11, 3313. [Google Scholar]
- Leggieri, M.C.; Lanubile, A.; Dall’Asta, C.; Pietri, A.; Battilani, P. The impact of seasonal weather variation on mycotoxins: Maize crop in 2014 in northern Italy as a case study. World Mycotoxin J. 2020, 13, 25–36. [Google Scholar] [CrossRef]
- Aydin, A.; Aksu, H.; Gunsen, U. Assessment. Mycotoxin levels and incidence of mould in Turkish rice. Environ. Monit. Assess. 2011, 178, 271–280. [Google Scholar] [CrossRef]
- Bansal, J.; Pantazopoulos, P.; Tam, J.; Cavlovic, P.; Kwong, K.; Turcotte, A.-M.; Lau, B.-Y.; Scott, P. Contaminants. Surveys of rice sold in Canada for aflatoxins, ochratoxin A and fumonisins. Food Addit. Contam. Part A Chem. Anal. Control Expo. Risk Assess. 2011, 28, 767–774. [Google Scholar] [CrossRef] [PubMed]
- Ferre, F.S. Worldwide occurrence of mycotoxins in rice. Food Control 2016, 62, 291–298. [Google Scholar] [CrossRef]
- Makun, H.A.; Dutton, M.F.; Njobeh, P.B.; Mwanza, M.; Kabiru, A.Y. Natural multi-occurrence of mycotoxins in rice from Niger State, Nigeria. Mycotoxin Res. 2011, 27, 97–104. [Google Scholar] [CrossRef] [PubMed]
- Fredlund, E.; Thim, A.-M.; Gidlund, A.; Brostedt, S.; Nyberg, M.; Olsen, M.; Contaminants. Moulds and mycotoxins in rice from the Swedish retail market. Food Addit. Contam. Part A Chem. Anal. Control. Expo. Risk Assess. 2009, 26, 527–533. [Google Scholar] [CrossRef] [PubMed]
- Park, J.W.; Choi, S.-Y.; Hwang, H.-J.; Kim, Y.-B. Fungal mycoflora and mycotoxins in Korean polished rice destined for humans. Int. J. Food Microbiol. 2005, 103, 305–314. [Google Scholar] [CrossRef] [PubMed]
- Sales, A.C.; Yoshizawa, T. Contaminants. Updated profile of aflatoxin and Aspergillus section Flavi contamination in rice and its byproducts from the Philippines. Food Addit. Contam. Part A Chem. Anal. Control Expo. Risk Assess. 2005, 22, 429–436. [Google Scholar] [CrossRef]
- Jettanajit, A.; Nhujak, T. Determination of mycotoxins in brown rice using QuEChERS sample preparation and UHPLC–MS-MS. Chromatogr. Sci. 2016, 54, 720–729. [Google Scholar] [CrossRef]
- Sinphithakkul, P.; Poapolathep, A.; Klangkaew, N.; Imsilp, K.; Logrieco, A.F.; Zhang, Z.; Poapolathep, S. Occurrence of multiple mycotoxins in various types of rice and barley samples in Thailand. J. Food Prot. 2019, 82, 1007–1015. [Google Scholar] [CrossRef]
- Suprasert, D. Contamination of aflatoxin in cereal grains. Health Prom. Environ. Health 2008, 31, 102–107. [Google Scholar]
- Limonard, T. A modified blotter test for seed health. Neth. J. Plant Pathol. 1966, 72, 319–321. [Google Scholar] [CrossRef]
- ISTA. International Rules for Seed Testing; Edition 2013; International Seed Testing Association: Bassersdorf, Switzerland, 2013. [Google Scholar]
- Mathur, S.; Kongsdal, O. Common Laboratory Seed Health Testing Methods for Detecting Fungi; International Seed Testing Association: Zürich, Switzerland, 2003. [Google Scholar]
- Butt, A.; Yaseen, S.; Javaid, A. Seed-borne mycoflora of stored rice grains and its [sic] chemical control. J. Anim. Plant Sci. 2011, 21, 193–196. [Google Scholar]
- Malone, J.P.; Muckett, A. Seed Borne Fungi: Description of 77 Fungus Species; International Seed Testing Association: Zürich, Switzerland, 1964. [Google Scholar]
- Watanabe, T. Pictorial Atlas of Soil and Seed Fungi: Morphologies of Cultured Fungi and Key to Species; CRC Press: Boca Raton, FL, USA, 2002. [Google Scholar]
- Valero, C.; de la Cruz-Villar, L.; Zaragoza, Ó.; Buitrago, M.J. New panfungal real-time PCR assay for diagnosis of invasive fungal infections. J. Clin. Microbiol. 2016, 54, 2910–2918. [Google Scholar] [CrossRef]
- Fredricks, D.N.; Smith, C.; Meier, A. Comparison of six DNA extraction methods for recovery of fungal DNA as assessed by quantitative PCR. Clin. Microbiol. Infect. 2005, 43, 5122–5128. [Google Scholar] [CrossRef]
- Raja, H.A.; Miller, A.N.; Pearce, C.J.; Oberlies, N.H. Fungal identification using molecular tools: A primer for the natural products research community. J. Nat. Prod. 2017, 80, 756–770. [Google Scholar] [CrossRef] [PubMed]
- Glass, N.L.; Donaldson, G.C. Development of primer sets designed for use with the PCR to amplify conserved genes from filamentous ascomycetes. Appl. Environ. Microbiol. Rep. 1995, 61, 1323–1330. [Google Scholar] [CrossRef]
- Liu, Y.J.; Whelen, S.; Hall, B.D. Evolution. Phylogenetic relationships among ascomycetes: Evidence from an RNA polymerse II subunit. Mol. Biol. Evol. 1999, 16, 1799–1808. [Google Scholar] [CrossRef] [PubMed]
- Hong, S.-B.; Go, S.-J.; Shin, H.-D.; Frisvad, J.C.; Samson, R.A. Polyphasic taxonomy of Aspergillus fumigatus and related species. Mycologia 2005, 97, 1316–1329. [Google Scholar] [CrossRef] [PubMed]
- Kerry, O.; Donnell, K.; Nirenberg, H.I.; Aoki, T.; Cigelnik, E. A multigene phylogeny of the Gibberella fujikuroi species complex: Detection of additional phylogenetically distinct species. Mycoscience 2000, 41, 61–78. [Google Scholar] [CrossRef]
- Lee, P.Y.; Costumbrado, J.; Hsu, C.-Y.; Kim, Y.H. Agarose gel electrophoresis for the separation of DNA fragments. JoVE 2012, 62, e3923. [Google Scholar] [CrossRef]
- Hall, T. BioEdit: Biological Sequence Alignment Editor for Win95; Ibis Biosciences: Carlsbad, CA, USA, 2007. [Google Scholar]
- Mount, D.W. Using the basic local alignment search tool (BLAST). Cold Spring Harb. Protoc. 2007, 2007, pdb.top17. [Google Scholar] [CrossRef]
- Samson, R.; Peterson, S.; Frisvad, J.C.; Varga, J. New species in Aspergillus section Terrei. Stud. Mycol. 2011, 69, 39–55. [Google Scholar] [CrossRef] [PubMed]
- Krimitzas, A.; Pyrri, I.; Kouvelis, V.N.; Kapsanaki-Gotsi, E.; Typas, M.A. A phylogenetic analysis of Greek isolates of Aspergillus species based on morphology and nuclear and mitochondrial gene sequences. BioMed Res. Int. 2013, 2013, 260395. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, V.D.; Pham, T.T. Penicillium vietnamense sp. nov., the first novel marine fungi species described from Vietnam with a unique conidiophore structure and molecular phylogeny of Penicillium section Charlesia. Mycobiology 2022, 50, 1–11. [Google Scholar] [CrossRef]
- Muñoz, K.; Vega, M.; Rios, G.; Geisen, R.; Degen, G.H. Mycotoxin production by different ochratoxigenic Aspergillus and Penicillium species on coffee-and wheat-based media. Mycotoxin Res. 2011, 27, 239–247. [Google Scholar] [CrossRef]
- Alkuwari, A.; Hassan, Z.U.; Zeidan, R.; Al-Thani, R.; Jaoua, S. Occurrence of mycotoxins and toxigenic fungi in cereals and application of yeast volatiles for their biological control. J. Toxins 2022, 14, 404. [Google Scholar] [CrossRef]
- Pongpraket, M.; Poapolathep, A.; Wongpanit, K.; Tanhan, P.; Giorgi, M.; Zhang, Z.; Li, P.; Poapolathep, S. Exposure assessment of multiple mycotoxins in black and white sesame seeds consumed in Thailand. J. Food Prot. 2020, 83, 1198–1207. [Google Scholar] [CrossRef]
- European Commission. Commission Regulation (EC) No 401/2006 of 23 February 2006 Laying Down the Methods of Sampling and Analysis for the Official Control of the Levels of Mycotoxins in Foodstuffs; European Commission: Brussels, Belgium, 2006. [Google Scholar]
- Pathmanathan, N.D.N.; Manamgoda, D.S.; Sandamali, T.G.I.; Munasinghe, M. Comparison of colonization, diversity, and molecular phylogeny of endophytic fungi in selected traditional and newly improved rice (Oryza sativa L.) varieties in Sri Lanka. Fungal Biol. 2022, 12, 147–169. [Google Scholar] [CrossRef]
- Shiratori, N.; Kobayashi, N.; Tulayakul, P.; Sugiura, Y.; Takino, M.; Endo, O.; Sugita-Konishi, Y. Occurrence of Penicillium brocae and Penicillium citreonigrum, which produce a mutagenic metabolite and a mycotoxin citreoviridin, respectively, in selected commercially available rice grains in Thailand. Toxins 2017, 9, 194. [Google Scholar] [CrossRef]
- Singh, S.; Sinha, A. Evaluation of seed-borne mycoflora of rice (Oryza sativa L.) by the effect of storage length on fungal invasion under different storage technique. Pure Appl. Microbiol. 2016, 10, 2753–2761. [Google Scholar] [CrossRef]
- Lücking, R.; Aime, M.C.; Robbertse, B.; Miller, A.N.; Aoki, T.; Ariyawansa, H.A.; Cardinali, G.; Crous, P.W.; Druzhinina, I.S.; Geiser, D.M. Fungal taxonomy and sequence-based nomenclature. Nat. Microbiol. 2021, 6, 540–548. [Google Scholar] [CrossRef]
- Houbraken, J.; Kocsubé, S.; Visagie, C.M.; Yilmaz, N.; Wang, X.-C.; Meijer, M.; Kraak, B.; Hubka, V.; Bensch, K.; Samson, R. Classification of Aspergillus, Penicillium, Talaromyces and related genera (Eurotiales): An overview of families, genera, subgenera, sections, series and species. Stud. Mycol. 2020, 96, 141–153. [Google Scholar] [CrossRef] [PubMed]
- Schoch, C.L.; Seifert, K.A.; Huhndorf, S.; Robert, V.; Spouge, J.L.; Levesque, C.A.; Chen, W.; Consortium, F.B.; List, F.B.C.A.; Bolchacova, E. Nuclear ribosomal internal transcribed spacer (ITS) region as a universal DNA barcode marker for Fungi. Proc. Natl. Acad. Sci. USA 2012, 109, 6241–6246. [Google Scholar] [CrossRef] [PubMed]
- Lücking, R.; Aime, M.C.; Robbertse, B.; Miller, A.N.; Ariyawansa, H.A.; Aoki, T.; Cardinali, G.; Crous, P.W.; Druzhinina, I.S.; Geiser, D.M. Unambiguous identification of fungi: Where do we stand and how accurate and precise is fungal DNA barcoding? IMA Fungus 2020, 11, 14. [Google Scholar] [CrossRef] [PubMed]
- Tekpinar, A.D.; Kalmer, A. Utility of various molecular markers in fungal identification and phylogeny. Nova Hedwig. 2019, 109, 187–224. [Google Scholar] [CrossRef]
- Kongkapan, J.; Poapolathep, S.; Isariyodom, S.; Kumagai, S.; Poapolathep, A. Simultaneous detection of multiple mycotoxins in broiler feeds using a liquid chromatography tandem-mass spectrometry. Vet. Med. Sci. 2016, 78, 259–264. [Google Scholar] [CrossRef]
- Jitjak, W.; Sanoamuang, N. The contamination of toxins produced by naturally occurring fungi in non-chemical rice products. Int. J. Agric. Tech. 2019, 15, 17–34. [Google Scholar]
- Mikušová, P.; Caboň, M.; Melichárková, A.; Urík, M.; Ritieni, A.; Slovák, M. Genetic diversity, ochratoxin A and fumonisin profiles of strains of Aspergillus section Nigri isolated from dried vine fruits. Toxins 2020, 12, 592. [Google Scholar] [CrossRef]
- Tavakol Noorabadi, M.; Babaeizad, V.; Zare, R.; Asgari, B.; Haidukowski, M.; Epifani, F.; Stea, G.; Moretti, A.; Logrieco, A.F.; Susca, A. Isolation, Molecular identification, and mycotoxin production of Aspergillus species isolated from the rhizosphere of sugarcane in the South of Iran. Toxins 2020, 12, 122. [Google Scholar] [CrossRef]
- Testempasis, S.I.; Kamou, N.N.; Papadakis, E.-N.; Menkissoglu-Spiroudi, U.; Karaoglanidis, G.S. Conventional vs. organic vineyards: Black Aspergilli population structure, mycotoxigenic capacity and mycotoxin contamination assessment in wines, using a new Q-TOF MS-MS detection method. Food Control 2022, 136, 108860. [Google Scholar] [CrossRef]
- Rastogi, M.; Shrivastava, S.; Shukla, P. Bioprospecting of xylanase producing fungal strains: Multilocus phylogenetic analysis and enzyme activity profiling. J. Basic Microbiol. 2022, 62, 150–161. [Google Scholar] [CrossRef]
- Jiménez-López, J.; Llorent-Martínez, E.; Ortega-Barrales, P.; Ruiz-Medina, A. Multi-commutated fluorometric optosensor for the determination of citrinin in rice and red yeast rice supplements. Food Addit. Contam. Part A Chem. Anal. Control Expo. Risk Assess. 2014, 31, 1744–1750. [Google Scholar] [CrossRef] [PubMed]
- El-Aziz, A.; Mahmoud, M.A.; Al-Sohaibani, S.A.; Al-Othman, M.R.; microbiology, a. Survey of postharvest fungi associated with wheat, rice and barley grains in Riyadh (Saudi Arabia). Pure Appl. Chem. 2013, 7, 45–55. [Google Scholar]
- Trung, T.S.; Bailly, J.; Querin, A.; Le Bars, P.; Guerre, P. Fungal contamination of rice from south Vietnam, mycotoxinogenesis of selected strains and residues in rice. Rev. Med. Vet. 2001, 152, 555–560. [Google Scholar]
- Ekpakpale, D.O.; Kraak, B.; Meijer, M.; Ayeni, K.I.; Houbraken, J.; Ezekiel, C.N. Fungal Diversity and Aflatoxins in Maize and Rice Grains and Cassava-Based Flour (Pupuru) from Ondo State, Nigeria. J. Fungi 2021, 7, 635. [Google Scholar] [CrossRef]
- Yun, T.-S.; Park, S.-Y.; Yu, J.; Hwang, Y.; Hong, K.-J. Isolation and identification of fungal species from the insect pest Tribolium castaneum in rice processing complexes in Korea. Plant Pathol. 2018, 34, 356. [Google Scholar] [CrossRef]
- Santos, A.R.; Carreiró, F.; Freitas, A.; Barros, S.; Brites, C.; Ramos, F.; Sanches Silva, A. Mycotoxins Contamination in Rice: Analytical Methods, Occurrence and Detoxification Strategies. J. Toxins 2022, 14, 647. [Google Scholar] [CrossRef]
- Olagunju, O.; Mchunu, N.; Venter, S.; Guibert, B.; Durand, N.; Metayer, I.; Montet, D.; Ijabadeniyi, O. Fungal contamination of food commodities in Durban, South Africa. J. Food Saf. 2018, 38, e12515. [Google Scholar] [CrossRef]
- Houbraken, J.; de Vries, R.P.; Samson, R.A. Modern taxonomy of biotechnologically important Aspergillus and Penicillium species. Adv. Appl. Microbiol. 2014, 86, 199–249. [Google Scholar]
- Egbuta, M.A.; Mwanza, M.; Njobeh, P.B.; Phoku, J.Z.; Chilaka, C.A.; Dutton, M.F. Isolation of filamentous fungi species contaminating some Nigerian food commodities. Food Res. 2015, 4, 38. [Google Scholar] [CrossRef]
- González-Jartín, J.M.; Ferreiroa, V.; Rodríguez-Cañás, I.; Alfonso, A.; Sainz, M.J.; Aguín, O.; Vieytes, M.R.; Gomes, A.; Ramos, I.; Botana, L.M. Occurrence of mycotoxins and mycotoxigenic fungi in silage from the north of Portugal at feed-out. Int. J. Food Microbiol. 2022, 365, 109556. [Google Scholar] [CrossRef]
- Chilaka, C.A.; De Kock, S.; Phoku, J.Z.; Mwanza, M.; Egbuta, M.A.; Dutton, M.F. Fungal and mycotoxin contamination of South African commercial maize. J. Food Agric. Environ. 2012, 10, 296–303. [Google Scholar]
- Aboagye-Nuamah, F.; Kwoseh, C.K.; Maier, D.E. Toxigenic mycoflora, aflatoxin and fumonisin contamination of poultry feeds in Ghana. Toxicon 2021, 198, 164–170. [Google Scholar] [CrossRef] [PubMed]
- Pasqualotto, A.C. Differences in pathogenicity and clinical syndromes due to Aspergillus fumigatus and Aspergillus flavus. Med. Mycol. J. 2009, 47, S261–S270. [Google Scholar] [CrossRef] [PubMed]
- Reddy, K.; Reddy, C.; Muralidharan, K. Detection of Aspergillus spp. and aflatoxin B1 in rice in India. Food Microbiol. 2009, 26, 27–31. [Google Scholar] [CrossRef] [PubMed]
- Panrapee, I.; Phakpoom, K.; Thanapoom, M.; Nampeung, A.; Warapa, M. Exposure to aflatoxin B1 in Thailand by consumption of brown and color rice. Mycotoxin Res. 2016, 32, 19–25. [Google Scholar] [CrossRef]
- Fraeyman, S.; Croubels, S.; Devreese, M.; Antonissen, G. Emerging Fusarium and Alternaria mycotoxins: Occurrence, toxicity and toxicokinetics. Toxins 2017, 9, 228. [Google Scholar] [CrossRef]
| Molecular Marker | Primer | Direction | Reference | PCR Amplification | ||
|---|---|---|---|---|---|---|
| Denature | Repeat Step | Extension | ||||
| Internal transcribed spacer (ITS) | ITS1 | Forward | [32,33] | 94 °C (2 min) | 35 cycles, 94 °C (60 s), 55 °C (60 s), 72 °C (2 min) | 72 °C (10 min) |
| ITS5 | ||||||
| ITS4 | Reverse | |||||
| β-Tubulin (BenA) | Bt2a | Forward | [33] | 94 °C (3 min) | 35 cycles, 94 °C (60 s), 57 °C (60 s), 72 °C (2 min) | 72 °C (10 min) |
| Bt2b | Reverse | |||||
| Calmodulin (CaM) | cmd5 | Forward | [35,36] | 94 °C (10 min) | 30 cycles, 94 °C (50 s), 55 °C (55 s), 72 °C (1 min) | 72 °C (7 min) |
| cmd6 | Reverse | |||||
| RNA polymerase II second largest subunit (RPB2) | 5F2 | Forward | [34] | 94 °C (2 min) | 35 cycles, 94 °C (1 min), 55 °C (1 min), 72 °C (2 min) | 72 °C (10 min) |
| 7cR | Reverse | |||||
| Variety of Rice Sample | Number of Rice Samples | Total Tested Rice Seeds | Number of Contaminated Rice Seeds | Fungal Contamination (%) |
|---|---|---|---|---|
| Rice berry | 13 | 5200 | 1291 | 24.83 |
| Red jasmine rice | 8 | 3200 | 1475 | 46.09 |
| Brown rice | 7 | 2800 | 405 | 14.18 |
| Germinated brown rice | 6 | 2400 | 1037 | 42.96 |
| White rice | 6 | 2400 | 77 | 3.21 |
| Total | 40 | 16,000 | 4285 | 26.78 |
| Variety of Rice Sample | Number of Rice Samples | Number of Fungal Isolates | Pathogen Incidence | |
|---|---|---|---|---|
| Aspergillus spp. | Penicillium spp./ Talaromyces spp. | |||
| Rice berry | 13 | 34 | 32 | 2 |
| Red jasmine rice | 8 | 20 | 14 | 6 |
| Brown rice | 7 | 12 | 9 | 3 |
| Germinated brown rice | 6 | 14 | 12 | 2 |
| White rice | 6 | 10 | 72 | 18 |
| Total | 40 | 90 | 72 | 18 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Laut, S.; Poapolathep, S.; Piasai, O.; Sommai, S.; Boonyuen, N.; Giorgi, M.; Zhang, Z.; Fink-Gremmels, J.; Poapolathep, A. Storage Fungi and Mycotoxins Associated with Rice Samples Commercialized in Thailand. Foods 2023, 12, 487. https://doi.org/10.3390/foods12030487
Laut S, Poapolathep S, Piasai O, Sommai S, Boonyuen N, Giorgi M, Zhang Z, Fink-Gremmels J, Poapolathep A. Storage Fungi and Mycotoxins Associated with Rice Samples Commercialized in Thailand. Foods. 2023; 12(3):487. https://doi.org/10.3390/foods12030487
Chicago/Turabian StyleLaut, Seavchou, Saranya Poapolathep, Onuma Piasai, Sujinda Sommai, Nattawut Boonyuen, Mario Giorgi, Zhaowei Zhang, Johanna Fink-Gremmels, and Amnart Poapolathep. 2023. "Storage Fungi and Mycotoxins Associated with Rice Samples Commercialized in Thailand" Foods 12, no. 3: 487. https://doi.org/10.3390/foods12030487
APA StyleLaut, S., Poapolathep, S., Piasai, O., Sommai, S., Boonyuen, N., Giorgi, M., Zhang, Z., Fink-Gremmels, J., & Poapolathep, A. (2023). Storage Fungi and Mycotoxins Associated with Rice Samples Commercialized in Thailand. Foods, 12(3), 487. https://doi.org/10.3390/foods12030487









