A Proteomics Data Mining Strategy for the Identification of Quinoa Grain Proteins with Potential Immunonutritional Bioactivities
Abstract
1. Introduction
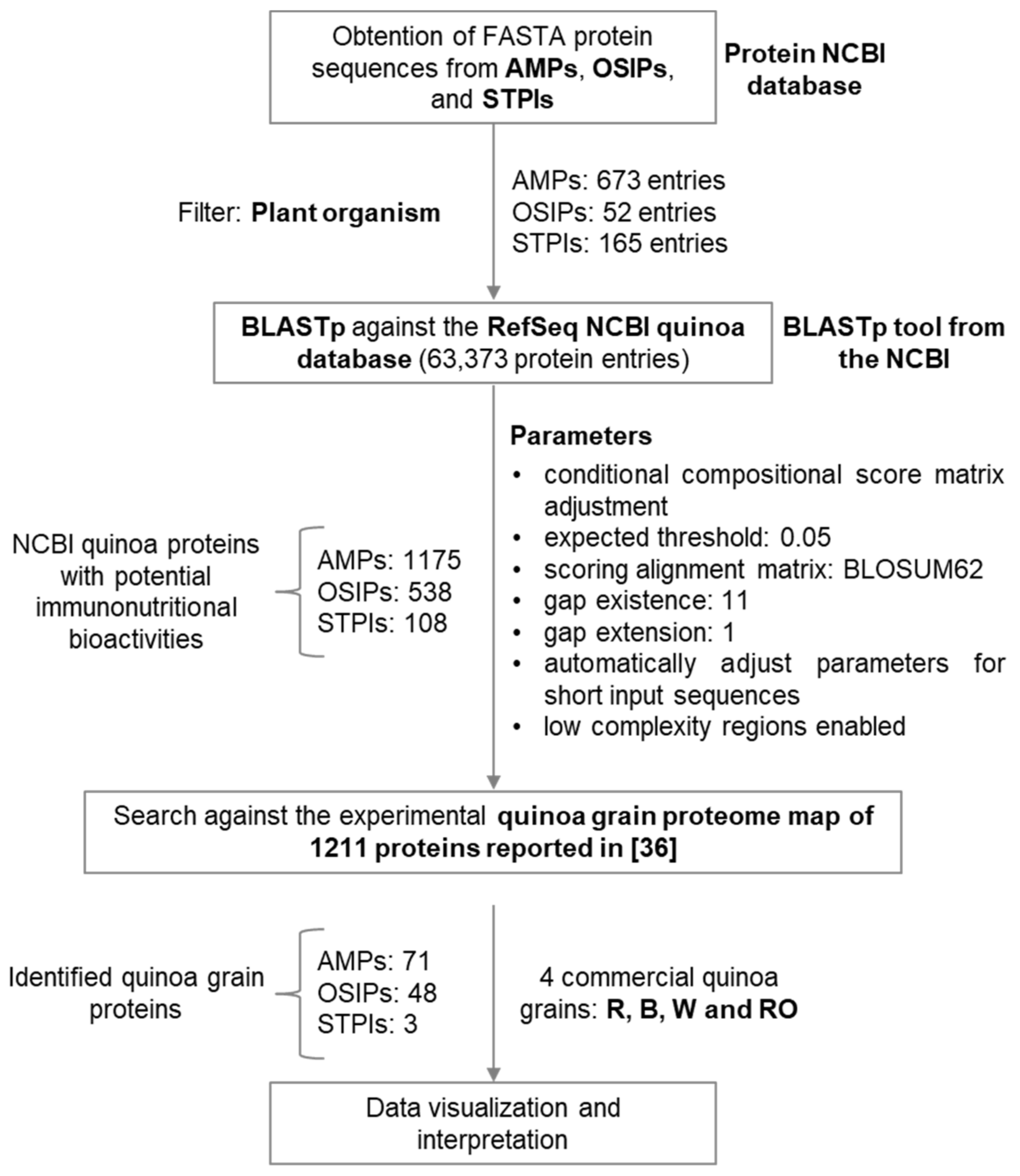
2. Materials and Methods
2.1. Obtaining FASTA Protein Sequences from Plant-Derived AMPs, OSIPs, and STPIs
2.2. Determination of Protein Sequence Similarities
2.3. Data Interpretation
3. Results
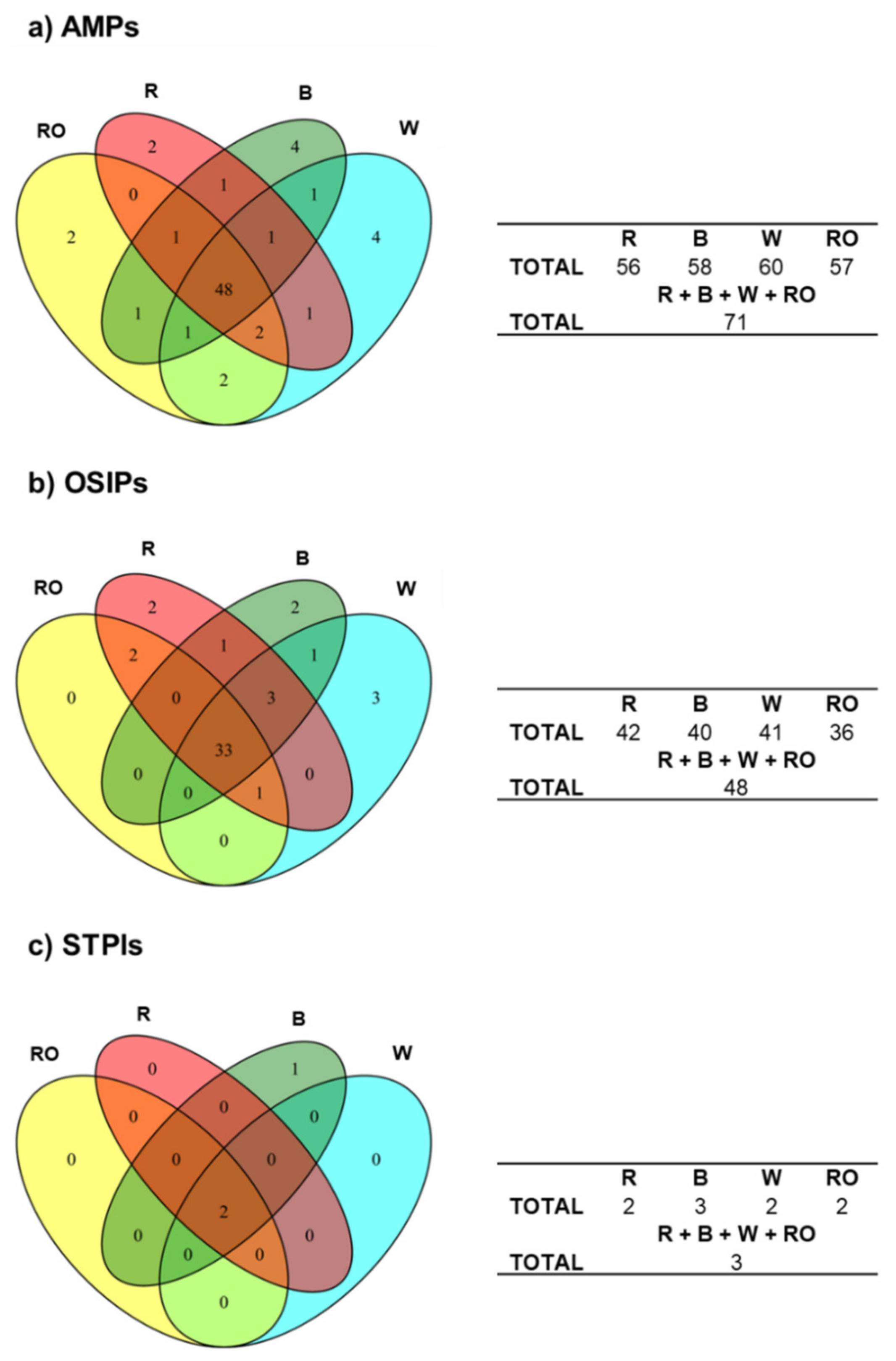
3.1. Identification of Quinoa Grain Proteins with Immunonutritional Bioactivities
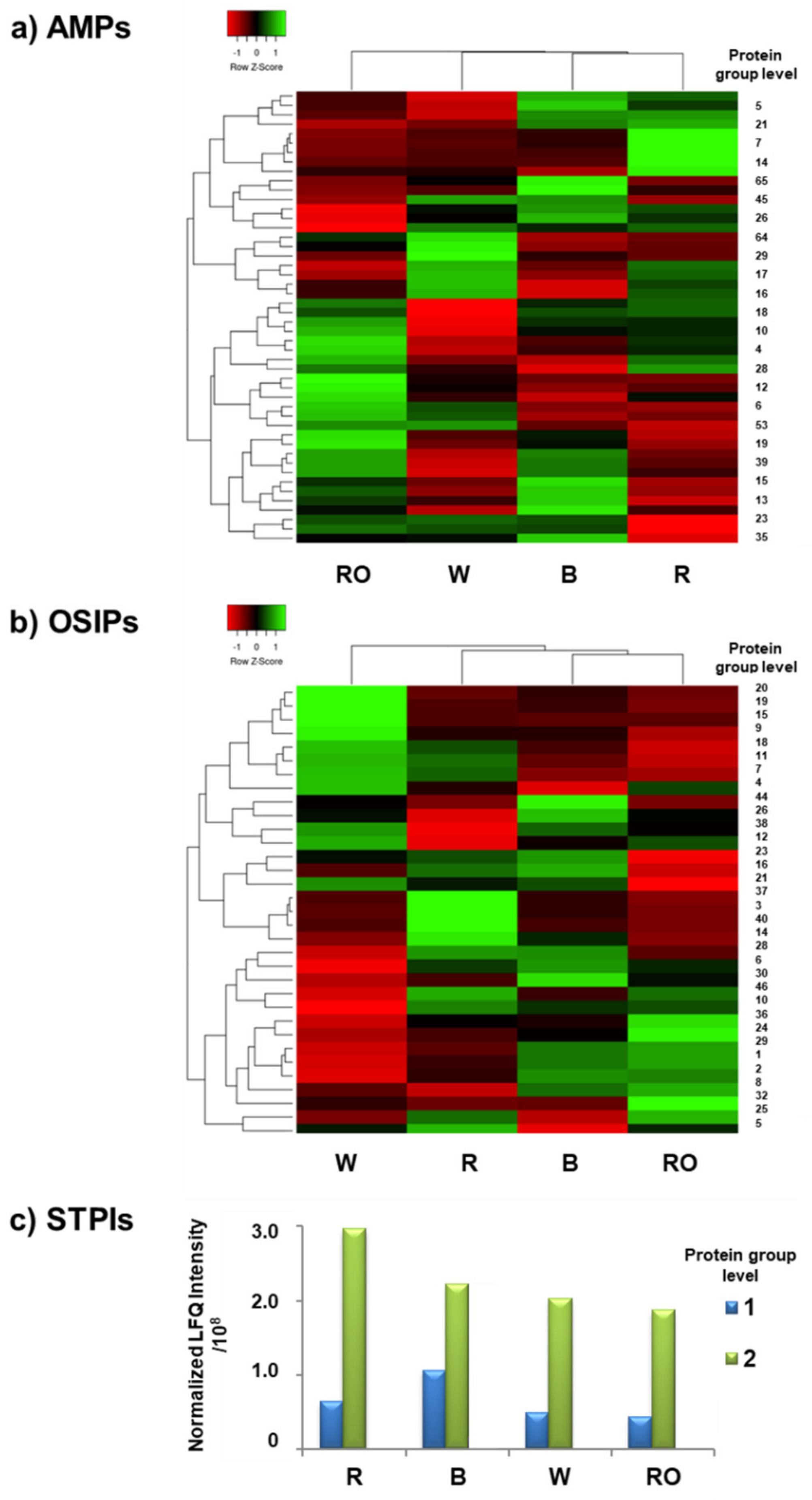
3.2. Determination of the Most Relevant Quinoa Grain Proteins with Immunonutritional Bioactivities
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bajpai, G.; Nahrendorf, M. Infectious and Lifestyle Modifiers of Immunity and Host Resilience. Immunity 2021, 54, 1110–1122. [Google Scholar] [CrossRef] [PubMed]
- Mazzucca, C.B.; Raineri, D.; Cappellano, G.; Chiocchetti, A. How to Tackle the Relationship between Autoimmune Diseases and Diet: Well Begun Is Half-Done. Nutrients 2021, 13, 3956. [Google Scholar] [CrossRef] [PubMed]
- Shurin, M.R. Cancer as an Immune-Mediated Disease. ImmunoTargets Ther. 2012, 1, 1–6. [Google Scholar] [CrossRef]
- Calder, P.C. Immunonutrition. BMJ 2003, 327, 117–118. [Google Scholar] [CrossRef]
- Santiago-López, L.; Hernández-Mendoza, A.; Vallejo-Cordoba, B.; Mata-Haro, V.; González-Córdova, A.F. Food-Derived Immunomodulatory Peptides. J. Sci. Food Agric. 2016, 96, 3631–3641. [Google Scholar] [CrossRef]
- Singh, B.P.; Bangar, S.P.; Albaloosh, M.; Ajayi, F.F.; Mudgil, P.; Maqsood, S. Plant-Derived Proteins as a Sustainable Source of Bioactive Peptides: Recent Research Updates on Emerging Production Methods, Bioactivities, and Potential Application. Crit. Rev. Food Sci. Nutr. 2022, 1–22. [Google Scholar] [CrossRef]
- Fan, H.; Liu, H.; Zhang, Y.; Zhang, S.; Liu, T.; Wang, D. Review on Plant-Derived Bioactive Peptides: Biological Activities, Mechanism of Action and Utilizations in Food Development. J. Futur. Foods 2022, 2, 143–159. [Google Scholar] [CrossRef]
- Amigo, L.; Hernández-Ledesma, B. Current Evidence on the Bioavailability of Food Bioactive Peptides. Molecules 2020, 25, 4479. [Google Scholar] [CrossRef]
- Guzmán-Rodríguez, J.J.; Ochoa-Zarzosa, A.; López-Gómez, R.; López-Meza, J.E. Plant Antimicrobial Peptides as Potential Anticancer Agents. Biomed Res. Int. 2015, 2015, 735087. [Google Scholar] [CrossRef]
- De Coninck, B.; Carron, D.; Tavormina, P.; Willem, L.; Craik, D.J.; Vos, C.; Thevissen, K.; Mathys, J.; Cammue, B.P.A. Mining the Genome of Arabidopsis Thaliana as a Basis for the Identification of Novel Bioactive Peptides Involved in Oxidative Stress Tolerance. J. Exp. Bot. 2013, 64, 5297–5307. [Google Scholar] [CrossRef]
- Laparra, J.M.; Haros, C.M. Plant Seed Protease Inhibitors Differentially Affect Innate Immunity in a Tumor Microenvironment to Control Hepatocarcinoma. Food Funct. 2019, 10, 4210–4219. [Google Scholar] [CrossRef] [PubMed]
- Barbosa Pelegrini, P.; Del Sarto, R.P.; Silva, O.N.; Franco, O.L.; Grossi-De-Sa, M.F. Antibacterial Peptides from Plants: What They Are and How They Probably Work. Biochem. Res. Int. 2011, 2011, 250349. [Google Scholar] [CrossRef] [PubMed]
- Stotz, H.U.; Waller, F.; Wang, K. Innate Immunity in Plants: The Role of Antimicrobial Peptides. In Antimicrobial Peptides and Innate Imunity; Hiemstra, P.S., Zaat, S.A.J., Eds.; Springer Science&Business Media: Berlin/Heidelberg, Germany, 2013; pp. 29–51. [Google Scholar]
- Luo, Y.; Song, Y. Mechanism of Antimicrobial Peptides: Antimicrobial, Anti-Inflammatory and Antibiofilm Activities. Int. J. Mol. Sci. 2021, 22, 11401. [Google Scholar] [CrossRef]
- Breen, S.; Solomon, P.S.; Bedon, F.; Vincent, D. Surveying the Potential of Secreted Antimicrobial Peptides to Enhance Plant Disease Resistance. Front. Plant Sci. 2015, 6, 900. [Google Scholar] [CrossRef] [PubMed]
- Brieger, K.; Schiavone, S.; Miller, F.J.; Krause, K.H. Reactive Oxygen Species: From Health to Disease. Swiss Med. Wkly. 2012, 142, w13659. [Google Scholar] [CrossRef]
- Chakrabarti, S.; Jahandideh, F.; Wu, J. Food-Derived Bioactive Peptides on Inflammation and Oxidative Stress. BioMed Res. Int. 2014, 2014, 608979. [Google Scholar] [CrossRef]
- Hazarika, R.R.; De Coninck, B.; Yamamoto, L.R.; Martin, L.R.; Cammue, B.P.A.; Van Noort, V. ARA-PEPs: A Repository of Putative SORF-Encoded Peptides in Arabidopsis Thaliana. BMC Bioinformatics 2017, 18, 37. [Google Scholar] [CrossRef]
- Srikanth, S.; Chen, Z. Plant Protease Inhibitors in Therapeutics-Focus on Cancer Therapy. Front. Pharmacol. 2016, 7, 470. [Google Scholar] [CrossRef]
- Laparra, J.; Fotschki, B.; Haros, C. Immunonutritional Consequences of Different Serine-Type Protease Inhibitors in a C57BL/6 Hepatocarcinoma Model. Oncotarget 2019, 10, 760–772. [Google Scholar] [CrossRef]
- Pereira, E.; Encina-Zelada, C.; Barros, L.; Gonzales-Barron, U.; Cadavez, V.; Ferreira, I.C.F.R. Chemical and Nutritional Characterization of Chenopodium Quinoa Willd (Quinoa) Grains: A Good Alternative to Nutritious Food. Food Chem. 2019, 280, 110–114. [Google Scholar] [CrossRef]
- Vega-Gálvez, A.; Miranda, M.; Vergara, J.; Uribe, E.; Puente, L.; Martínez, E.A. Nutrition Facts and Functional Potential of Quinoa (Chenopodium Quinoa Willd.), an Ancient Andean Grain: A Review. J. Sci. Food Agric. 2010, 90, 2541–2547. [Google Scholar] [CrossRef]
- Ruales, J.; Nair, B. Nutritional Quality of the Protein in (Chenopodium Quinoa, Willd) Quinoa Seeds. Plant Foods Hum. Nutr. 1992, 42, 1–11. [Google Scholar] [CrossRef]
- Navruz-Varli, S.; Sanlier, N. Nutritional and Health Benefits of Quinoa (Chenopodium Quinoa Willd.). J. Cereal Sci. 2016, 69, 371–376. [Google Scholar] [CrossRef]
- You, H.; Wu, T.; Wang, W.; Li, Y.; Liu, X.; Ding, L. Preparation and Identification of Dipeptidyl Peptidase IV Inhibitory Peptides from Quinoa Protein. Food Res. Int. 2022, 156, 111176. [Google Scholar] [CrossRef] [PubMed]
- González-Muñoz, A.; Valle, M.; Aluko, R.E.; Bazinet, L.; Enrione, J. Production of Antihypertensive and Antidiabetic Peptide Fractions from Quinoa (Chenopodium Quinoa Willd.) by Electrodialysis with Ultrafiltration Membranes. Food Sci. Hum. Wellness 2022, 11, 1650–1659. [Google Scholar] [CrossRef]
- Vilcacundo, R.; Miralles, B.; Carrillo, W.; Hernández-Ledesma, B. In Vitro Chemopreventive Properties of Peptides Released from Quinoa (Chenopodium Quinoa Willd.) Protein under Simulated Gastrointestinal Digestion. Food Res. Int. 2018, 105, 403–411. [Google Scholar] [CrossRef]
- Wong, F.C.; Ong, J.H.; Kumar, D.T.; Chai, T.T. In Silico Identification of Multi-Target Anti-SARS-CoV-2 Peptides from Quinoa Seed Proteins. Int. J. Pept. Res. Ther. 2021, 27, 1837–1847. [Google Scholar] [CrossRef] [PubMed]
- Capraro, J.; De Benedetti, S.; Di Dio, M.; Bona, E.; Abate, A.; Corsetto, P.A.; Scarafoni, A. Characterization of Chenopodin Isoforms from Quinoa Seeds and Assessment of Their Potential Anti-Inflammatory Activity in Caco-2 Cells. Biomolecules 2020, 10, 795. [Google Scholar] [CrossRef] [PubMed]
- Srdić, M.; Ovčina, I.; Fotschki, B.; Haros, C.M.; Llopis, J.M.L. C. Quinoa and S. Hispanica L. Seeds Provide Immunonutritional Agonists to Selectively Polarize Macrophages. Cells 2020, 9, 593. [Google Scholar] [CrossRef]
- Laparra, J.M.; Aguilar-Aguilar, E.; Haros, C.M. Chenopodium Quinoa to Modulate Innate Myeloid Cells in the Induction of Obesity. Biol. Life Sci. Forum 2022, 8, 13. [Google Scholar] [CrossRef]
- Selma-Gracia, R.; Megušar, P.; Haros, C.M.; Llopis, J.M.L. Immunonutritional Bioactives from Chenopodium Quinoa and Salvia Hispanica l. Flour Positively Modulate Insulin Resistance and Preserve Alterations in Peripheral Myeloid Population. Nutrients 2021, 13, 1537. [Google Scholar] [CrossRef] [PubMed]
- Di Silvestre, D.; Passignani, G.; Rossi, R.; Ciuffo, M.; Turina, M.; Vigani, G.; Mauri, P.L. Presence of a Mitovirus Is Associated with Alteration of the Mitochondrial Proteome, as Revealed by Protein-Protein Interaction (PPI) and Co-Expression Network Models in Chenopodium Quinoa Plants. Biology 2022, 11, 95. [Google Scholar] [CrossRef] [PubMed]
- Derbali, W.; Manaa, A.; Spengler, B.; Goussi, R.; Abideen, Z.; Ghezellou, P.; Abdelly, C.; Forreiter, C.; Koyro, H.W. Comparative Proteomic Approach to Study the Salinity Effect on the Growth of Two Contrasting Quinoa Genotypes. Plant Physiol. Biochem. 2021, 163, 215–229. [Google Scholar] [CrossRef] [PubMed]
- Rasouli, F.; Kiani-Pouya, A.; Shabala, L.; Li, L.; Tahir, A.; Yu, M.; Hedrich, R.; Chen, Z.; Wilson, R.; Zhang, H.; et al. Salinity Effects on Guard Cell Proteome in Chenopodium Quinoa. Int. J. Mol. Sci. 2021, 22, 428. [Google Scholar] [CrossRef]
- Galindo-Luján, R.; Pont, L.; Minic, Z.; Berezovski, M.V.; Sanz-Nebot, V.; Benavente, F. Characterization and Differentiation of Quinoa Seed Proteomes by Label-Free Mass Spectrometry-Based Shotgun Proteomics. Food Chem. 2021, 363, 130250. [Google Scholar] [CrossRef]
- Yu, Y.K.; Wootton, J.C.; Altschul, S.F. The Compositional Adjustment of Amino Acid Substitution Matrices. Proc. Natl. Acad. Sci. USA 2003, 100, 15688–15693. [Google Scholar] [CrossRef]
- Kanduc, D. Homology, Similarity, and Identity in Peptide Epitope Immunodefinition. J. Pept. Sci. 2012, 18, 487–494. [Google Scholar] [CrossRef]
- Chen, H. Package “VennDiagram”, Generate High-Resolution Venn and Euler Plot; CRAN, 2022. [Google Scholar]
- Haarman, B.C.M.; der Lek, R.F.R.-V.; Nolen, W.A.; Mendes, R.; Drexhage, H.A.; Burger, H. Feature-Expression Heat Maps-A New Visual Method to Explore Complex Associations between Two Variable Sets. J. Biomed. Inform. 2015, 53, 156–161. [Google Scholar] [CrossRef]
- Jack, H.W.; Tzi, B.N. Sesquin, a Potent Defensin-like Antimicrobial Peptide from Ground Beans with Inhibitory Activities toward Tumor Cells and HIV-1 Reverse Transcriptase. Peptides 2005, 26, 1120–1126. [Google Scholar] [CrossRef]
- Wong, J.H.; Ng, T.B. Gymnin, a Potent Defensin-like Antifungal Peptide from the Yunnan Bean (Gymnocladus Chinensis Baill). Peptides 2003, 24, 963–968. [Google Scholar] [CrossRef]
- Sweetlove, L.J.; Heazlewood, J.L.; Herald, V.; Holtzapffel, R.; Day, D.A.; Leaver, C.J.; Millar, A.H. The Impact of Oxidative Stress on Arabidopsis Mitochondria. Plant J. 2002, 32, 891–904. [Google Scholar] [CrossRef] [PubMed]
- Broin, M.; Cuiné, S.; Eymery, F.; Rey, P. The Plastidic 2-Cysteine Peroxiredoxin Is a Target for a Thioredoxin Involved in the Protection of the Photosynthetic Apparatus against Oxidative Damage. Plant Cell 2002, 14, 1417–1432. [Google Scholar] [CrossRef] [PubMed]
- Guo, X.L.; Cao, Y.R.; Cao, Z.Y.; Zhao, Y.X.; Zhang, H. Molecular Cloning and Characterization of a Stress-Induced Peroxiredoxin Q Gene in Halophyte Suaeda Salsa. Plant Sci. 2004, 167, 969–975. [Google Scholar] [CrossRef]
- Moeder, W.; del Pozo, O.; Navarre, D.A.; Martin, G.B.; Klessig, D.F. Aconitase Plays a Role in Regulating Resistance to Oxidative Stress and Cell Death in Arabidopsis and Nicotiana Benthamiana. Plant Mol. Biol. 2007, 63, 273–287. [Google Scholar] [CrossRef]
- Sugimoto, M.; Sakamoto, W. Putative Phospholipid Hydroperoxide Glutathione Peroxidase Gene from Arabidopsis Thaliana Induced by Oxidative Stress. Genes Genet. Syst. 1997, 72, 311–316. [Google Scholar] [CrossRef]
- Mousavi, A.; Hotta, Y. Glycine-rich Proteins: A Class of Novel Proteins. Appl. Biochem. Biotechnol. 2005, 120, 169–174. [Google Scholar] [CrossRef]
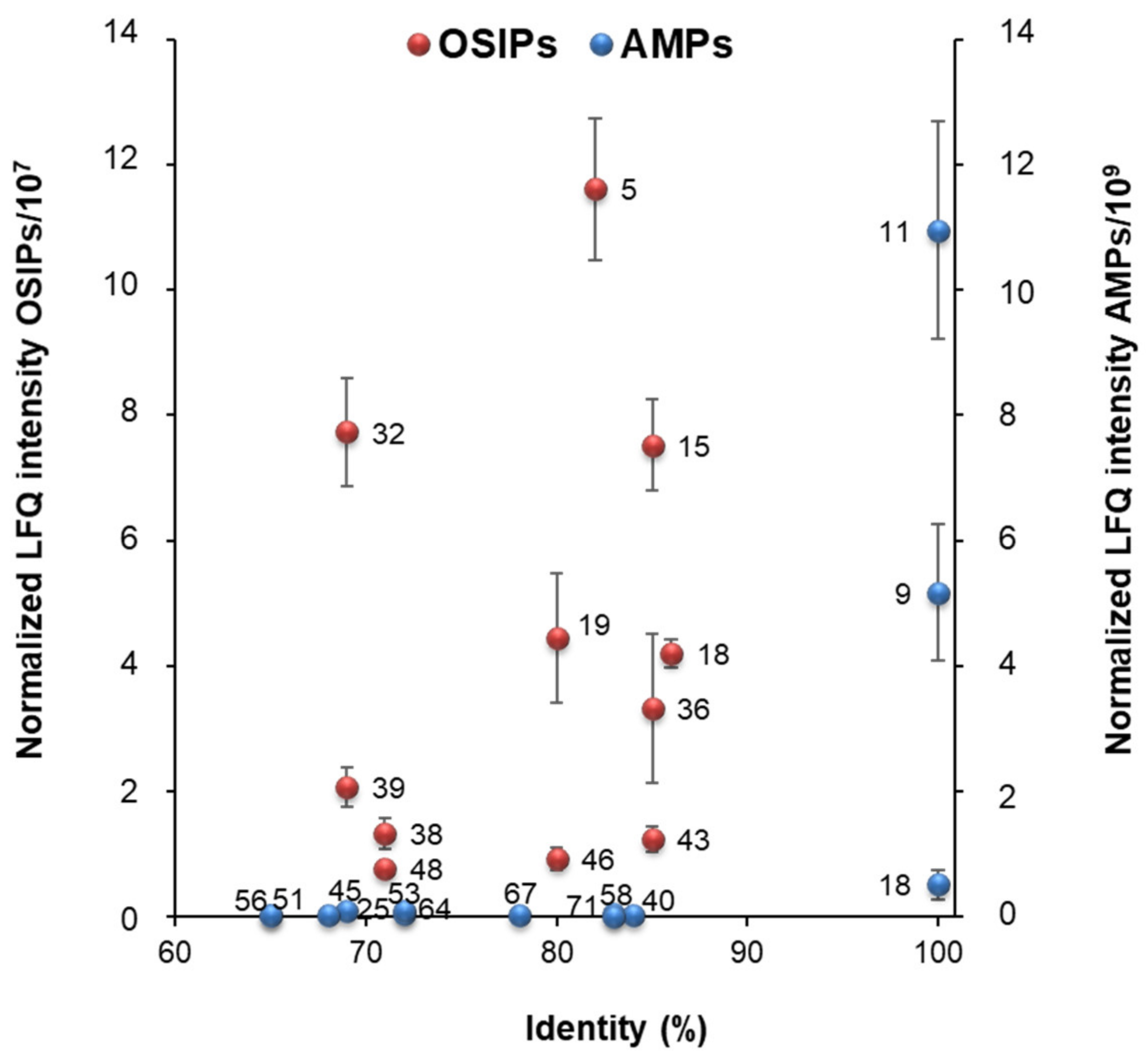
| Chenopodium quinoa | Plant Sequence Similarity | |||||||
|---|---|---|---|---|---|---|---|---|
| 1 Protein Group Level | ID | Protein Name | Mr | Plant Organism | ID | Protein Name | Mr | 2 Identity (%) |
| AMPs | ||||||||
| 9 | XP_021768828.1 | legumin A-like | 53,642 | Vigna unguiculata subsp. sesquipedalis | P84868.1 | defensin-like protein | 1157 | 100 |
| Gymnocladus chinensis | P84200.1 | gymnin | 1171 | |||||
| 11 | XP_021770181.1 | legumin A-like | 53,576 | Vigna unguiculata subsp. sesquipedalis | P84868.1 | defensin-like protein | 1157 | 100 |
| Gymnocladus chinensis | P84200.1 | gymnin | 1171 | |||||
| 18 | XP_021717953.1 | antimicrobial peptide 2-like | 12,973 | Chenopodium quinoa | XP_021717953.1 | antimicrobial peptide 2-like | 12,973 | 100 |
| 25 | XP_021746256.1 | protein disulfide isomerase-like 1–4 | 64,057 | Oldenlandia affinis | ABS11216.1 | protein disulfide isomerase precursor, partial | 58,968 | 69 |
| 40 | XP_021768967.1 | caffeine synthase 1-like | 43,964 | Vigna unguiculata subsp. sesquipedalis | P84868.1 | defensin-like protein | 1157 | 83 |
| 45 | XP_021728344.1 | protein disulfide isomerase-like 1–4 | 64,758 | Oldenlandia affinis | ABS11216.1 | protein disulfide isomerase precursor, partial | 58,968 | 68 |
| 51 | XP_021760599.1; XP_021725721.1 | defensin-like protein 2; defensin Ec-AMP-D2-like | 8588 | Solanum lycopersicum var. Cerasiforme | ADK36631.1 | defensin-like protein, partial | 5351 | 69 |
| 53 | XP_021714401.1; XP_021735018.1 | peamaclein-like | 10,270 | Solanum tuberosum | AAD01518.1 | snakin-1, partial | 5491 | 72 |
| 56 | XP_021749254.1 | peamaclein-like | 10,287 | Solanum tuberosum | AAD01518.1 | snakin-1, partial | 5491 | 65 |
| 58 | XP_021771951.1; XP_021771949.1; XP_021728837.1 | serine/threonine-protein phosphatase PP2A-2 catalytic subunit isoform X2; isoform X1; subunit-like | 30,928 | Peltophorum dubium | AWY94151.1 | protein phosphatase 2A, partial | 10,719 | 84 |
| 64 | XP_021735074.1; XP_021714402.1 | peamaclein-like | 10,322 | Solanum tuberosum | AAD01518.1 | snakin-1, partial | 5491 | 72 |
| 67 | XP_021714548.1; XP_021718041.1 | gamma carbonic anhydrase 1, mitochondrial-like | 28,557 | Viola odorata | P58434.1 | cycloviolacin-O2 | 3165 | 78 |
| Viola odorata | 2KNM_A | chain A, cycloviolacin-O2 | 3165 | |||||
| Viola odorata | 2KCG_A | chain A, cycloviolacin-O2 | 3165 | |||||
| 71 | XP_021773131.1; XP_021742272.1 | probable prefoldin subunit 4 | 14,794 | Vigna unguiculata subsp. sesquipedalis | P84868.1 | defensin-like protein | 1157 | 83 |
| Gymnocladus chinensis | P84200.1 | gymnin | 1171 | |||||
| OSIPs | ||||||||
| 5 | XP_021774578.1 | putative aconitate hydratase, cytoplasmic | 108,650 | Arabidopsis thaliana | Q9SIB9.2 | aconitate hydratase 3 | 108,200 | 82 |
| 15 | XP_021764772.1 | fructose-bisphosphate aldolase 3, chloroplastic | 43,229 | Arabidopsis thaliana | Q9ZU52.1 | fructose-bisphosphate aldolase 3 | 42,327 | 85 |
| 18 | XP_021774583.1 | putative aconitate hydratase, cytoplasmic | 97,932 | Arabidopsis thaliana | Q9SIB9.2 | aconitate hydratase 3 | 108,200 | 86 |
| 19 | XP_021765715.1; XP_021766570.1; XP_021765707.1 | 2-Cys peroxiredoxin BAS1, chloroplastic-like isoform X2; chloroplastic; chloroplastic-like isoform X1 | 29,900 | Arabidopsis thaliana | Q96291.2 | 2-Cys peroxiredoxin BAS1 | 29,092 | 80 |
| 32 | XP_021754079.1 | probable phospholipid hydroperoxide glutathione peroxidase | 26,199 | Arabidopsis thaliana | O48646.2 | probable phospholipid hydroperoxide glutathione peroxidase 6 | 25,584 | 69 |
| 36 | XP_021756717.1 | peroxiredoxin Q, chloroplastic-like | 23,654 | Suaeda salsa | Q6UBI3.1 | peroxiredoxin Q | 23,589 | 85 |
| 38 | XP_021746311.1; XP_021746313.1 | probable phospholipid hydroperoxide glutathione peroxidase isoform X1; isoform X2 | 26,468 | Arabidopsis thaliana | O22850.1 | probable glutathione peroxidase 3 | 23,258 | 71 |
| 39 | XP_021773039.1 | superoxide dismutase [Cu-Zn] | 15,262 | Arabidopsis thaliana | O78310.2 | superoxide dismutase [Cu-Zn] 2 | 22,244 | 69 |
| 43 | XP_021728703.1 | fructose-bisphosphate aldolase 3, chloroplastic-like | 43,043 | Arabidopsis thaliana | Q9ZU52.1 | fructose-bisphosphate aldolase 3 | 42,327 | 85 |
| 46 | XP_021776210.1; XP_021717201.1 | peptide methionine sulfoxide reductase B5-like | 24,740 | Arabidopsis thaliana | Q9M0Z6.2 | peptide methionine sulfoxide reductase B3 | 18,847 | 80 |
| 48 | XP_021770821.1; XP_021770820.1 | acylpyruvase FAHD1, mitochondrial-like isoform X2; isoform X1 | 22,257 | Arabidopsis thaliana | Q93ZE5.1 | probable acylpyruvase FAHD1 | 24,283 | 71 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Galindo-Luján, R.; Pont, L.; Sanz-Nebot, V.; Benavente, F. A Proteomics Data Mining Strategy for the Identification of Quinoa Grain Proteins with Potential Immunonutritional Bioactivities. Foods 2023, 12, 390. https://doi.org/10.3390/foods12020390
Galindo-Luján R, Pont L, Sanz-Nebot V, Benavente F. A Proteomics Data Mining Strategy for the Identification of Quinoa Grain Proteins with Potential Immunonutritional Bioactivities. Foods. 2023; 12(2):390. https://doi.org/10.3390/foods12020390
Chicago/Turabian StyleGalindo-Luján, Rocío, Laura Pont, Victoria Sanz-Nebot, and Fernando Benavente. 2023. "A Proteomics Data Mining Strategy for the Identification of Quinoa Grain Proteins with Potential Immunonutritional Bioactivities" Foods 12, no. 2: 390. https://doi.org/10.3390/foods12020390
APA StyleGalindo-Luján, R., Pont, L., Sanz-Nebot, V., & Benavente, F. (2023). A Proteomics Data Mining Strategy for the Identification of Quinoa Grain Proteins with Potential Immunonutritional Bioactivities. Foods, 12(2), 390. https://doi.org/10.3390/foods12020390







