Multi-Target Detection of Nuts and Peanuts as Hidden Allergens in Bakery Products through Bottom-Up Proteomics and High-Resolution Mass Spectrometry
Abstract
1. Introduction
2. Materials and Methods
2.1. Chemicals, Materials, and Allergenic Ingredients
2.2. Isotopically Labelled Peptides
2.3. Incurred and Allergen-Free Cookies Production
2.4. Sample Preparation
2.4.1. Extraction and Tryptic Digestion of Proteins from Raw and Roasted Allergenic Ingredients
2.4.2. Extraction and Tryptic Digestion of Proteins from Incurred and Allergen-Free Cookies
2.5. Liquid Chromatography—Mass Spectrometry: Instrumentation and Conditions
2.6. Identification of Allergenic Proteins and Peptides Using Bioinformatics
2.7. Evaluation of the Performance of the Developed Method
2.7.1. Sensitivity: Calibration and Calculation of LOD and LOQ Values
2.7.2. Evaluation of the LC-MS Analysis Precision
3. Results and Discussion
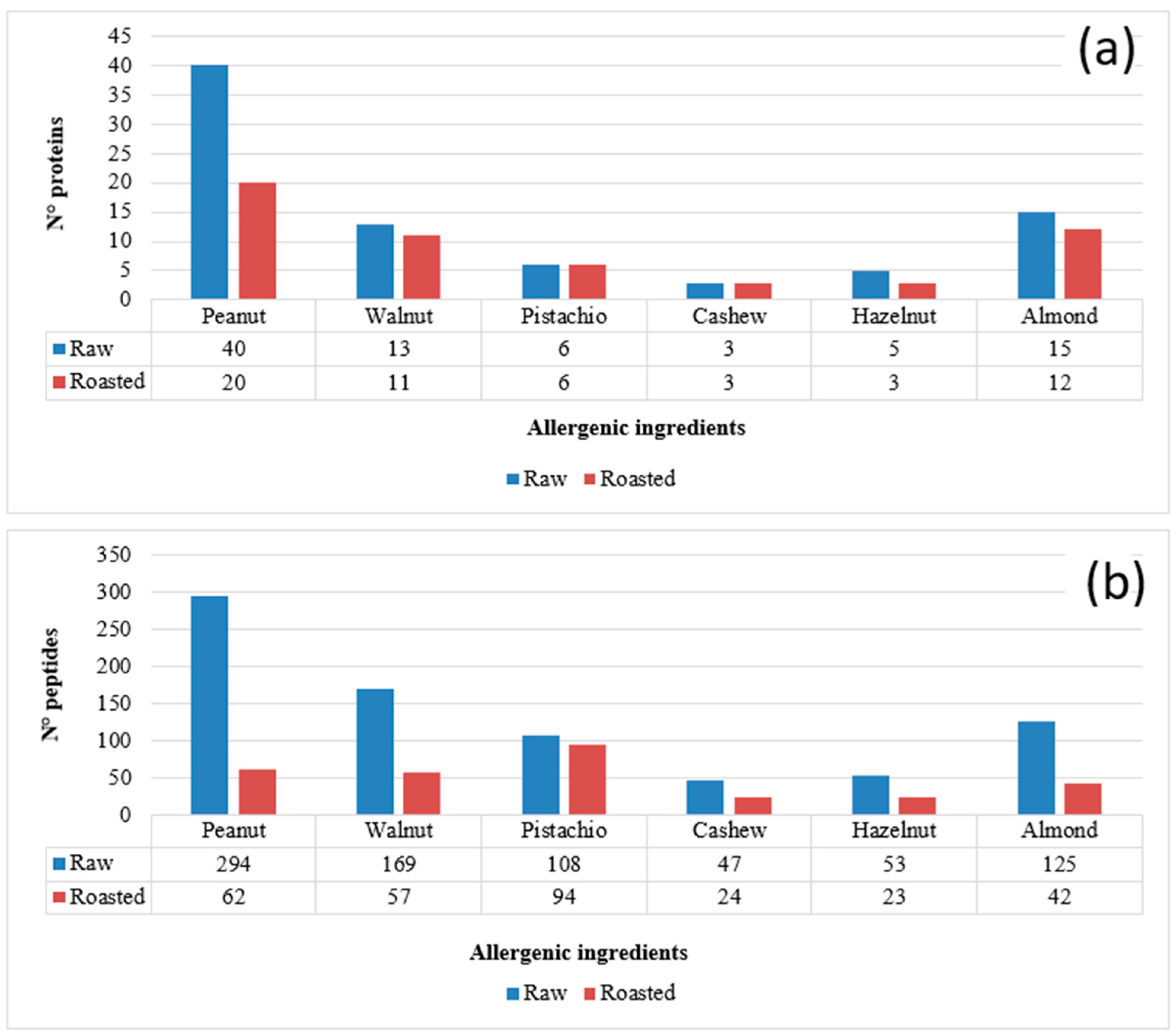
3.1. Protein Identification and Marker Peptide Selection for Raw and Roasted Allergenic Ingredients (Almond, Walnut, Cashew, Hazelnut, Pistachio and Peanut)
3.2. Selection of Marker Peptides for Nuts and Peanuts Quantification in Incurred Cookies
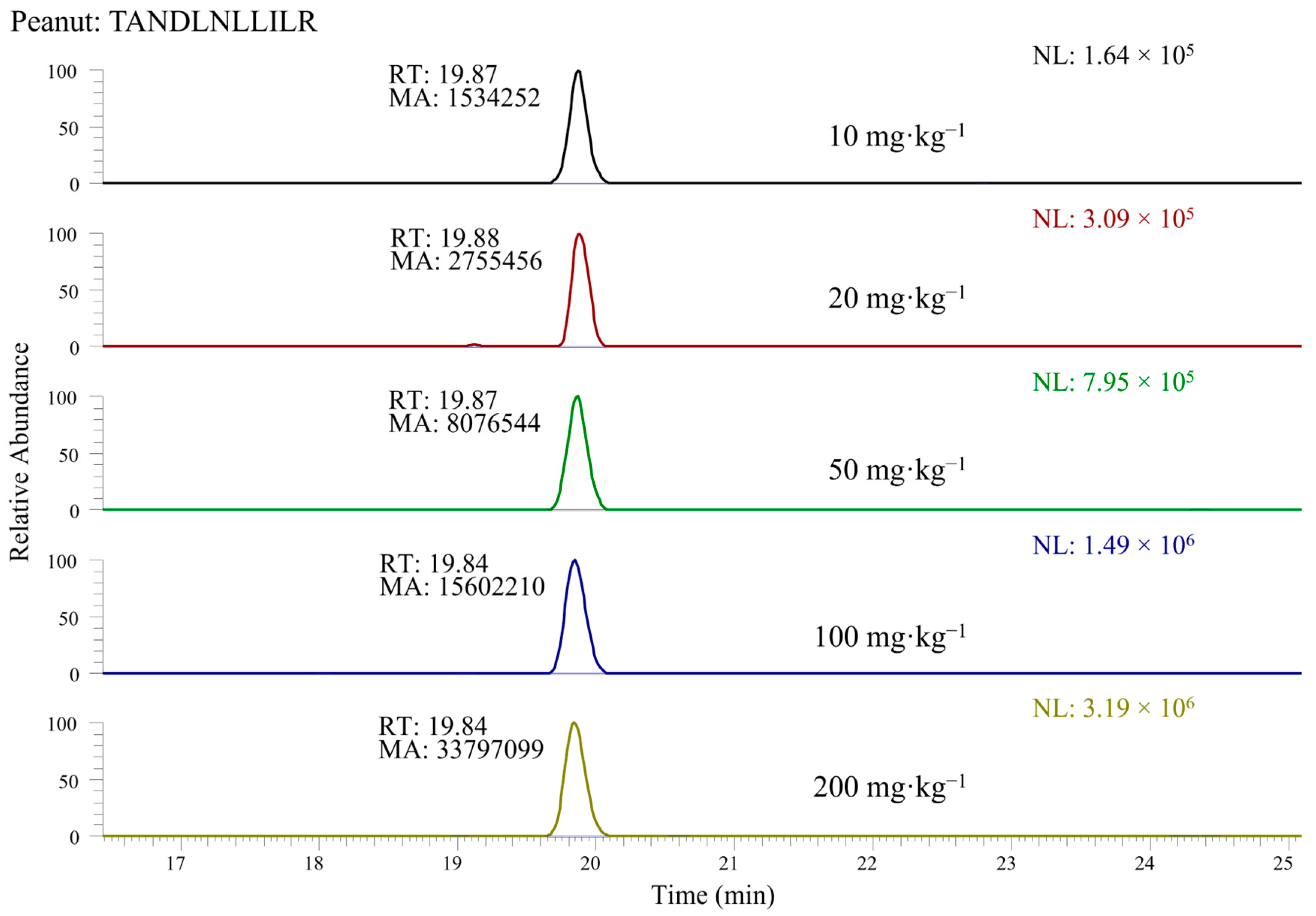
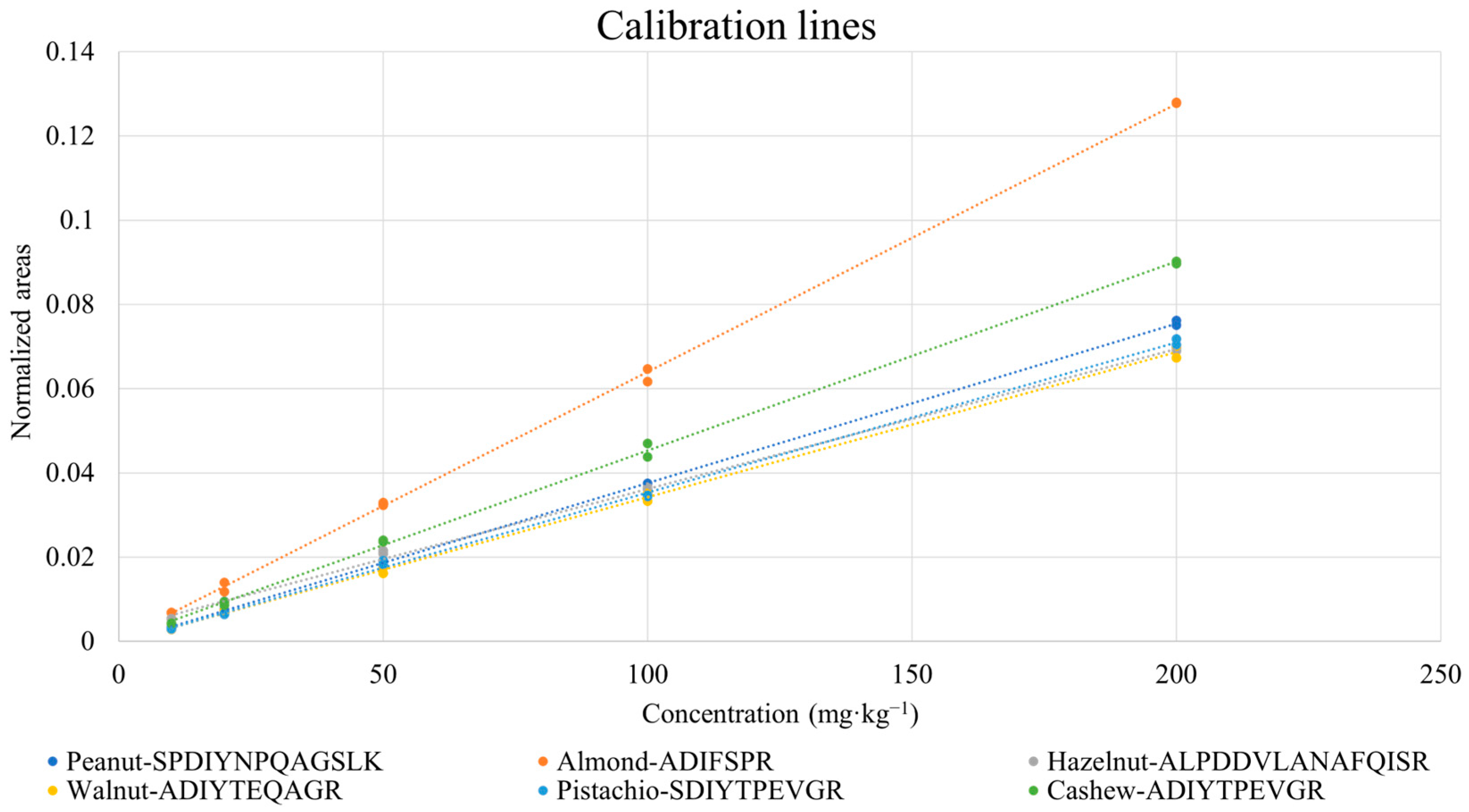
3.3. Quantification of Nuts and Peanuts in the Incurred Cookie Based on Marker Peptides: Evaluation of Linearity and Sensitivity
3.4. Comparison of the Method Performance with the Thresholds for Nuts and Peanuts Reported in the VITAL® Program
3.5. Evaluation of the Precision of the Analytical Method
4. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sicherer, S.H.; Sampson, H.A. Food allergy: A review and update on epidemiology, pathogenesis, diagnosis, prevention, and management. J. Allergy Clin. Immunol. 2018, 141, 41–58. [Google Scholar] [CrossRef]
- Laubach, S.S. The public health impact of parent-reported childhood food allergies in the United States. Pediatrics 2019, 144, S28. [Google Scholar] [CrossRef]
- Verhoeckx, K.; Bøgh, K.L.; Constable, A.; Epstein, M.M.; Sommergruber, K.H.; Holzhauser, T.; Houben, G.; Kuehn, A.; Roggen, E.; O’Mahony, L.; et al. COST Action “ImpARAS”: What have we learnt to improve food allergy risk assessment. A summary of a 4 year networking consortium. Clin. Transl. Allergy 2020, 10, 13. [Google Scholar] [CrossRef]
- Bilaver, L.A.; Chadha, A.S.; Doshi, P.; O’Dwyer, L.; Gupta, R.S. Economic burden of food allergy: A systematic review. Ann. Allergy Asthma Immunol. 2019, 122, 373–380.e1. [Google Scholar] [CrossRef]
- Taylor, S.L.; Baumert, J.L. Cross-Contamination of Foods and Implications for Food Allergic Patients. Curr. Allergy Asthma Rep. 2010, 10, 265–270. [Google Scholar] [CrossRef]
- Skypala, I.J. Food-induced anaphylaxis: Role of hidden allergens and cofactors. Front. Immunol. 2019, 10, 673. [Google Scholar] [CrossRef]
- McWilliam, V.; Koplin, J.; Lodge, C.; Tang, M.; Dharmage, S.; Allen, K. The Prevalence of Tree Nut Allergy: A Systematic Review. Curr. Allergy Asthma Rep. 2015, 15, 54. [Google Scholar] [CrossRef]
- McWilliam, V.L.; Perrett, K.P.; Dang, T.; Peters, R.L. Prevalence and natural history of tree nut allergy. Ann. Allergy Asthma Immunol. 2020, 124, 466–472. [Google Scholar] [CrossRef]
- Cetinkaya, P.G.; Buyuktiryaki, B.; Soyer, O.; Sahiner, U.M.; Sackesen, C.; Sekerel, B.E. Phenotypical characterization of tree nuts and peanut allergies in east Mediterranean children. Allergol. Immunopathol. 2020, 48, 316–322. [Google Scholar] [CrossRef]
- Weinberger, T.; Sicherer, S. Current perspectives on tree nut allergy: A review. J. Asthma Allergy 2018, 11, 41–51. [Google Scholar] [CrossRef]
- Vanga, S.K.; Raghavan, V. Processing effects on tree nut allergens: A review. Crit. Rev. Food Sci. Nutr. 2017, 57, 3794–3806. [Google Scholar] [CrossRef] [PubMed]
- Ros, E.; Singh, A.; O’keefe, J.H. Nuts: Natural pleiotropic nutraceuticals. Nutrients 2021, 13, 3269. [Google Scholar] [CrossRef] [PubMed]
- Geiselhart, S.; Hoffmann-Sommergruber, K.; Bublin, M. Tree nut allergens. Mol. Immunol. 2018, 100, 71–81. [Google Scholar] [CrossRef] [PubMed]
- Luparelli, A.; Losito, I.; De Angelis, E.; Pilolli, R.; Lambertini, F.; Monaci, L. Tree Nuts and Peanuts as a Source of Beneficial Compounds and a Threat for Allergic Consumers: Overview on Methods for Their Detection in Complex Food Products. Foods 2022, 11, 728. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Jin, T. Almond allergens: Update and perspective on identification and characterization. J. Sci. Food Agric. 2020, 100, 4657–4663. [Google Scholar] [CrossRef] [PubMed]
- Costa, J.; Mafra, I.; Carrapatoso, I.; Oliveira, M.B.P.P. Hazelnut allergens: Molecular characterization, detection, and clinical relevance. Crit. Rev. Food Sci. Nutr. 2016, 56, 2579–2605. [Google Scholar] [CrossRef]
- Costa, J.; Carrapatoso, I.; Oliveira, M.B.P.P.; Mafra, I. Walnut allergens: Molecular characterization, detection and clinical relevance. Clin. Exp. Allergy 2014, 44, 319–341. [Google Scholar] [CrossRef]
- Mendes, C.; Costa, J.; Vicente, A.A.; Oliveira, M.B.P.P.; Mafra, I. Cashew Nut Allergy: Clinical Relevance and Allergen Characterisation. Clin. Rev. Allergy Immunol. 2019, 57, 1–22. [Google Scholar] [CrossRef]
- Costa, J.; Silva, I.; Vicente, A.A.; Oliveira, M.B.P.P.; Mafra, I. Pistachio nut allergy: An updated overview. Crit. Rev. Food Sci. Nutr. 2019, 59, 546–562. [Google Scholar] [CrossRef]
- Palladino, C.; Breiteneder, H. Peanut allergens. Mol. Immunol. 2018, 100, 58–70. [Google Scholar] [CrossRef]
- Roux, K.H.; Teuber, S.S.; Sathe, S.K. Tree nut allergens. Int. Arch. Allergy Immunol. 2003, 131, 234–244. [Google Scholar] [CrossRef] [PubMed]
- Radauer, C.; Breiteneder, H. Current perspectives Evolutionary biology of plant food allergens. J. Allergy Clin. Immunol. 2007, 120, 518–525. [Google Scholar] [CrossRef] [PubMed]
- European Union Commission Notice C 428/1 of 13 July 2017 Commission Notice of 13 July 2017 relating to the provision of information on substances or products causing allergies or intolerances as listed in Annex II to Regulation (EU) No 1169/2011 of the European Parliament and of the Council on the provision of. Off. J. Eur. Union 2017, 2017, 11–15.
- Planque, M.; Arnould, T.; Dieu, M.; Delahaut, P.; Renard, P.; Gillard, N. Liquid chromatography coupled to tandem mass spectrometry for detecting ten allergens in complex and incurred foodstuffs. J. Chromatogr. A 2017, 1530, 138–151. [Google Scholar] [CrossRef]
- Allen, K.J.; Remington, B.C.; Baumert, J.L.; Crevel, R.W.R.; Houben, G.F.; Brooke-Taylor, S.; Kruizinga, A.G.; Taylor, S.L. Allergen reference doses for precautionary labeling (VITAL 2.0): Clinical implications. J. Allergy Clin. Immunol. 2014, 133, 156–164. [Google Scholar] [CrossRef]
- DunnGalvin, A.; Chan, C.H.; Crevel, R.; Grimshaw, K.; Poms, R.; Schnadt, S.; Taylor, S.L.; Turner, P.; Allen, K.J.; Austin, M.; et al. Precautionary allergen labelling: Perspectives from key stakeholder groups. Allergy 2015, 70, 1039–1051. [Google Scholar] [CrossRef]
- Taylor, S.L.; Baumert, J.L.; Kruizinga, A.G.; Remington, B.C.; Crevel, R.W.R.; Brooke-Taylor, S.; Allen, K.J.; Houben, G. Establishment of Reference Doses for residues of allergenic foods: Report of the VITAL Expert Panel. Food Chem. Toxicol. 2014, 63, 9–17. [Google Scholar] [CrossRef]
- Cuadrado, C.; Sanchiz, Á.; Linacero, R. Nut Allergenicity: Effect of Food Processing. Allergies 2021, 1, 150–162. [Google Scholar] [CrossRef]
- Maleki, S.J. Food processing: Effects on allergenicity. Curr. Opin. Allergy Clin. Immunol. 2004, 4, 241–245. [Google Scholar] [CrossRef]
- Downs, M.L.; Simpson, A.; Custovic, A.; Semic-Jusufagic, A.; Bartra, J.; Fernandez-Rivas, M.; Taylor, S.L.; Baumert, J.L.; Mills, E.N.C. Insoluble and soluble roasted walnut proteins retain antibody reactivity. Food Chem. 2016, 194, 1013–1021. [Google Scholar] [CrossRef]
- Gou, J.; Liang, R.; Huang, H.; Ma, X. Maillard Reaction Induced Changes in Allergenicity of Food. Foods 2022, 11, 530. [Google Scholar] [CrossRef]
- Barazorda-Ccahuana, H.L.; Theiss-De-Rosso, V.; Valencia, D.E.; Gómez, B. Heat-stable hazelnut profilin: Molecular dynamics simulations and immunoinformatics analysis. Polymers 2020, 12, 1742. [Google Scholar] [CrossRef]
- De Angelis, E.; Di Bona, D.; Pilolli, R.; Loiodice, R.; Luparelli, A.; Giliberti, L.; D’uggento, A.M.; Rossi, M.P.; Macchia, L.; Monaci, L. In Vivo and In Vitro Assessment and Proteomic Analysis of the Effectiveness of Physical Treatments in Reducing Allergenicity of Hazelnut Proteins. Nutrients 2022, 14, 874. [Google Scholar] [CrossRef]
- Cuadrado, C.; Cabanillas, B.; Pedrosa, M.M.; Varela, A.; Guillamón, E.; Muzquiz, M.; Crespo, J.F.; Rodriguez, J.; Burbano, C. Influence of thermal processing on IgE reactivity to lentil and chickpea proteins. Mol. Nutr. Food Res. 2009, 53, 1462–1468. [Google Scholar] [CrossRef]
- Sanchiz, A.; Cuadrado, C.; Dieguez, M.C.; Ballesteros, I.; Rodríguez, J.; Crespo, J.F.; de las Cuevas, N.; Rueda, J.; Linacero, R.; Cabanillas, B.; et al. Thermal processing effects on the IgE-reactivity of cashew and pistachio. Food Chem. 2018, 245, 595–602. [Google Scholar] [CrossRef]
- Su, M.; Venkatachalam, M.; Teuber, S.S.; Roux, K.H.; Sathe, S.K. Impact of γ-irradiation and thermal processing on the antigenicity of almond, cashew nut and walnut proteins. J. Sci. Food Agric. 2004, 84, 1119–1125. [Google Scholar] [CrossRef]
- Zhou, H.; Wu, Z.; Chang, X.; Tang, Y.; Yuan, J.; Li, X.; Yang, A.; Tong, P.; Chen, H. The effect of roasting on peanut allergens’ digestibility, allergenicity, and structure. Food Biosci. 2021, 44, 101454. [Google Scholar] [CrossRef]
- Villa, C.; Costa, J.; Mafra, I. Detection and quantification of milk ingredients as hidden allergens in meat products by a novel specific real-time PCR method. Biomolecules 2019, 9, 804. [Google Scholar] [CrossRef] [PubMed]
- Yu, Z.; Wang, Y.; Li, Z.; Pramod, S.N.; Zhang, L.; Lin, H. Development of ELISA Method for Detecting Crustacean Major Allergen Tropomyosin in Processed Food Samples. Food Anal. Methods 2019, 12, 2719–2729. [Google Scholar] [CrossRef]
- Sena-Torralba, A.; Pallás-Tamarit, Y.; Morais, S.; Maquieira, Á. Recent advances and challenges in food-borne allergen detection. TrAC Trends Anal. Chem. 2020, 132, 116050. [Google Scholar] [CrossRef]
- Sena-Torralba, A.; Gabaldón-Atienza, J.; Cubells-Gómez, A.; Casino, P.; Maquieira, Á.; Morais, S. Lateral Flow Microimmunoassay (LFµIA) for the Reliable Quantification of Allergen Traces in Food Consumables. Biosensors 2022, 12, 980. [Google Scholar] [CrossRef]
- Xu, J.; Ye, Y.; Ji, J.; Sun, J.; Sun, X. Advances on the rapid and multiplex detection methods of food allergens. Crit. Rev. Food Sci. Nutr. 2022, 62, 6887–6907. [Google Scholar] [CrossRef] [PubMed]
- Monaci, L.; De Angelis, E.; Guagnano, R.; Ganci, A.P.; Garaguso, I.; Fiocchi, A.; Pilolli, R. Validation of a MS based proteomics method for milk and egg quantification in cookies at the lowest VITAL levels: An alternative to the use of precautionary labeling. Foods 2020, 9, 1489. [Google Scholar] [CrossRef]
- Henrottin, J.; Pilolli, R.; Huet, A.C.; van Poucke, C.; Nitride, C.; De Loose, M.; Tranquet, O.; Larré, C.; Adel-Patient, K.; Bernard, H.; et al. Optimization of a sample preparation workflow based on UHPLC-MS/MS method for multi-allergen detection in chocolate: An outcome of the ThRAll project. Food Control 2023, 143, 109256. [Google Scholar] [CrossRef]
- Torii, A.; Seki, Y.; Arimoto, C.; Hojo, N.; Iijima, K.; Nakamura, K.; Ito, R.; Yamakawa, H.; Akiyama, H. Development of a simple and reliable LC-MS/MS method to simultaneously detect walnut and almond as specified in food allergen labelling regulations in processed foods. Curr. Res. Food Sci. 2023, 6, 100444. [Google Scholar] [CrossRef]
- Lu, Y.; Ji, H.; Chen, Y.; Li, Z.; Timira, V. A systematic review on the recent advances of wheat allergen detection by mass spectrometry: Future prospects. Crit. Rev. Food Sci. Nutr. 2022, 1–17. [Google Scholar] [CrossRef]
- Monaci, L.; De Angelis, E.; Montemurro, N.; Pilolli, R. Comprehensive overview and recent advances in proteomics MS based methods for food allergens analysis. TrAC Trends Anal. Chem. 2018, 106, 21–36. [Google Scholar] [CrossRef]
- Pilolli, R.; Nitride, C.; Gillard, N.; Huet, A.C.; van Poucke, C.; de Loose, M.; Tranquet, O.; Larré, C.; Adel-Patient, K.; Bernard, H.; et al. Critical review on proteotypic peptide marker tracing for six allergenic ingredients in incurred foods by mass spectrometry. Food Res. Int. 2020, 128, 108747. [Google Scholar] [CrossRef]
- Pilolli, R.; De Angelis, E.; Monaci, L. In house validation of a high resolution mass spectrometry Orbitrap-based method for multiple allergen detection in a processed model food. Anal. Bioanal. Chem. 2018, 410, 5653–5662. [Google Scholar] [CrossRef]
- Pilolli, R.; De Angelis, E.; Monaci, L. Streamlining the analytical workflow for multiplex MS/MS allergen detection in processed foods. Food Chem. 2017, 221, 1747–1753. [Google Scholar] [CrossRef]
- Cabanillas, B.; Novak, N. Effects of daily food processing on allergenicity. Crit. Rev. Food Sci. Nutr. 2019, 59, 31–42. [Google Scholar] [CrossRef]
- Yan, X.; Wang, Y.; Chen, Y.; Xie, J.; Yu, Q. Effect of roasting duration on the solubility, structure, and IgE-binding capacity of cashew nut proteins. Innov. Food Sci. Emerg. Technol. 2021, 68, 102635. [Google Scholar] [CrossRef]
- Verhoeckx, K.C.M.; Vissers, Y.M.; Baumert, J.L.; Faludi, R.; Feys, M.; Flanagan, S.; Herouet-Guicheney, C.; Holzhauser, T.; Shimojo, R.; van der Bolt, N.; et al. Food processing and allergenicity. Food Chem. Toxicol. 2015, 80, 223–240. [Google Scholar] [CrossRef]
- Polenta, G.A.; Weber, D.; Godefroy-Benrejeb, S.; Abbott, M. Effect of Processing on the Detectability of Pecan Proteins Assessed by Immunological and Proteomic Tools. Food Anal. Methods 2012, 5, 216–225. [Google Scholar] [CrossRef]
- Besler, M.; Steinhart, H.; Paschke, A. Stability of food allergens and allergenicity of processed foods. J. Chromatogr. B Biomed. Sci. Appl. 2001, 756, 207–228. [Google Scholar] [CrossRef]
- Pekar, J.; Ret, D.; Untersmayr, E. Stability of allergens. Mol. Immunol. 2018, 100, 14–20. [Google Scholar] [CrossRef]
- Korte, R.; Oberleitner, D.; Brockmeyer, J. Determination of food allergens by LC-MS: Impacts of sample preparation, food matrix, and thermal processing on peptide detectability and quantification. J. Proteom. 2019, 196, 131–140. [Google Scholar] [CrossRef]
- Li, B.; Schowen, R.L.; Topp, E.M.; Borchardt, R.T. Effect of N-1 and N-2 residues on peptide deamidation rate in solution and solid state. AAPS J. 2006, 8, 166–173. [Google Scholar] [CrossRef]
- Bignardi, C.; Mattarozzi, M.; Penna, A.; Sidoli, S.; Elviri, L.; Careri, M.; Mangia, A. A Rapid Size-Exclusion Solid-Phase Extraction Step for Enhanced Sensitivity in Multi-Allergen Determination in Dark Chocolate and Biscuits by Liquid Chromatography-Tandem Mass Spectrometry. Food Anal. Methods 2013, 6, 1144–1152. [Google Scholar] [CrossRef]
- Zhang, J.; Hong, Y.; Cai, Z.; Huang, B.; Wang, J.; Ren, Y. Simultaneous determination of major peanut allergens Ara h1 and Ara h2 in baked foodstuffs based on their signature peptides using ultra-performance liquid chromatography coupled to tandem mass spectrometry. Anal. Methods 2019, 11, 1689–1696. [Google Scholar] [CrossRef]
- Korte, R.; Lepski, S.; Brockmeyer, J. Comprehensive peptide marker identification for the detection of multiple nut allergens using a non-targeted LC-HRMS multi-method. Anal. Bioanal. Chem. 2016, 408, 3059–3069. [Google Scholar] [CrossRef] [PubMed]
- New, L.S.; Schreiber, A.; Stahl-Zeng, J.; Liu, H.F. Simultaneous analysis of multiple allergens in food products by LC-MS/MS. J. AOAC Int. 2018, 101, 132–145. [Google Scholar] [CrossRef] [PubMed]
| Allergenic Ingredient | Protein | Peptide Sequence | Charge | m/z (Theoretical Values) |
|---|---|---|---|---|
| Peanut | Ara h 3—Cupin | SPDIYNPQAGSL(K) | +2 | 699.3612 |
| TANDLNLLIL(R) | +2 | 633.3764 | ||
| Hazelnut | Cor a 9—11S Seed Storage Globulin | ADIYTEQVG(R) | +2 | 581.2925 |
| ALPDDVLANAFQIS(R) | +2 | 820.4373 | ||
| Almond | Pru du 6—Amandin, 11S Globulin | TEENAFINTLAG(R) | +2 | 723.3666 |
| ADIFSP(R) | +2 | 408.2180 |
| Allergenic Ingredient | Peptide Sequence | Protein | Accessions UniProt Database | Exact m/z ratio for the Singly Charged Peptide |
|---|---|---|---|---|
| Peanut | ADFYNPAAGR | Legumin | A0A445AEY9 | 1081.5061 |
| ANLDAFTR | Desiccation-related protein PCC13-62-like | A0A445AWA2 | 907.4632 | |
| EGEQEWGTPGSHVR | Ara h 1 | N1NG13 | 1568.7088 | |
| FFVPPFQQSPR | Arachin 6 | A1DZF0 | 1349.7001 | |
| FNLAGNHEQEFLR | Arachin 6 | A1DZF0 | 1574.771 | |
| GTGNLELVAVR | Ara h 1 | B3IXL2 | 1128.6372 | |
| LNALTPDNR | Legumin | A0A445AEY9 | 1013.5374 | |
| NALFVPHYNTNAHSIIYALR | Arachin 6 | A1DZF0 | 2314.2091 | |
| NNPFYFPSR | Ara h 1 | B3IXL2 | 1141.5425 | |
| QILQNLR | Arachin 6 | A0A444YLX0 | 884.5312 | |
| QIVQNLR | Glycinin | O82580 | 870.5156 | |
| SFNLDEGHALR | Ara h 1 | B3IXL2 | 1258.6175 | |
| SPDIYNPQAGSLK | Glycinin | O82580 | 1389.7009 | |
| SSDNEGVIVK | Ara h 1 | B3IXL2 | 1047.5317 | |
| SSNPDIYNPQAGSLR | Ara h 3 | E5G077 | 1618.782 | |
| TANDLNLLILR | Glycinin | O82580 | 1255.7369 | |
| TPQEILR | Desiccation-related protein PCC13-62-like | A0A445AWA2 | 856.4887 | |
| VGDVFFVPR | Cupin | A0A445AA44 | 1035.5622 | |
| VPGGFFPR | Desiccation-related protein PCC13-62-like | A0A445EM48 | 876.4727 | |
| WGPAEPR | Ara h 1 | B3IXL2 | 812.405 | |
| WGPAGPR | Ara h 1 | N1NG13 | 740.3838 | |
| WLGLSAEYGNLYR | Arachin 6 | A1DZF0 | 1541.7747 | |
| Walnut | ADIYTEEAGR | Jug r 4 | Q2TPW5 | 1124.5218 |
| ADIYTEQAGR | 11S globulin-like | A0A2I4F6R4 | 1123.5378 | |
| AIPEEVLANAFQIPR | 11S globulin-like | A0A2I4F6R4 | 1667.9115 | |
| ALPEDVLINAYR | legumin B-like | A0A2I4GEH1 | 1373.7423 | |
| ALPEEVLATAFQIPR | Jug r 4 | Q2TPW5 | 1654.9163 | |
| ATLTLVSQETR | Jug r 2.0101 | Q9SEW4 | 1218.6688 | |
| EGDVFAVPR | vicilin-like seed storage protein | A0A2I4F3W3 | 989.5051 | |
| ELAFNFPAR | Jug r 6.0101 | A0A2I4E5L6 | 1064.5524 | |
| FYLAGNPHQQQQGGR | legumin B-like | A0A2I4GEH1 | 1700.8252 | |
| GIIVTVEDELR | legumin B-like | A0A2I4GEH1 | 1243.6892 | |
| HNLDTQTESDVFSR | Jug r 6.0101 | A0A2I4EG83 | 1648.7562 | |
| INALAGR | legumin B-like | A0A2I4GEH1 | 714.4257 | |
| INNLNAQEPGR | legumin B-like | A0A2I4GEH1 | 1225.6284 | |
| ISTVNSQNLPILR | 11S globulin-like | A0A2I4F6R4 | 1454.8326 | |
| ITSLNSFNLPILR | legumin B-like | A0A2I4GEH1 | 1487.858 | |
| LDALEPTNR | Jug r 4 | Q2TPW5 | 1028.5371 | |
| LVALEPSNR | 11S globulin-like | A0A2I4F6R4 | 998.5629 | |
| LVYVVQGR | legumin B-like | A0A2I4GEH1 | 933.5516 | |
| NEGFEWVSFK | Jug r 4 | -Q2TPW5 | 1242.579 | |
| NNIVNEFEK | Jug r 6.0101 | A0A2I4E5L6 | 1106.5477 | |
| QETFLAR | 11S globulin-like | A0A2I4F6R4 | 864.4574 | |
| SFFLAGGEPR | 11S globulin seed storage protein 2-like | A0A2I4F669 | 1080.5473 | |
| SFLLAGGEPR | Jug r 6.0101 | A0A2I4EG83 | 1046.5629 | |
| SGPSYQQIR | Jug r 6.0101 | A0A2I4E5L6 | 1035.5218 | |
| SPDQSYLR | Jug r 2.0101 | Q9SEW4 | 965.4687 | |
| SSGGPISLK | Jug r 2.0101 | Q9SEW4 | 845.4727 | |
| VFSNDILVAALNTPR | Jug r 2.0101 | Q9SEW4 | 1629.8959 | |
| WLQLSAER | Jug r 4 | Q2TPW5 | 1002.5367 | |
| YIQLSAER | legumin B-like | A0A2I4GEH1 | 979.5207 | |
| Pistachio | ADVYNPR | Pis v 2.0101 | B7P073 | 834.4104 |
| ALPLDVIK | Pis v 2.0101 | B7P073 | 868.5502 | |
| DTDILAAFR | Ribulose bisphosphate carboxylase large | A0A1 × 9ZER6 | 1021.5313 | |
| EDAWNLK | Pis v 2.0101 | B7P074 | 875.4258 | |
| EVLEAALK | Pis v 3.0101 | B4 × 640 | 872.5088 | |
| FEWISFK | Pis v 5.0101 | B7SLJ1 | 956.4876 | |
| FEWVSFK | Pis v 2.0101 | B7P073 | 942.472 | |
| FLQLSAK | Pis v 2.0101 | B7P073 | 806.4771 | |
| FLQLSVEK | Pis v 2.0101 | B7P074 | 963.551 | |
| FVLGGSPQQEIQGSGQSR | Pis v 2.0101 | B7P073 | 1874.9355 | |
| GDLQVIRPPR | Pis v 5.0101 | B7SLJ1 | 1150.6691 | |
| GFESEEESEYER | Pis v 5.0101 | B7SLJ1 | 1490.59177 | |
| GLPLDVIQNSFDISR | Pis v 2.0101 | B7P074 | 1673.8857 | |
| IAIVVSGEGR | Pis v 3.0101 | B4 × 640 | 1000.5786 | |
| ILAEVFQVEQSLVK | Pis v 5.0101 | B7SLJ1 | 1602.9101 | |
| IPTAYTK | Ribulose bisphosphate carboxylase large | A0A1 × 9ZER6 | 793.4454 | |
| ISQLAGR | Pis v 2.0101 | B7P074 | 744.4363 | |
| ITSLNSLNLPILK | Pis v 5.0101 | B7SLJ1 | 1425.8675 | |
| LNINDPSR | Pis v 2.0101 | B7P074 | 928.4847 | |
| LQELYETASELPR | Pis v 1 | B7P072 | 1548.7904 | |
| SDIYTPEVGR | Pis v 5.0101 | B7SLJ1 | 1136.5582 | |
| SETTIFAPGSSSQR | Pis v 2.0101 | B7P073 | 1467.7074 | |
| STGTFNLFK | Pis v 3.0101 | B4 × 640 | 1142.6204 | |
| TFQGPPHGIQVER | Ribulose bisphosphate carboxylase large | A0A1 × 9ZER6 | 1465.7546 | |
| VQEDLEVLSPHR | Pis v 2.0101 | B7P073 | 1421.7383 | |
| VTSINALNLPILR | Pis v 2.0101 | B7P074 | 1423.8631 | |
| WLQLSAER | Pis v 5.0101 | B7SLJ1 | 1002.5367 | |
| YNINDPSR | Pis v 2.0101 | B7P073 | 978.4639 | |
| Cashew | ADIYTPEVGR | Ana o 2.0101 | Q8GZP6 | 1120.5633 |
| DVFQQQQQHQSR | Ana o 2.0101 | Q8GZP6 | 1528.7251 | |
| ELYETASELPR | 2s albumin | Q8H2B8 | 1307.6478 | |
| FEWISFK | Ana o 2.0101 | Q8GZP6 | 956.4876 | |
| FHLAGNPK | Ana o 2.0101 | Q8GZP6 | 883.4785 | |
| IDYPPLEK | Vicilin-like protein | Q8L5L6 | 974.5193 | |
| LTTLNSLNLPILK | Ana o 2.0101 | Q8GZP6 | 1439.8832 | |
| VFDGEVR | Ana o 2.0101 | Q8GZP6 | 821.4152 | |
| WLQLSVEK | Ana o 2.0101 | Q8GZP6 | 1002.5619 | |
| YGQLFEAER | Vicilin-like protein | Q8L5L6 | 1112.5371 | |
| Hazelnut | ADIYTEQVGR | 11S globulin-like protein | A0A0A0P7E3 | 1151.5691 |
| ALPDDVLANAFQISR | 11S globulin-like protein | A0A0A0P7E3 | 1629.8595 | |
| ALSQHEEGPPR | Cor a 11.0101 | Q8S4P9 | 1220.6018 | |
| ELAFNLPSR | Cor a 11.0101 | Q8S4P9 | 1046.5629 | |
| GNIVNEFER | Cor a 11.0101 | Q8S4P9 | 1077.5324 | |
| HFYLAGNPDDEHQR | 11S globulin-like protein | A0A0A0P7E3 | 1698.7619 | |
| INTVNSNTLPVLR | 11S globulin-like protein | A0A0A0P7E3 | 1440.8169 | |
| IWPFGGESSGPINLLHK | Cor a 11.0101 | Q8S4P9 | 1851.9752 | |
| LLSGIENFR | Cor a 11.0101 | Q8S4P9 | 1048.5786 | |
| NIVKVEGR | 11S globulin-like protein | A0A0A0P7E3 | 914.5418 | |
| QGQQQFGQR | 11S globulin-like protein | A0A0A0P7E3 | 1076.5232 | |
| VQVLENFTK | Cor a 11.0101 | Q8S4P9 | 1077.5939 | |
| WLQLSAER | 11S globulin-like protein | A0A0A0P7E3 | 1002.5367 | |
| Almond | ADFYNPQGGR | Prunin 2 | A0A5E4FK23 | 1124.512 |
| ADIFSPR | Pru du 6.0101 | A0A5E4FFS0 | 805.4203 | |
| ALPDEVLANAYQISR | Pru du 6.0101 | A0A5E4FFS0 | 1659.8701 | |
| ALPDEVLQNAFR | Prunin 2 | A0A5E4FK23 | 1372.7219 | |
| ELAFNVPAR | Vicilin | A0A5E4EE27 | 1016.5524 | |
| FEEFFPAGSR | Vicilin | A0A5E4EZP4 | 1186.5528 | |
| FVSEDGIDNVR | (R)-mandelonitrile lyase | A0A5E4GEN6 | 1250.6012 | |
| FYEASPQEFK | Vicilin | A0A5E4F2T7 | 1245.5786 | |
| GNLDFVQPPR | Pru du 6.0101 | A0A5E4FFS0 | 1142.5953 | |
| LGFSSSLLFR | Gamma conglutin 1 | P82952 | 1126.6255 | |
| LKENIGNPER | Pru du 6.0101 | A0A5E4FFS0 | 1169.6273 | |
| LLSATSPPR | Prunin 2 | A0A5E4FK23 | 941.5415 | |
| LSQNIGDPSR | Prunin 2 | A0A5E4FK23 | 1086.5538 | |
| NQIIQVR | Pru du 6.0101 | A0A5E4FFS0 | 870.5156 | |
| QAYPWWR | Vicilin | A0A5E4F2T7 | 1006.4894 | |
| QSYFVPASR | Vicilin | A0A5E4F2T7 | 1054.5316 | |
| SLIGLAGTTPDR | Non-specific lipid-transfer protein | A0A4Y1RRI6 | 1200.6583 | |
| TEENAFINTLAGR | Pru du 6.0101 | A0A5E4FFS0 | 1435.7176 | |
| VQGQLDFVSPFSR | Prunin 2 | A0A5E4FK23 | 1479.7591 |
| Allergenic Species | Marker Peptides | Labelled Peptides Used for Signal Normalization | ||
|---|---|---|---|---|
| Sequence | m/z (+2) | Sequence | m/z (+2) | |
| Peanut | GTGNLELVAVR | 564.8222 | ADIYTEQVG(R) | 581.2925 |
| SPDIYNPQAGSLK | 695.3541 | SPDIYNPQAGSL(K) | 699.3612 | |
| TANDLNLLILR | 628.3721 | TANDLNLLIL(R) | 633.3764 | |
| Walnut | ADIYTEQAGR | 562.2726 | ADIYTEQVG(R) | 581.2925 |
| INNLNAQEPGR | 613.3178 | TANDLNLLIL(R) | 633.3764 | |
| SFLLAGGEPR | 523.7851 | ADIYTEQVG(R) | 581.2925 | |
| Pistachio | EDAWNLK | 438.2165 | ADIYTEQVG(R) | 581.2925 |
| SDIYTPEVGR | 568.7828 | ADIYTEQVG(R) | 581.2925 | |
| SETTIFAPGSSSQR | 734.3573 | TEENAFINTLAG(R) | 723.3666 | |
| Cashew | ADIYTPEVGR | 560.7853 | ADIYTEQVG(R) | 581.2925 |
| ELYETASELPR | 654.3275 | ADIYTEQVG(R) | 581.2925 | |
| LTTLNSLNLPILK | 720.4452 | TEENAFINTLAG(R) | 723.3666 | |
| Hazelnut | ADIYTEQVGR | 576.2882 | ADIYTEQVG(R) | 581.2925 |
| ALPDDVLANAFQISR | 815.4334 | ALPDDVLANAFQIS(R) | 820.4373 | |
| INTVNSNTLPVLR | 720.9121 | TEENAFINTLAG(R) | 723.3666 | |
| Almond | ADIFSPR | 403.2138 | ADIFSP(R) | 408.218 |
| LLSATSPPR | 471.2744 | ADIFSP(R) | 408.218 | |
| TEENAFINTLAGR | 718.3624 | TEENAFINTLAG(R) | 723.3666 | |
| Species | Peptides | R2 | LOD (mg·kg−1) * | LOQ (mg·kg−1) * |
|---|---|---|---|---|
| Peanut | GTGNLELVAVR | 0.9984 | 4.4 | 15 |
| SPDIYNPQAGSLK | 0.9995 | 2.4 | 8 | |
| TANDLNLLILR | 0.9977 | 5.2 | 17 | |
| Walnut | ADIYTEQAGR | 0.9987 | 3.9 | 13 |
| INNLNAQEPGR | 0.9987 | 3.8 | 13 | |
| SFLLAGGEPR | 0.9969 | 6.1 | 20 | |
| Pistachio | EDAWNLK | 0.9981 | 4.7 | 16 |
| SDIYTPEVGR | 0.999 | 3.4 | 11 | |
| SETTIFAPGSSSQR | 0.9989 | 3.6 | 12 | |
| Cashew | ADIYTPEVGR | 0.9991 | 3.2 | 11 |
| ELYETASELPR | 0.9957 | 7.1 | 24 | |
| LTTLNSLNLPILK | 0.9945 | 8.1 | 27 | |
| Hazelnut | ADIYTEQVGR | 0.9981 | 4.7 | 16 |
| ALPDDVLANAFQISR | 0.9982 | 4.5 | 15 | |
| INTVNSNTLPVLR | 0.9987 | 3.9 | 13 | |
| Almond | ADIFSPR | 0.9995 | 2.3 | 7.8 |
| LLSATSPPR | 0.9838 | 14 | 47 | |
| TEENAFINTLAGR | 0.9996 | 2.3 | 7.6 |
| Species | Reference Dose (mg Proteins) | Action Level 1 (mg·kg−1) | Action Level 2 (mg·kg−1) |
|---|---|---|---|
| Cashew | 0.05 | <1.0 | ≥1.0 |
| Pistachio | 0.05 | <1.0 | ≥1.0 |
| Almond | 0.10 | <2.0 | ≥2.0 |
| Hazelnut | 0.10 | <2.0 | ≥2.0 |
| Walnut | 0.03 | <0.6 | ≥0.6 |
| Peanut | 0.20 | <4.0 | ≥4.0 |
| Species | Selected Peptides | Protein Content (%) * | LOD (mg·kg−1) | LOQ (mg·kg−1) | Thresholds (mg·kg−1) for PAL Labelling According to VITAL® 3.0 |
|---|---|---|---|---|---|
| Peanut | GTGNLELVAVR | 25.2 | 1.1 | 3.7 | |
| SPDIYNPQAGSLK | 0.6 | 2 | 4 | ||
| TANDLNLLILR | 1.3 | 4.3 | |||
| Walnut | ADIYTEQAGR | 15.9 | 0.6 | 2 | |
| INNLNAQEPGR | 0.6 | 2 | 0.6 | ||
| SFFLAGGEPR | 0.9 | 3.2 | |||
| Pistachio | EDAWNLK | 23.8 | 1.1 | 3.7 | |
| SDIYTPEVGR | 0.8 | 2.7 | 1 | ||
| SETTIFAPGSSSQR | 0.8 | 2.9 | |||
| Cashew | ADIYTPEVGR | 19 | 0.6 | 2 | |
| ELYETASELPR | 1.3 | 4.5 | 1 | ||
| LTTLNSLNLPILK | 1.5 | 5.1 | |||
| Hazelnut | ADIYTEQVGR | 16.4 | 0.8 | 2.6 | |
| ALPDDVLANAFQISR | 0.7 | 2.5 | 2 | ||
| INTVNSNTLPVLR | 0.6 | 2.2 | |||
| Almond | ADIFSPR | 21.7 | 0.5 | 1.7 | |
| LLSATSPPR | 3.0 | 10 | 2 | ||
| TEENAFINTLAGR | 0.5 | 1.6 |
| Species | Selected Peptides | Intra-Day | Inter-Day | ||
|---|---|---|---|---|---|
| CV% | CV% | CV% | CV% | ||
| DAY 1 | DAY 2 | DAY 3 | DAYS 1,2,3 | ||
| Peanut | GTGNLELVAVR | 7.8 | 2.8 | 8.7 | 6.2 |
| SPDIYNPQAGSLK | 1.6 | 1.7 | 1.6 | 2.1 | |
| TANDLNLLILR | 3.2 | 3.7 | 3.0 | 5.5 | |
| Walnut | ADIYTEQAGR | 2.4 | 2.3 | 0.8 | 3.0 |
| INNLNAQEPGR | 5.2 | 3.5 | 1.0 | 4.9 | |
| SFFLAGGEPR | 9.6 | 4.5 | 1.3 | 7.1 | |
| Pistachio | EDAWNLK | 3.6 | 6.4 | 7.1 | 4.6 |
| SDIYTPEVGR | 3.0 | 1.9 | 3.9 | 2.5 | |
| SETTIFAPGSSSQR | 7.5 | 9.8 | 5.5 | 7.5 | |
| Cashew | ADIYTPEVGR | 2.5 | 3 | 4.4 | 3.7 |
| ELYETASELPR | 1.1 | 0.5 | 0.5 | 3.5 | |
| LTTLNSLNLPILK | 3.7 | 4.4 | 8.7 | 6.3 | |
| Hazelnut | ADIYTEQVGR | 6.7 | 5.6 | 0.5 | 19 |
| ALPDDVLANAFQISR | 6.4 | 3.2 | 8.7 | 6.1 | |
| INTVNSNTLPVLR | 5.6 | 1.8 | 3.1 | 4.1 | |
| Almond | ADIFSPR | 3.7 | 2.6 | 1.6 | 3.3 |
| LLSATSPPR | 2.1 | 1.8 | 3.5 | 2.6 | |
| TEENAFINTLAGR | 3.1 | 3.9 | 0.7 | 3.2 | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Luparelli, A.; Losito, I.; De Angelis, E.; Pilolli, R.; Monaci, L. Multi-Target Detection of Nuts and Peanuts as Hidden Allergens in Bakery Products through Bottom-Up Proteomics and High-Resolution Mass Spectrometry. Foods 2023, 12, 726. https://doi.org/10.3390/foods12040726
Luparelli A, Losito I, De Angelis E, Pilolli R, Monaci L. Multi-Target Detection of Nuts and Peanuts as Hidden Allergens in Bakery Products through Bottom-Up Proteomics and High-Resolution Mass Spectrometry. Foods. 2023; 12(4):726. https://doi.org/10.3390/foods12040726
Chicago/Turabian StyleLuparelli, Anna, Ilario Losito, Elisabetta De Angelis, Rosa Pilolli, and Linda Monaci. 2023. "Multi-Target Detection of Nuts and Peanuts as Hidden Allergens in Bakery Products through Bottom-Up Proteomics and High-Resolution Mass Spectrometry" Foods 12, no. 4: 726. https://doi.org/10.3390/foods12040726
APA StyleLuparelli, A., Losito, I., De Angelis, E., Pilolli, R., & Monaci, L. (2023). Multi-Target Detection of Nuts and Peanuts as Hidden Allergens in Bakery Products through Bottom-Up Proteomics and High-Resolution Mass Spectrometry. Foods, 12(4), 726. https://doi.org/10.3390/foods12040726











