Quantification of Pork, Chicken, Beef, and Sheep Contents in Meat Products Using Duplex Real-Time PCR
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Preparation
2.2. DNA Extraction
2.3. Primers and Probes
2.4. Real-Time PCR Procedure
2.5. Amplification Efficiency and Quantification
2.6. Sensitivity, Accuracy, and Precision
2.7. Limit of Detection (LOD) and Limit of Quantification (LOQ)
2.8. Quantification with Calibration Factors
2.9. Meat Species Identification and Quantification Using Commercial Kits
3. Results and Discussion
3.1. Design of the Duplex Real-Time PCR System
3.2. Specificity of the Duplex Primer/Probe Systems
3.3. Sensitivity, Precision, and Accuracy
3.4. Determination of Calibration Factors with Reference Materials
3.5. Analysis of Proficiency Test Samples and Market Monitoring Samples
3.6. Future Perspectives
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Bonnet, C.; Bouamra-Mechemache, Z.; Réquillart, V.; Treich, N. Viewpoint: Regulating meat consumption to improve health, the environment and animal welfare. Food Policy 2020, 97, 101847. [Google Scholar] [CrossRef]
- Ali, S.; Alsayeqh, A.F. Review of major meat-borne zoonotic bacterial pathogens. Front. Public Health 2022, 10, 1045599. [Google Scholar] [CrossRef]
- Das, A.K.; Nanda, P.K.; Das, A.; Biswas, S. Chapter 6-Hazards and Safety Issues of Meat and Meat Products. In Food Safety and Human Health; Singh, R.L., Mondal, S., Eds.; Academic Press: Cambridge, MA, USA, 2019; pp. 145–168. [Google Scholar]
- WHO. Zoonotic Disease: Emerging Public Health Threats in the Region. Available online: https://www.emro.who.int/about-who/rc61/zoonotic-diseases.html (accessed on 1 April 2023).
- Rahman, T.; Sobur, A.; Islam, S.; Ievy, S.; Hossain, J.; El Zowalaty, M.E.; Rahman, A.T.; Ashour, H.M. Zoonotic Diseases: Etiology, Impact, and Control. Microorganisms 2020, 8, 1405. [Google Scholar] [CrossRef]
- EU. Regulation (EU) 2017/625 of the European Parliament and of the Council of 15 March 2017 on Official Controls and Other Official Activities Performed to Ensure the Application of Food and Feed Law, Rules on Animal Health and Welfare, Plant Health and Plant Protection Products. Available online: https://eur-lex.europa.eu/legal-content/EN/TXT/?uri=CELEX%3A02017R0625-20191214&qid=1617129031298 (accessed on 1 April 2023).
- Commission, E. EU Entry Conditions for Composite Products. Available online: https://food.ec.europa.eu/horizontal-topics/international-affairs/eu-entry-conditions/composite-products_en (accessed on 1 April 2023).
- USDA-FSIS. Import Guidance. Available online: https://www.fsis.usda.gov/inspection/import-export/import-guidance (accessed on 1 April 2023).
- SFA (Singapore Food Agency). Food Import & Export. In Conditions for Specific Types of Food for Import; 2023. Available online: https://www.sfa.gov.sg/food-import-export/conditions-for-specific-types-of-food-for-import (accessed on 1 April 2023).
- SFA (Singapore Food Agency). Wholesome Meat and Fish Act 1999. 2020. Available online: https://www.sfa.gov.sg/docs/default-source/default-document-library/wholesome-meat-and-fish-act-1999.pdf?sfvrsn=726a849b_0 (accessed on 1 April 2023).
- SFA (Singapore Food Agency). Food Import & Export. In Seeking Accreditation of Overseas Food Sources; 2023. Available online: https://www.sfa.gov.sg/food-import-export/seeking-accreditation-of-overseas-food-sources (accessed on 1 April 2023).
- Abbas, O.; Zadravec, M.; Baeten, V.; Mikuš, T.; Lešić, T.; Vulić, A.; Prpić, J.; Jemeršić, L.; Pleadin, J. Analytical methods used for the authentication of food of animal origin. Food Chem. 2018, 246, 6–17. [Google Scholar] [CrossRef]
- Li, Y.; Liu, S.; Meng, F.; Liu, D.; Zhang, Y.; Wang, W.; Zhang, J. Comparative review and the recent progress in detection technologies of meat product adulteration. Compr. Rev. Food Sci. Food Saf. 2020, 19, 2256–2296. [Google Scholar] [CrossRef]
- Zia, Q.; Alawami, M.; Mokhtar, N.F.K.; Nhari, R.M.H.R.; Hanish, I. Current analytical methods for porcine identification in meat and meat products. Food Chem. 2020, 324, 126664. [Google Scholar] [CrossRef]
- Xu, H.; Ma, X.; Ye, Z.; Yu, X.; Liu, G.; Wang, Z. A Droplet Digital PCR Based Approach for Identification and Quantification of Porcine and Chicken Derivatives in Beef. Foods 2022, 11, 3265. [Google Scholar] [CrossRef]
- Köppel, R.; Ganeshan, A.; Weber, S.; Pietsch, K.; Graf, C.; Hochegger, R.; Griffiths, K.; Burkhardt, S. Duplex digital PCR for the determination of meat proportions of sausages containing meat from chicken, turkey, horse, cow, pig and sheep. Eur. Food Res. Technol. 2019, 245, 853–862. [Google Scholar] [CrossRef]
- Böhme, K.; Calo-Mata, P.; Barros-Velázquez, J.; Ortea, I. Review of recent DNA-based methods for main food-authentication Topics. J. Agric. Food Chem. 2019, 67, 3854–3864. [Google Scholar] [CrossRef]
- El Sheikha, A.F.; Mokhtar, N.F.K.; Amie, C.; Lamasudin, D.U.; Isa, N.M.; Mustafa, S. Authentication technologies using DNA-based approaches for meats and halal meats determination. Food Biotechnol. 2017, 31, 281–315. [Google Scholar] [CrossRef]
- Köppel, R.; Ruf, J.; Rentsch, J. Multiplex real-time PCR for the detection and quantification of DNA from beef, pork, horse and sheep. Eur. Food Res. Technol. 2010, 232, 151–155. [Google Scholar] [CrossRef]
- Dolch, K.; Andrée, S.; Schwägele, F. Comparison of real-time PCR quantification methods in the identification of poultry species in meat products. Foods 2020, 9, 1049. [Google Scholar] [CrossRef]
- Eugster, A.; Ruf, J.; Rentsch, J.; Köppel, R. Quantification of beef, pork, chicken and turkey proportions in sausages: Use of matrix-adapted standards and comparison of single versus multiplex PCR in an interlaboratory trial. Eur. Food Res. Technol. 2009, 230, 55–61. [Google Scholar] [CrossRef]
- Köppel, R.; Eugster, A.; Ruf, J.; Rentsch, J. Quantification of meat proportions by measuring DNA contents in raw and boiled sausages using matrix-adapted calibrators and multiplex real-time PCR. J. AOAC Int. 2012, 95, 494–499. [Google Scholar] [CrossRef][Green Version]
- Köppel, R.; Ruf, J.; Zimmerli, F.; Breitenmoser, A. Multiplex real-time PCR for the detection and quantification of DNA from beef, pork, chicken and turkey. Eur. Food Res. Technol. 2008, 227, 1199–1203. [Google Scholar] [CrossRef]
- Köppel, R.; Daniels, M.; Felderer, N.; Brünen-Nieweler, C. Multiplex real-time PCR for the detection and quantification of DNA from duck, goose, chicken, turkey and pork. Eur. Food Res. Technol. 2013, 236, 1093–1098. [Google Scholar] [CrossRef]
- Köppel, R.; van Velsen, F.; Ganeshan, A.; Pietsch, K.; Weber, S.; Graf, C.; Murmann, P.; Hochegger, R.; Licina, A. Multiplex real-time PCR for the detection and quantification of DNA from chamois, roe, deer, pork and beef. Eur. Food Res. Technol. 2020, 246, 1007–1015. [Google Scholar] [CrossRef]
- SFA (Singapore Food Agency). SFA Singapore Food Statistics. 2021. Available online: https://www.sfa.gov.sg/publications/sgfs (accessed on 1 April 2023).
- ISO 21571:2005; Foodstuffs-Methods of Analysis for the Detection of Genetically Modified Organisms and Derived Products-Nucleic Acid Extraction. International Organization for Standardization: Geneva, Switzerland, 2005.
- Meira, L.; Costa, J.; Villa, C.; Ramos, F.; Oliveira, M.B.P.; Mafra, I. EvaGreen real-time PCR to determine horse meat adulteration in processed foods. LWT 2017, 75, 408–416. [Google Scholar] [CrossRef]
- Amaral, J.S.; Santos, G.; Oliveira, M.B.P.; Mafra, I. Quantitative detection of pork meat by EvaGreen real-time PCR to assess the authenticity of processed meat products. Food Control 2017, 72, 53–61. [Google Scholar] [CrossRef]
- Broeders, S.; Huber, I.; Grohmann, L.; Berben, G.; Taverniers, I.; Mazzara, M.; Roosens, N.; Morisset, D. Guidelines for validation of qualitative real-time PCR methods. Trends Food Sci. Technol. 2014, 37, 115–126. [Google Scholar] [CrossRef]
- ISO 20395:2019; IBiotechnology—Requirements for Evaluating the Performance of Quantification Methods for Nucleic Acid Target Sequences—qPCR and dPCR. International Standard: Geneva, Switzerland, 2019.
- ENGL. Definition of Minimum Performance Requirements for Analytical Methods of GMO Testing; European Network of GMO Laboratories, Publications Office of the European Union: Luxembourg, 2015; Available online: http://gmo-crl.jrc.ec.europa.eu/guidancedocs.htm (accessed on 1 April 2023).
- ISO 5725-1:2003; Accuracy (Trueness and Precision) of Measurement Methods and Results—Part 1: General Principles and Definitions. International Organization for Standardization: Geneva, Switzerland, 2003.
- Kim, M.; Yoo, I.; Lee, S.-Y.; Hong, Y.; Kim, H.-Y. Quantitative detection of pork in commercial meat products by TaqMan® real-time PCR assay targeting the mitochondrial D-loop region. Food Chem. 2016, 210, 102–106. [Google Scholar] [CrossRef] [PubMed]
- Sarlak, Z.; Shojaee-Aliabadi, S.; Rezvani, N.; Hosseini, H.; Rouhi, M.; Dastafkan, Z. Development and validation of TaqMan real-time PCR assays for quantification of chicken adulteration in hamburgers. J. Food Compos. Anal. 2022, 106, 104302. [Google Scholar] [CrossRef]
- Iwobi, A.; Sebah, D.; Kraemer, I.; Losher, C.; Fischer, G.; Busch, U.; Huber, I. A multiplex real-time PCR method for the quantification of beef and pork fractions in minced meat. Food Chem. 2014, 169, 305–313. [Google Scholar] [CrossRef] [PubMed]
- Rentsch, J.; Weibel, S.; Ruf, J.; Eugster, A.; Beck, K.; Köppel, R. Interlaboratory validation of two multiplex quantitative real-time PCR methods to determine species DNA of cow, sheep and goat as a measure of milk proportions in cheese. Eur. Food Res. Technol. 2012, 236, 217–227. [Google Scholar] [CrossRef]
- Kang, T.S. Basic principles for developing real-time PCR methods used in food analysis: A review. Trends Food Sci. Technol. 2019, 91, 574–585. [Google Scholar] [CrossRef]
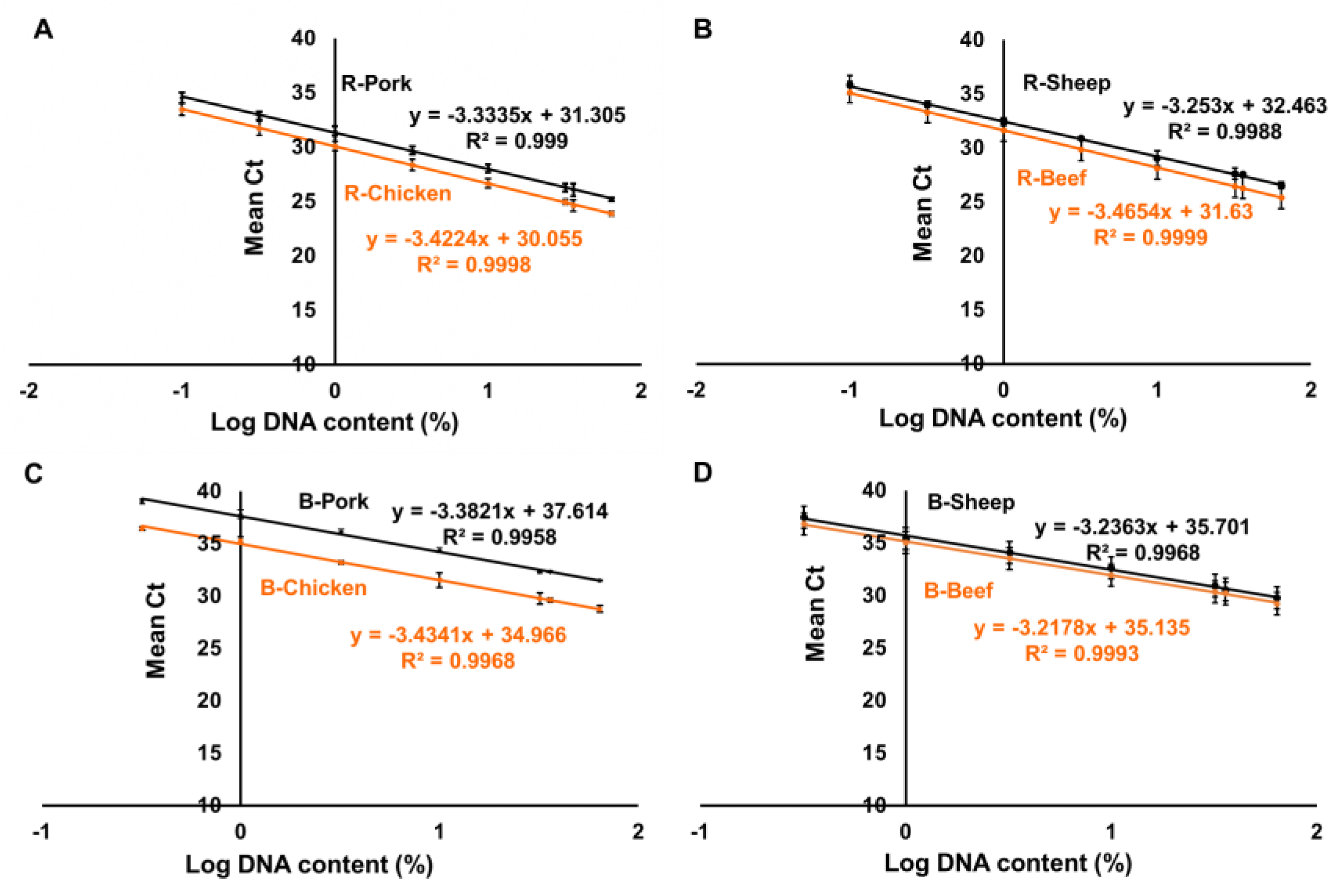
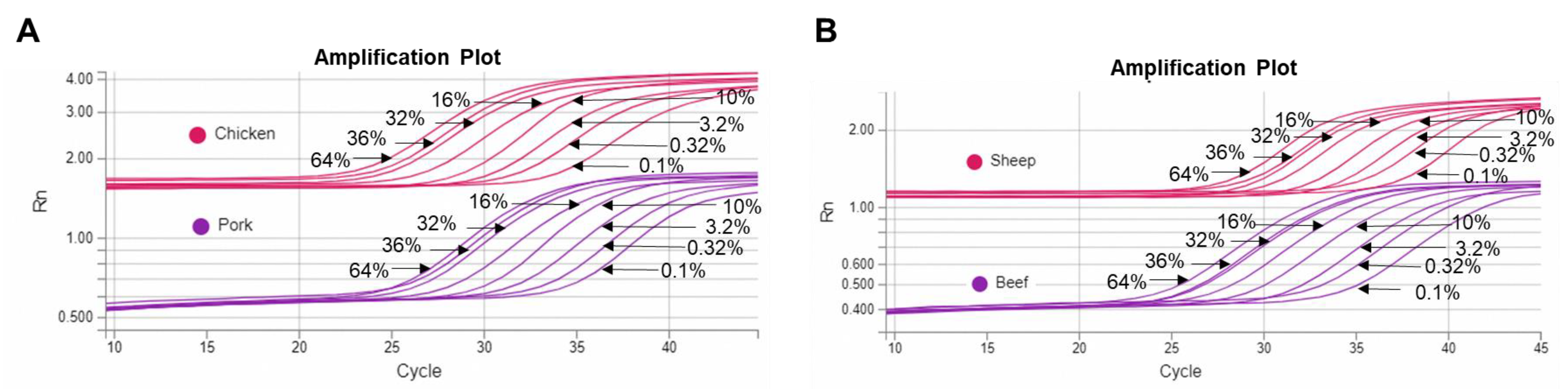
| Reference Materials Composition | Pork (%) | Beef (%) | Chicken (%) | Sheep (%) |
|---|---|---|---|---|
| RefA | 1 | 9 | 15 | 75 |
| RefB | 9 | 1 | 75 | 15 |
| RefC | 15 | 75 | 1 | 9 |
| RefD | 25 | 25 | 25 | 25 |
| RefE | 50 | 0 | 50 | 0 |
| RefF | 0 | 50 | 0 | 50 |
| RefG | 75 | 15 | 9 | 1 |
| Species | Primer/Probe | Final Conc. (nM) | Sequence | Amplicon | Gene |
|---|---|---|---|---|---|
| Pork | Sus1 F | 200 | CGAGAGGCTGCCGTAAAGG | 80 | Beta-actin-gene DQ452569 [23] |
| Sus1 R | 200 | TGCAAGGAACACGGCTAAGTG | |||
| Sus1-VIC | 100 | VIC-TCTGACGTGACTCCCCGACCTGG-BHQ1 | |||
| Chicken | Gallus1 F | 200 | CAGCTGGCCTGCCGG | 76 | TF-GB3 X6009 [23] |
| Gallus1 R | 200 | CCCAGTGGAATGTGGTATTCA | |||
| Gallus1-FAM | 100 | FAM-TCTGCCACTCCTCTGCACCCAGT-BHQ1 | |||
| Sheep | OA-PRLR-F | 200 | CCAACATGCCTTTAAACCCTCAA | 88 | Prolactin receptor [19] |
| OA-PRLR-R | 200 | GGAACTGTAGCCTTCTGACTCG | |||
| OA-PRLR FAM | 100 | FAM-TGCCTTTCCTTCCCCGCCAGTCTC-BHQ1 | |||
| Beef | Rd 1 F | 200 | GTAGGTGCACAGTACGTTCTGAAG | 96 | Beta-actin-gene EH170825 [19] |
| Rd 1 R | 200 | GGCCAGACTGGGCACATG | |||
| Rd 1-VIC | 100 | VIC-GAACCTCATTCTGGGGCCCCG-BHQ1 | |||
| 18S | 18S-F | 200 | CTGCCCTATCAACTTTCGATGGTA | 113 | 18S rRNA [28,29] |
| 18S-R | 200 | TTGGATGTGGTAGCCGTTTCTCA | |||
| 18S-FAM | 100 | FAM-ACGGGTAACGGGGAATCAGGGTTCGATT-BHQ1 |
| STD | Duplex Chicken and Pork qPCR | Duplex Sheep and Beef qPCR | ||
|---|---|---|---|---|
| Chicken % | Pork % | Beef % | Sheep % | |
| 1 | 64% | 36% | 36% | 64% |
| 2 | 32% | 32% | 32% | 32% |
| 3 | 10% | 10% | 10% | 10% |
| 4 | 3.2% | 3.2% | 3.2% | 3.2% |
| 5 | 1% | 1% | 1% | 1% |
| 6 | 0.32% | 0.32% | 0.32% | 0.32% |
| 7 | 0.1% | 0.1% | 0.1% | 0.1% |
| 8 | 36% | 64% | 64% | 36% |
| Duplex PCR | Chicken | Pork | Sheep | Beef |
|---|---|---|---|---|
| Amplification efficiencies % | 99.11 | 98.70 | 104.54 | 103.70 |
| Correlation R2 | 0.9998 | 0.9945 | 0.9969 | 0.9985 |
| RSD (%) | 5.23 | 8.54 | 8.75 | 11.4 |
| Bias (%) | 2.98 | 4.12 | 3.4 | 3.1 |
| Meat Compositions (%) | 1 | 9 | 15 | 25 | 50 | 75 | |
|---|---|---|---|---|---|---|---|
| Raw Pork | Md (%) | 1.24 | 9.89 | 16.32 | 25.04 | 48.85 | 72.02 |
| RSD (%) | 20.42 | 7.49 | 7.62 | 13.24 | 7.6 | 14.01 | |
| Bias (%) | 24.00 | 9.89 | 8.80 | 0.16 | −2.30 | −3.97 | |
| Boiled Pork | Md (%) | 0.91 | 9.93 | 16.52 | 23.68 | 45.89 | 70.76 |
| RSD (%) | 24.58 | 22.48 | 10.85 | 17.16 | 14.47 | 8.03 | |
| Bias (%) | −9.00 | 10.33 | 10.13 | −5.28 | −8.22 | −5.65 | |
| Raw Chicken | Md (%) | 1.23 | 9.74 | 12.18 | 27.59 | 51.15 | 81.25 |
| RSD (%) | 5.09 | 18.31 | 6.57 | 13.84 | 5.56 | 6.15 | |
| Bias (%) | 23.00 | 8.22 | −18.80 | 10.36 | 2.30 | 8.33 | |
| Boiled Chicken | Md (%) | 1.21 | 11.09 | 14.90 | 27.89 | 54.10 | 78.47 |
| RSD (%) | 22.41 | 9.41 | 7.14 | 4.38 | 2.45 | 7.12 | |
| Bias (%) | 21.00 | 23.22 | −0.67 | 11.56 | 8.20 | 4.63 | |
| Raw Beef | Md (%) | 1.06 | 10.25 | 17.33 | 28.35 | 51.31 | 72.22 |
| RSD (%) | 15.72 | 8.92 | 11.79 | 8.60 | 11.67 | 8.81 | |
| Bias (%) | 6.00 | 13.89 | 15.53 | 13.40 | 2.62 | −3.71 | |
| Boiled Beef | Md (%) | 1.15 | 9.89 | 17.33 | 29.20 | 51.45 | 72.47 |
| RSD (%) | 15.72 | 8.92 | 11.79 | 8.60 | 11.67 | 8.81 | |
| Bias (%) | 15.00 | 9.89 | 15.53 | 16.80 | 2.90 | −3.37 | |
| Raw Sheep | Md (%) | 0.91 | 9.68 | 15.03 | 25.28 | 49.69 | 75.74 |
| RSD (%) | 22.42 | 8.60 | 13.08 | 16.88 | 19.48 | 7.68 | |
| Bias (%) | −9.00 | 7.56 | 0.20 | 1.12 | −0.62 | 0.99 | |
| Boiled Sheep | Md (%) | 0.96 | 9.71 | 16.60 | 25.86 | 48.51 | 73.64 |
| RSD (%) | 23.02 | 8.60 | 11.79 | 16.88 | 24.98 | 10.16 | |
| Bias (%) | −4.00 | 7.89 | 10.67 | 3.44 | −2.98 | −1.81 | |
| Raw | Boiled | |||||||
|---|---|---|---|---|---|---|---|---|
| Chicken | Pork | Sheep | Beef | Chicken | Pork | Sheep | Beef | |
| 1 | 0.87 | 1.04 | 0.84 | 0.97 | 0.91 | 0.99 | 0.88 | 0.88 |
| 2 | 0.84 | 0.97 | 0.79 | 0.82 | 0.82 | 0.91 | 0.86 | 0.92 |
| 3 | 0.89 | 0.86 | 0.98 | 0.95 | 0.86 | 0.97 | 0.94 | 0.85 |
| 4 | 0.79 | 0.99 | 0.89 | 0.89 | 0.87 | 0.98 | 0.89 | 0.99 |
| Mean | 0.85 | 0.97 | 0.88 | 0.91 | 0.87 | 0.96 | 0.89 | 0.91 |
| RSD | 5.12 | 7.83 | 9.21 | 7.42 | 4.27 | 3.73 | 3.81 | 6.65 |
| Sample Name | Target | True Values (%) | Duplex PCR Results (Meat %) | RapidFinder™ Kit Results (DNA %) | ||||
|---|---|---|---|---|---|---|---|---|
| Md (%) | RSD (%) | Bias (%) | Md (%) | RSD (%) | Bias (%) | |||
| FAPAS-PT1 | Pork | 3 | 3.70 | 12.64 | 23.33 | 4.26 | 16.1 | 42 |
| Chicken | 0 | 0 | - | - | 0 | - | - | |
| Beef | 0 | 0 | - | - | 0 | - | - | |
| Sheep | 94 | 90.32 | 5.39 | −3.91 | 87.24 | 10.02 | −7.19 | |
| FAPAS-PT2 | Pork | 0 | 0 | - | - | 0 | - | - |
| Chicken | 0 | 0 | - | - | 0 | - | - | |
| Beef | 90 | 89.39 | 17.46 | −0.68 | 93.7 | 15.1 | 4.11 | |
| Sheep | 5 | 3.81 | 13.84 | −23.8 | 7.02 | 14.6 | 40.40 | |
| FAPAS-PT3 | Pork | 3 | 2.41 | 15.27 | −24.48 | 4.6 | 7.5 | 53.33 |
| Chicken | 92 | 88.79 | 4.56 | −3.49 | 87.7 | 6.58 | −4.67 | |
| Beef | 0 | 0 | - | - | 0 | - | - | |
| Sheep | 0 | 0 | - | - | 0 | - | - | |
| Study Type | S/N | Sample Name | Duplex PCR Results (Meat %) | |||
|---|---|---|---|---|---|---|
| Beef | Sheep | Chicken | Pork | |||
| Meat fraud market monitoring | 1 | RTE Mutton Briyani | 71.4 | 28.6 | 0 | 0 |
| 2 | Raw Minced Mutton | 0 | 99.8 | 0 | 0 | |
| 3 | RTE Mutton Briyani | 0 | 95.3 | 0 | 0 | |
| 4 | RTE Mutton Curry (Boneless) | 0 | 98.2 | 0 | 0 | |
| 5 | RTE Keema (minced mutton) | 0 | 94.9 | 0 | 0 | |
| 6 | Raw Mutton (with bones) | 0 | 98.5 | 0 | 0 | |
| 7 | Raw Mutton Minced Meat (Fine) | 0 | 99.5 | 0 | 0 | |
| 8 | Raw Mutton Cube for Mutton Briyani Curry | 72.7 | 27.3 | 0 | 0 | |
| 9 | Raw Mutton Bone Cut for Mutton Curry (prata) | 0 | 97.5 | 0 | 0 | |
| 10 | RTE Mutton Curry | 63.9 | 36.1 | 0 | 0 | |
| Meat permit compliance check | 11 | Chilli Pork Flavor Instant Bowl Noodles | 0 | 0 | 0 | 10.8 |
| 12 | Chilli Pork Flavor Instant Noodles | 0 | 0 | 0 | 10.9 | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, Y.; Teo, E.; Lin, K.J.; Wu, Y.; Chan, J.S.H.; Tan, L.K. Quantification of Pork, Chicken, Beef, and Sheep Contents in Meat Products Using Duplex Real-Time PCR. Foods 2023, 12, 2971. https://doi.org/10.3390/foods12152971
Wang Y, Teo E, Lin KJ, Wu Y, Chan JSH, Tan LK. Quantification of Pork, Chicken, Beef, and Sheep Contents in Meat Products Using Duplex Real-Time PCR. Foods. 2023; 12(15):2971. https://doi.org/10.3390/foods12152971
Chicago/Turabian StyleWang, Yanwen, Emily Teo, Kung Ju Lin, Yuansheng Wu, Joanne Sheot Harn Chan, and Li Kiang Tan. 2023. "Quantification of Pork, Chicken, Beef, and Sheep Contents in Meat Products Using Duplex Real-Time PCR" Foods 12, no. 15: 2971. https://doi.org/10.3390/foods12152971
APA StyleWang, Y., Teo, E., Lin, K. J., Wu, Y., Chan, J. S. H., & Tan, L. K. (2023). Quantification of Pork, Chicken, Beef, and Sheep Contents in Meat Products Using Duplex Real-Time PCR. Foods, 12(15), 2971. https://doi.org/10.3390/foods12152971






