Enhanced Production of β-Nicotinamide Mononucleotide with Exogenous Nicotinamide Addition in Saccharomyces boulardii-YS01
Abstract
1. Introduction
2. Materials and Methods
2.1. Chemicals and Culture Medium
2.2. Sample Preparation
2.3. UPLC-ESI-QqQ-MS/MS Analysis
2.4. Method Validation
2.5. Genome Scanning Sequencing and Analysis of Strain-YS01
2.6. Data Processing and Statistical Analysis
3. Results and Discussion
3.1. Establishment and Validation of the Analytical Method
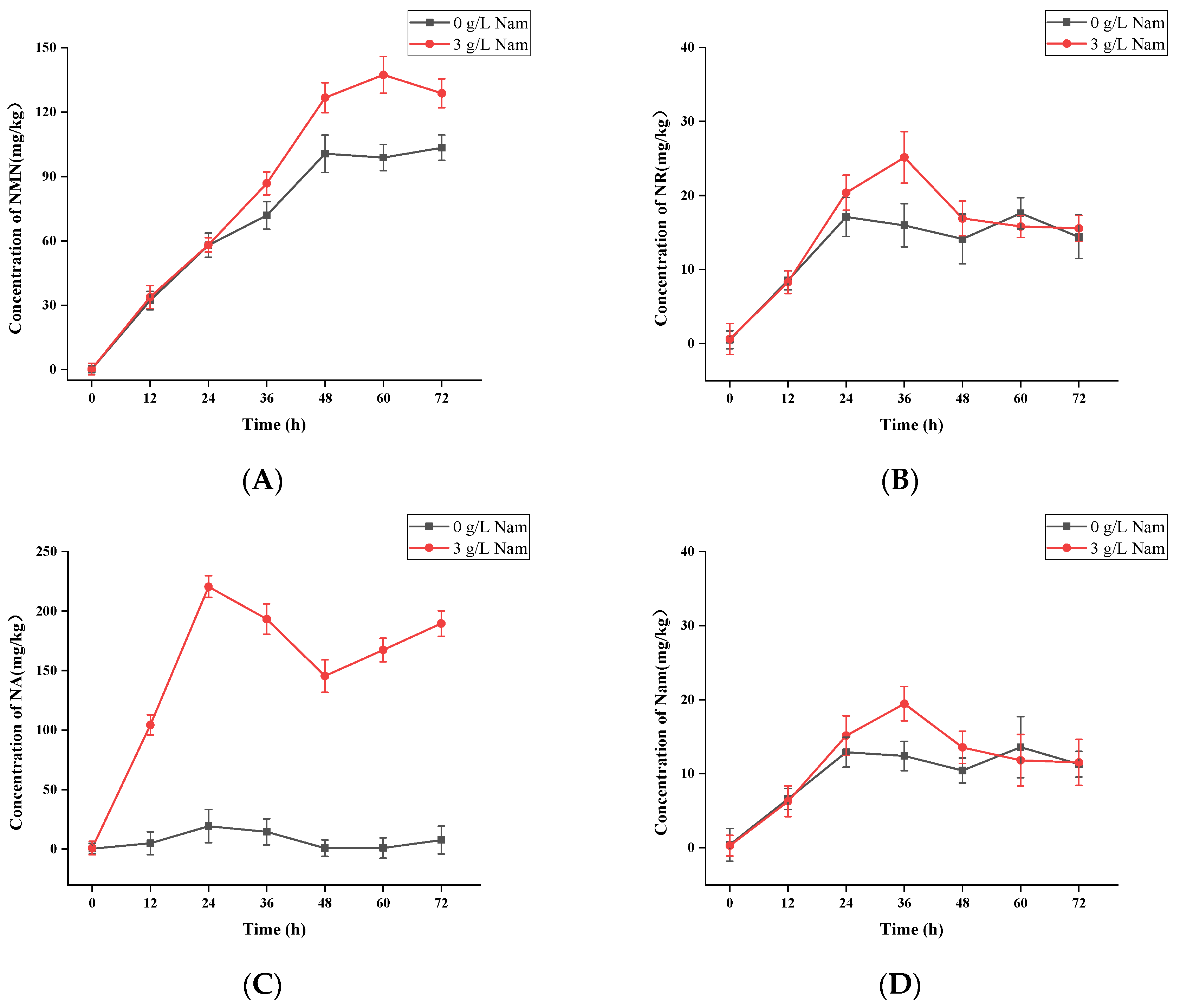
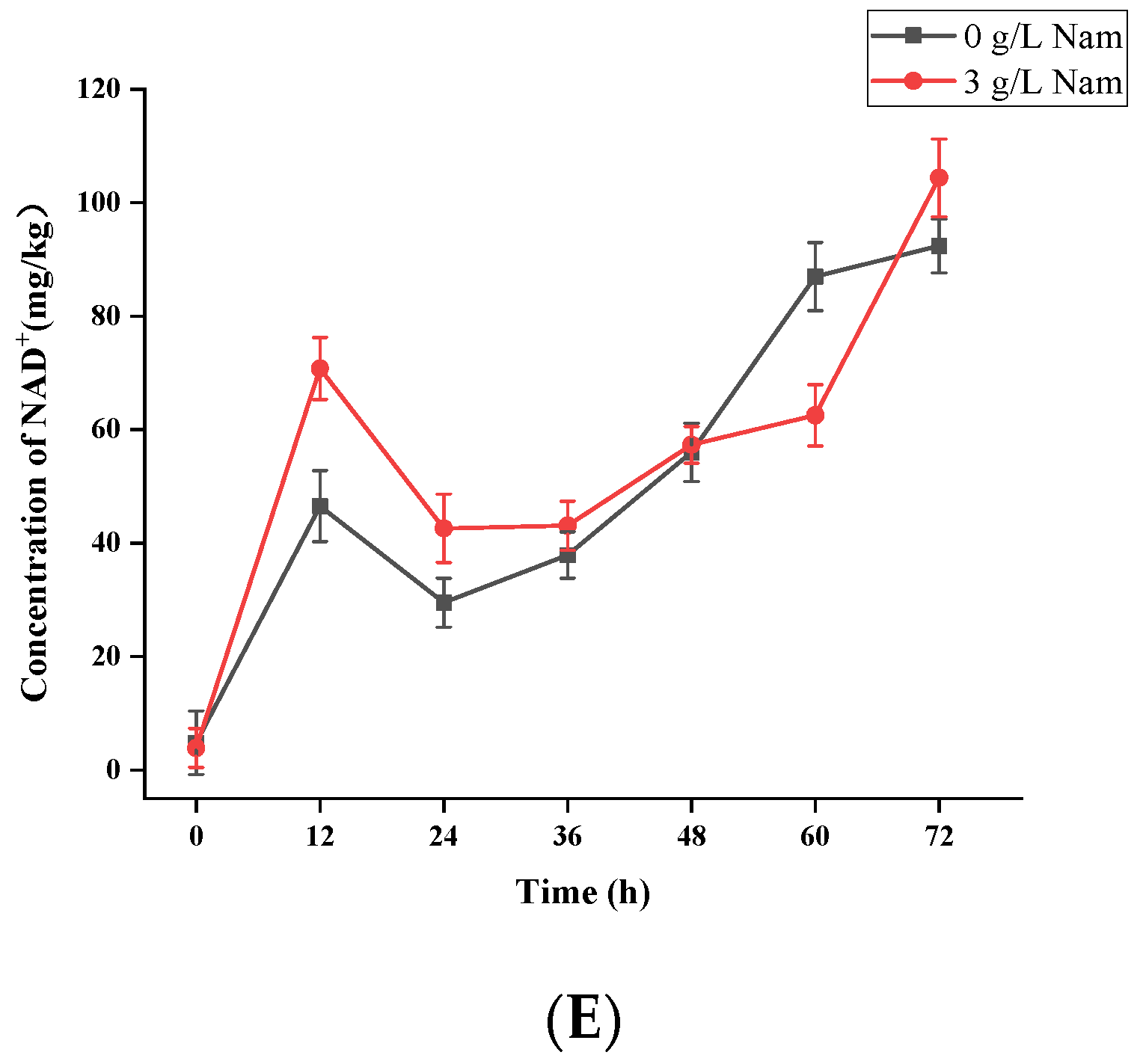
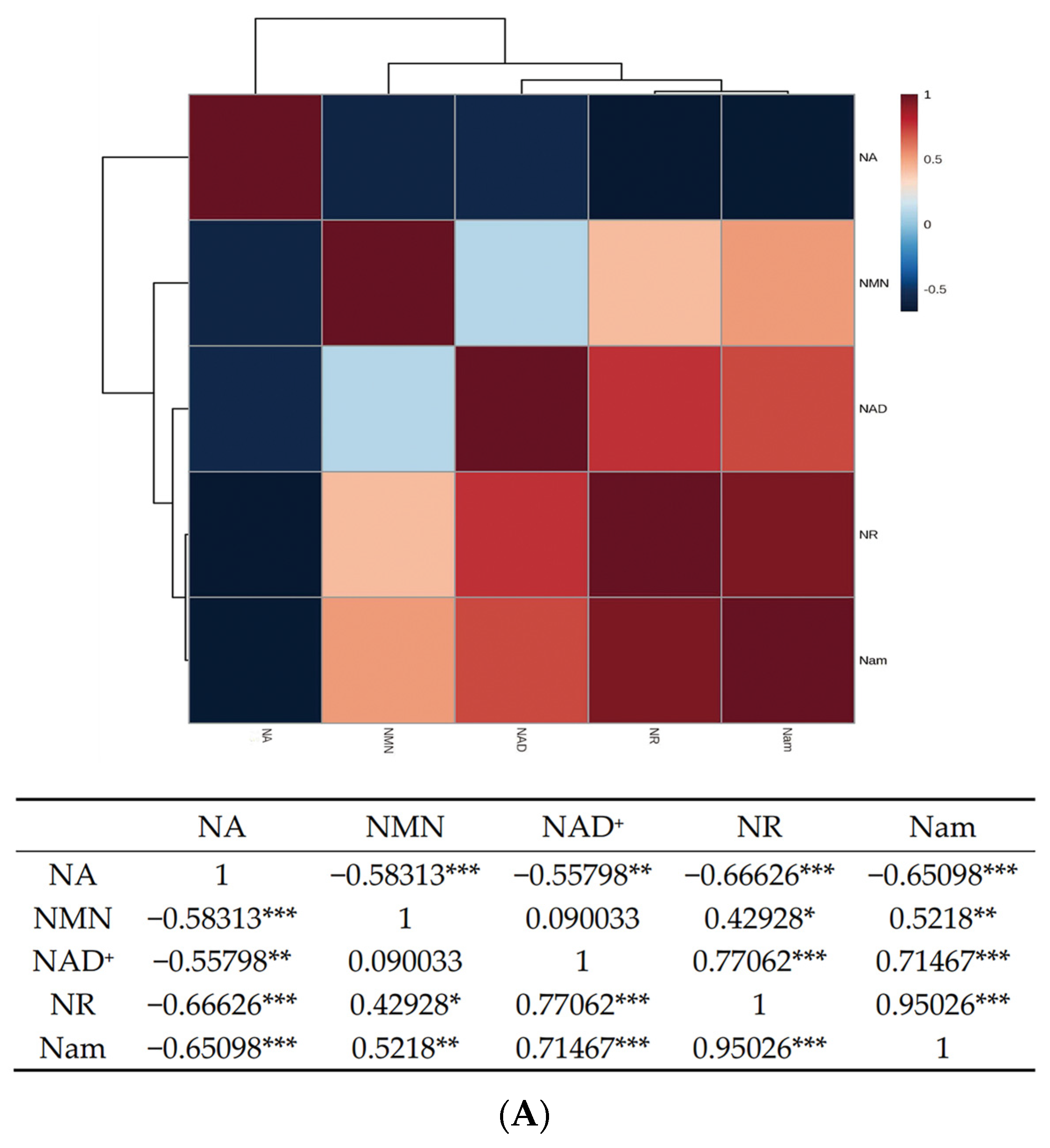
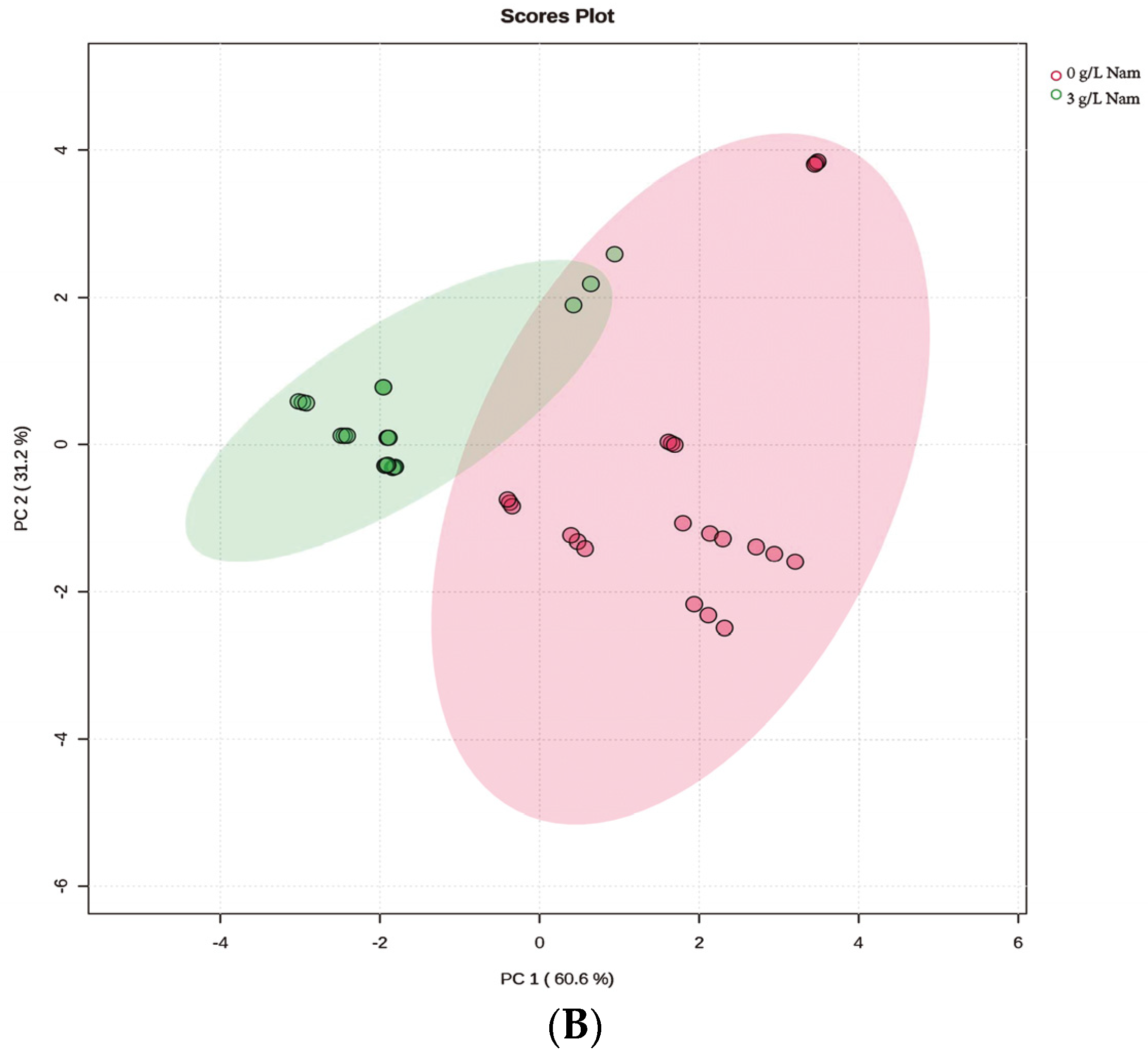
3.2. UPLC-ESI-QqQ-MS/MS Quantitation of NMN, NR, NA, Nam and NAD+ in S. boulardii-YS01
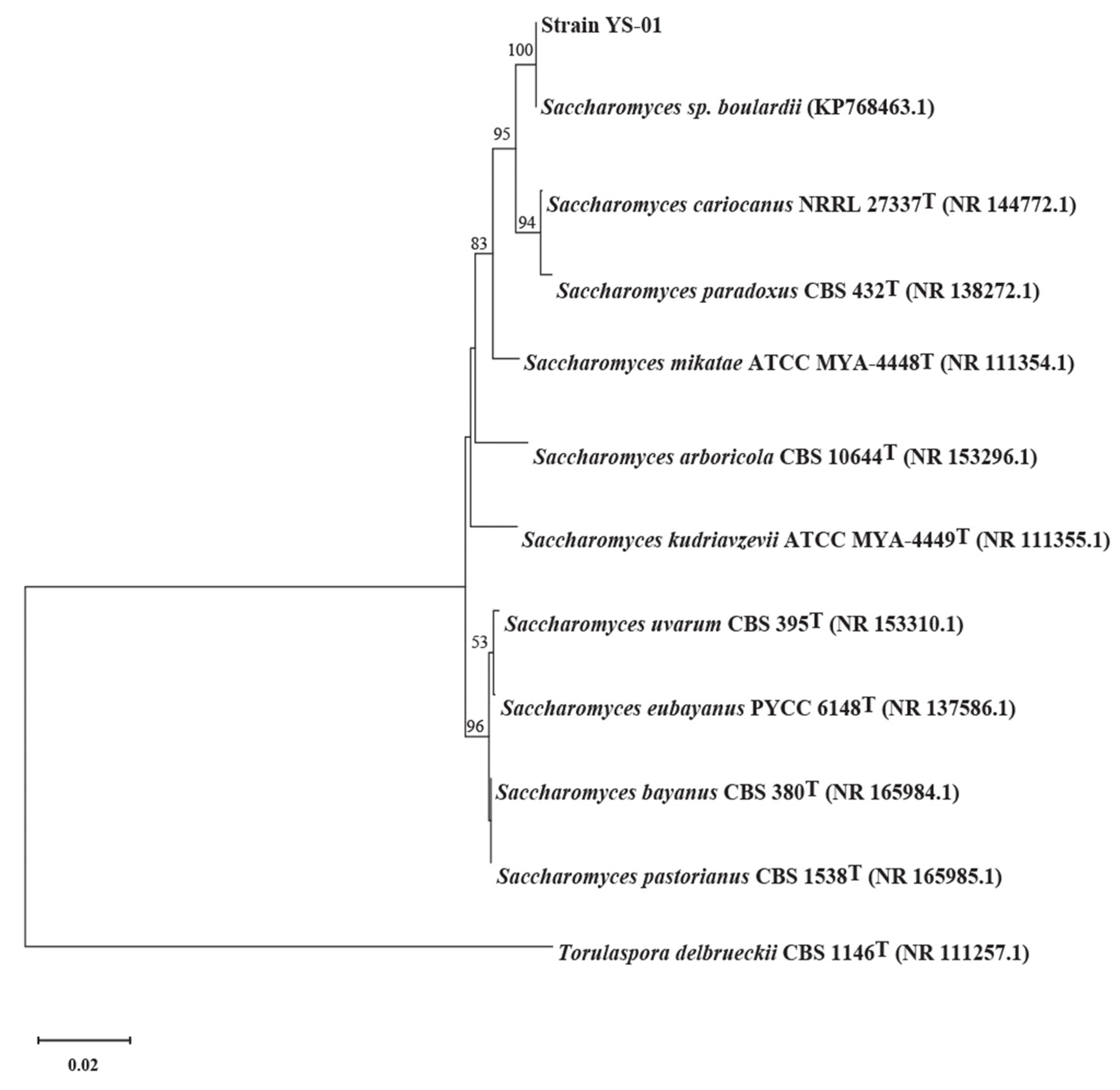
3.3. Overview of the S. boulardii Strain-YS01 Genome
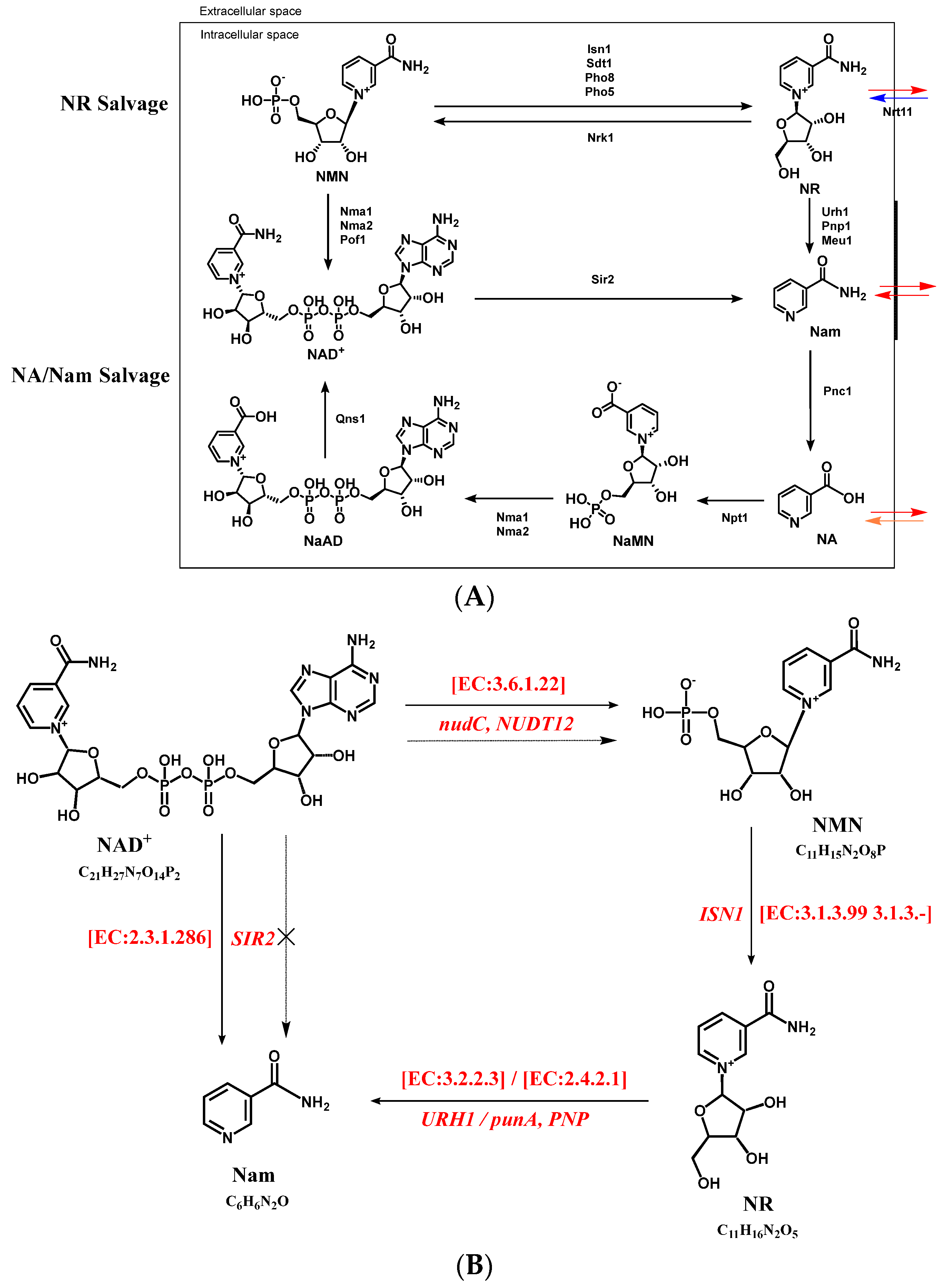
3.4. Targeted Metabolic Analysis Combined with Gene Anlysis for Proposing the Formation Pathway of NMN in S. boulardii-YS01
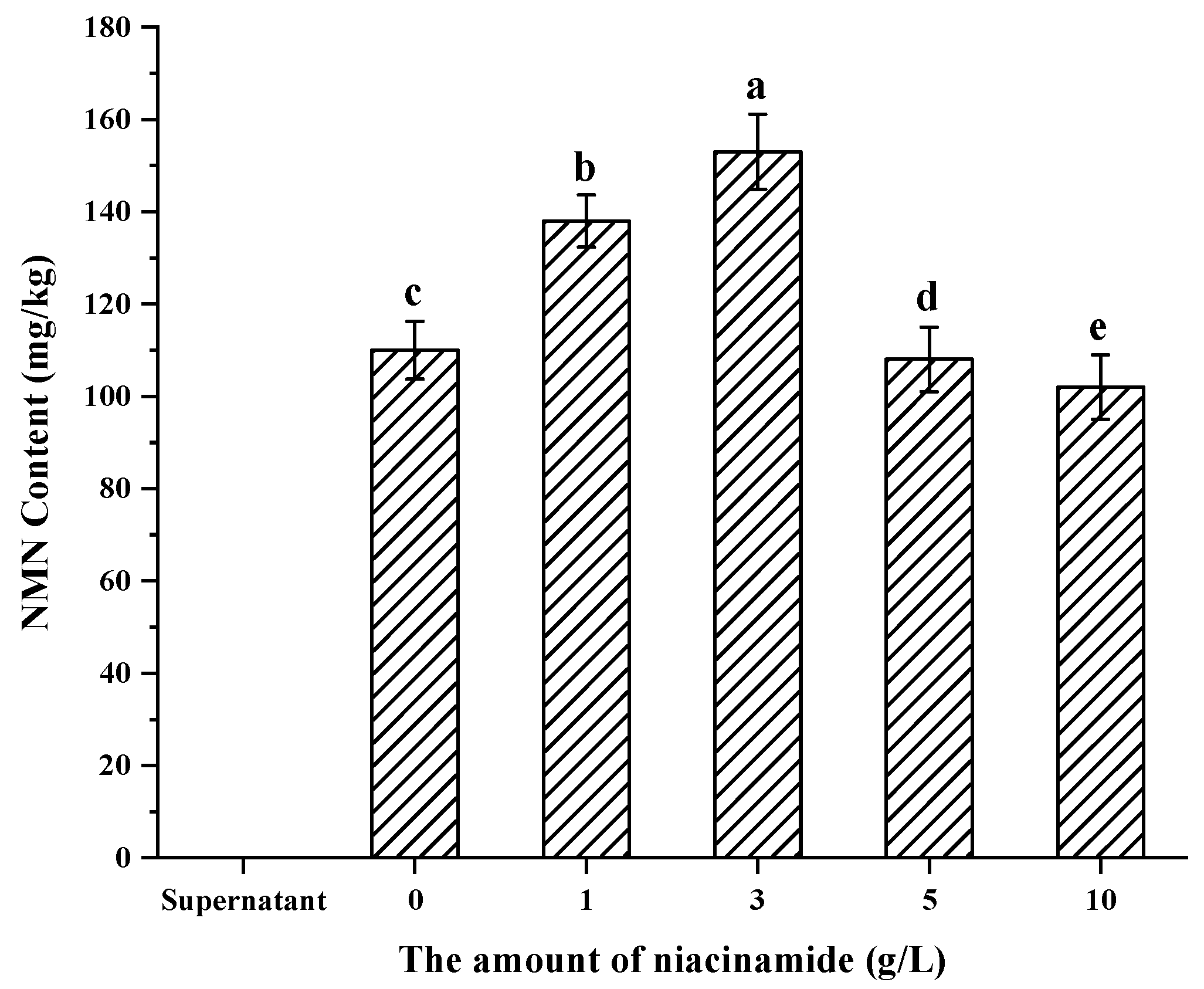
3.5. Confirmation of Appropriate Nam Addition Concentration for NMN Generation
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
Abbreviations
References
- Bieganowski, P.; Brenner, C. Discoveries of Nicotinamide Riboside as a Nutrient and Conserved NRKGenes Establish a Preiss-Handler Independent Route to NAD+ in Fungi and Humans. Cell 2004, 117, 495–502. [Google Scholar] [CrossRef] [PubMed]
- Nadeeshani, H.; Li, J.; Ying, T.; Zhang, B.; Lu, J. Nicotinamide mononucleotide (NMN) as an anti-aging health product—Promises and safety concerns. J. Adv. Res. 2022, 37, 267–278. [Google Scholar] [CrossRef] [PubMed]
- Alegre, G.F.S.; Pastore, G.M. NAD+ Precursors Nicotinamide Mononucleotide (NMN) and Nicotinamide Riboside (NR): Potential Dietary Contribution to Health. Curr. Nutr. Rep. 2023, 1–20. [Google Scholar] [CrossRef] [PubMed]
- Grozio, A.; Mills, K.F.; Yoshino, J.; Bruzzone, S.; Sociali, G.; Tokizane, K.; Lei, H.C.; Cunningham, R.; Sasaki, Y.; Migaud, M.E.; et al. Slc12a8 is a nicotinamide mononucleotide transporter. Nat. Metab. 2019, 1, 47–57. [Google Scholar] [CrossRef]
- Palikhe, S.; Nakagawa, T. NAD+ Metabolism in Aging. In Aging Mechanisms II: Longevity, Metabolism, and Brain Aging; Mori, N., Ed.; Springer Nature Singapore: Singapore, 2022; pp. 141–156. [Google Scholar]
- Liu, Y.; Gong, J.-S.; Marshall, G.; Su, C.; Shi, J.-S.; Xu, Z.-H. Technology and functional insights into the nicotinamide mononucleotide for human health. Appl. Microbiol. Biotechnol. 2023, 107, 4759–4775. [Google Scholar] [CrossRef]
- Yoshino, J.; Mills, K.F.; Yoon, M.J.; Imai, S. Nicotinamide mononucleotide, a key NAD(+) intermediate, treats the pathophysiology of diet- and age-induced diabetes in mice. Cell Metab. 2011, 14, 528–536. [Google Scholar] [CrossRef]
- Hosseini, L.; Vafaee, M.S.; Badalzadeh, R. Melatonin and Nicotinamide Mononucleotide Attenuate Myocardial Ischemia/Reperfusion Injury via Modulation of Mitochondrial Function and Hemodynamic Parameters in Aged Rats. J. Cardiovasc. Pharmacol. Ther. 2020, 25, 240–250. [Google Scholar] [CrossRef]
- Kiss, T.; Giles, C.B.; Tarantini, S.; Yabluchanskiy, A.; Balasubramanian, P.; Gautam, T.; Csipo, T.; Nyul-Toth, A.; Lipecz, A.; Szabo, C.; et al. Nicotinamide mononucleotide (NMN) supplementation promotes anti-aging miRNA expression profile in the aorta of aged mice, predicting epigenetic rejuvenation and anti-atherogenic effects. Geroscience 2019, 41, 419–439. [Google Scholar] [CrossRef]
- Guan, Y.; Wang, S.R.; Huang, X.Z.; Xie, Q.H.; Xu, Y.Y.; Shang, D.; Hao, C.M. Nicotinamide Mononucleotide, an NAD(+) Precursor, Rescues Age-Associated Susceptibility to AKI in a Sirtuin 1-Dependent Manner. J. Am. Soc. Nephrol. 2017, 28, 2337–2352. [Google Scholar] [CrossRef]
- Klimova, N.; Long, A.; Kristian, T. Nicotinamide mononucleotide alters mitochondrial dynamics by SIRT3-dependent mechanism in male mice. J. Neurosci. Res. 2019, 97, 975–990. [Google Scholar] [CrossRef]
- Li, H.R.; Liu, Q.; Zhu, C.L.; Sun, X.Y.; Sun, C.Y.; Yu, C.M.; Li, P.; Deng, X.M.; Wang, J.F. beta-Nicotinamide mononucleotide activates NAD+/SIRT1 pathway and attenuates inflammatory and oxidative responses in the hippocampus regions of septic mice. Redox Biol. 2023, 63, 102745. [Google Scholar] [CrossRef] [PubMed]
- Marinescu, G.C.; Popescu, R.G.; Stoian, G.; Dinischiotu, A. beta-nicotinamide mononucleotide (NMN) production in Escherichia coli. Sci. Rep. 2018, 8, 12278. [Google Scholar] [CrossRef]
- Shoji, S.; Yamaji, T.; Makino, H.; Ishii, J.; Kondo, A. Metabolic design for selective production of nicotinamide mononucleotide from glucose and nicotinamide. Metab. Eng. 2021, 65, 167–177. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Yasawong, M.; Yu, B. Metabolic engineering of Escherichia coli for biosynthesis of beta-nicotinamide mononucleotide from nicotinamide. Microb. Biotechnol. 2021, 14, 2581–2591. [Google Scholar] [CrossRef]
- Maharjan, A.; Singhvi, M.; Kafle, S.R.; Kim, B.S. Enhanced production of nicotinamide mononucleotide by high cell density culture of engineered Escherichia coli. Process Biochem. 2023, 131, 264–271. [Google Scholar] [CrossRef]
- Kato, M.; Lin, S.J. Regulation of NAD+ metabolism, signaling and compartmentalization in the yeast Saccharomyces cerevisiae. DNA Repair 2014, 23, 49–58. [Google Scholar] [CrossRef] [PubMed]
- Odoh, C.K.; Guo, X.; Arnone, J.T.; Wang, X.; Zhao, Z.K. The role of NAD and NAD precursors on longevity and lifespan modulation in the budding yeast, Saccharomyces cerevisiae. Biogerontology 2022, 23, 169–199. [Google Scholar] [CrossRef]
- He, Z.; Yang, X.; Tian, X.; Li, L.; Liu, M. Yeast Cell Surface Engineering of a Nicotinamide Riboside Kinase for the Production of beta-Nicotinamide Mononucleotide via Whole-Cell Catalysis. ACS Synth. Biol. 2022, 11, 3451–3459. [Google Scholar] [CrossRef]
- Goktas, H.; Dertli, E.; Sagdic, O. Comparison of functional characteristics of distinct Saccharomyces boulardii strains isolated from commercial food supplements. LWT 2021, 136, 110340. [Google Scholar] [CrossRef]
- Pais, P.; Almeida, V.; Yilmaz, M.; Teixeira, M.C. Saccharomyces boulardii: What Makes It Tick as Successful Probiotic? J. Fungi 2020, 6, 78. [Google Scholar] [CrossRef]
- Edwards-Ingram, L.; Gitsham, P.; Burton, N.; Warhurst, G.; Clarke, I.; Hoyle, D.; Oliver, S.G.; Stateva, L. Genotypic and physiological characterization of Saccharomyces boulardii, the probiotic strain of Saccharomyces cerevisiae. Appl. Environ. Microbiol. 2007, 73, 2458–2467. [Google Scholar] [CrossRef] [PubMed]
- Łukaszewicz, M. Saccharomyces cerevisiae var. boulardii–Probiotic Yeast. In Probiotics; IntechOpen: London, UK, 2012. [Google Scholar]
- Evans, C.; Bogan, K.L.; Song, P.; Burant, C.F.; Kennedy, R.T.; Brenner, C. NAD+ metabolite levels as a function of vitamins and calorie restriction: Evidence for different mechanisms of longevity. BMC Chem. Biol. 2010, 10, 2. [Google Scholar] [CrossRef] [PubMed]
- Trammell, S.A.; Brenner, C. Targeted, LCMS-based Metabolomics for Quantitative Measurement of NAD(+) Metabolites. Comput. Struct. Biotechnol. J. 2013, 4, e201301012. [Google Scholar] [CrossRef] [PubMed]
- Yamada, K.; Hara, N.; Shibata, T.; Osago, H.; Tsuchiya, M. The simultaneous measurement of nicotinamide adenine dinucleotide and related compounds by liquid chromatography/electrospray ionization tandem mass spectrometry. Anal. Biochem. 2006, 352, 282–285. [Google Scholar] [CrossRef]
- Mashabela, M.D.; Masamba, P.; Kappo, A.P. Metabolomics and Chemoinformatics in Agricultural Biotechnology Research: Complementary Probes in Unravelling New Metabolites for Crop Improvement. Biology 2022, 11, 1156. [Google Scholar] [CrossRef]
- Utpott, M.; Rodrigues, E.; Rios, A.O.; Mercali, G.D.; Flores, S.H. Metabolomics: An analytical technique for food processing evaluation. Food Chem. 2022, 366, 130685. [Google Scholar] [CrossRef]
- Fan, T.; Qu, J.; Wang, L.; Zhang, J.; Yang, X.; Zhang, H.; Qin, Y.; Tao, Y.; Jin, G. Genome sequencing, assembly, and characterization of Pichia fermentans Z9Y-3 as a non-Saccharomyces yeast with aroma enhancing potential. Food Biosci. 2023, 53, 102701. [Google Scholar] [CrossRef]
- Lopez-Varea, A.; Vega-Cuesta, P.; Ruiz-Gomez, A.; Ostale, C.M.; Molnar, C.; Hevia, C.F.; Martin, M.; Organista, M.F.; de Celis, J.; Culi, J.; et al. Genome-wide phenotypic RNAi screen in the Drosophila wing: Phenotypic description of functional classes. G3 (Bethesda) 2021, 11, jkab349. [Google Scholar] [CrossRef]
- Marinescu, G.C.; Popescu, R.G.; Dinischiotu, A. Size Exclusion Chromatography Method for Purification of Nicotinamide Mononucleotide (NMN) from Bacterial Cells. Sci. Rep. 2018, 8, 4433. [Google Scholar] [CrossRef]
- Ummarino, S.; Mozzon, M.; Zamporlini, F.; Amici, A.; Mazzola, F.; Orsomando, G.; Ruggieri, S.; Raffaelli, N. Simultaneous quantitation of nicotinamide riboside, nicotinamide mononucleotide and nicotinamide adenine dinucleotide in milk by a novel enzyme-coupled assay. Food Chem. 2017, 221, 161–168. [Google Scholar] [CrossRef]
- Jiao, W.; Xiao, Y.; Qian, X.; Tong, M.; Hu, Y.; Hou, R.; Hua, R. Optimized combination of dilution and refined QuEChERS to overcome matrix effects of six types of tea for determination eight neonicotinoid insecticides by ultra performance liquid chromatography-electrospray tandem mass spectrometry. Food Chem. 2016, 210, 26–34. [Google Scholar] [CrossRef] [PubMed]
- Stahnke, H.; Kittlaus, S.; Kempe, G.; Alder, L. Reduction of matrix effects in liquid chromatography-electrospray ionization-mass spectrometry by dilution of the sample extracts: How much dilution is needed? Anal. Chem. 2012, 84, 1474–1482. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Yang, H.; Jin, H.; Turkez, H.; Ozturk, G.; Doganay, H.L.; Zhang, C.; Nielsen, J.; Uhlen, M.; Boren, J.; et al. The acute effect of different NAD(+) precursors included in the combined metabolic activators. Free. Radic. Biol. Med. 2023, 205, 77–89. [Google Scholar] [CrossRef]
- Gasperi, V.; Sibilano, M.; Savini, I.; Catani, M.V. Niacin in the Central Nervous System: An Update of Biological Aspects and Clinical Applications. Int. J. Mol. Sci. 2019, 20, 974. [Google Scholar] [CrossRef] [PubMed]
- Fletcher, R.S.; Lavery, G.G. The emergence of the nicotinamide riboside kinases in the regulation of NAD+ metabolism. J. Mol. Endocrinol. 2018, 61, R107–R121. [Google Scholar] [CrossRef]
- Tempel, W.; Rabeh, W.M.; Bogan, K.L.; Belenky, P.; Wojcik, M.; Seidle, H.F.; Nedyalkova, L.; Yang, T.; Sauve, A.A.; Park, H.W.; et al. Nicotinamide riboside kinase structures reveal new pathways to NAD+. PLoS Biol. 2007, 5, e263. [Google Scholar] [CrossRef]
- Gallo, C.M.; Smith, D.L., Jr.; Smith, J.S. Nicotinamide clearance by Pnc1 directly regulates Sir2-mediated silencing and longevity. Mol. Cell. Biol. 2004, 24, 1301–1312. [Google Scholar] [CrossRef]
| Compound | Precursor Ion [M + H]+ (m/z) | Product Ions (m/z) | Fragmentor Voltage (V) | Collision Energy (eV) |
|---|---|---|---|---|
| NMN | 335.1 | 123.1 */97.2 | 380 | 12/22 |
| NR | 255.1 | 123.2 */80.1 | 380 | 10/45 |
| NA | 124.0 | 80.1 */78.1 | 380 | 25/25 |
| Nam | 123.1 | 80.1 */53.3 | 380 | 23/42 |
| NAD+ | 664.1 | 136 */427.9 | 380 | 50/30 |
| Analytes | Linear Equation | R2 | Recoveries,% (RSD,%) n = 6 Spiked Level (mg/kg) | LOQ (μg/kg) | LOD (μg/kg) | ||
|---|---|---|---|---|---|---|---|
| 10 | 50 | 100 | |||||
| NMN | Y = 287.42X + 1536.15 | 0.9994 | 76% (4.6) | 78% (6.2) | 89% (7.1) | 0.055 | 0.016 |
| NR | Y = 21,162.59X + 3489.69 | 0.9999 | 70% (3.4) | 82% (5.7) | 75% (4.9) | 0.003 | 0.001 |
| NA | Y = 7151.59X + 11687.73 | 0.9995 | 72% (3.9) | 84% (4.5) | 80% (2.7) | 0.034 | 0.010 |
| Nam | Y = 29,987.60X + 12,387.99 | 0.9996 | 71% (4.1) | 73% (5.6) | 82% (3.9) | 0.082 | 0.025 |
| NAD+ | Y = 90.48X + 199.94 | 0.9989 | 85% (5.4) | 79% (6.3) | 74% (4.8) | 0.016 | 0.005 |
| Features | Description |
|---|---|
| Clean reads bases | 1,083,269,987 bp |
| Scaffolds | 153 |
| Total bases in scaffold (Genome Size) | 11,541,211 bp |
| Contigs | 374 |
| Total bases in contigs | 11,538,623 bp |
| N50 | 100,937 bp |
| GC content | 38.05% |
| Gene coding (CDS) | 4275 |
| Gene total length | 9,367,056 bp |
| Gene average length | 2191.12 bp |
| Gene/Genome | 81.16% |
| tRNA | 282 |
| rRNA | 2 |
| SINEs a | 18 |
| LINEs b | 348 |
| LTR c | 701 |
| Simple repeat | 2544 |
| Genes of KEGG | 2388 |
| Genes of COG | 1369 |
| Gene ID | Location | Start | End | Length (bp) | Non-Redundant Protein Sequence Database Description | Gene Name | Swiss-Prot Description | GO ID | COG ID | COG Type | KO ID | Pfam ID |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| gene0375 | Scaffold3 | 97834 | 99978 | 2145 | Qns1p | nadE | Glutamine-dependent NAD(+) synthetase OS = Saccharomyces cerevisiae (strain ATCC 204,508/S288c) | GO:0005488; GO:0003824; GO:0110165; GO:0008152; GO:0009987 | COG0171 | H | K01950 | PF00795; PF02540 |
| gene1306 | Scaffold10 | 253886 | 252160 | 1727 | Npy1p | nudC | NAD-capped RNA hydrolase NPY1 OS = Saccharomyces cerevisiae (strain ATCC 204,508/S288c) | GO:0005488; GO:0003824; GO:0110165; GO:0008152; GO:0009987 | COG2816 | F | K03426 | PF00293 |
| gene1408 | Scaffold11 | 233570 | 232548 | 1023 | Urh1p | - | Uridine nucleosidase OS = Saccharomyces cerevisiae (strain ATCC 204,508/S288c) | GO:0003824; GO:0008152; GO:0009987 | COG1957 | F | K01240 | PF01156 |
| gene1812 | Scaffold16 | 58366aa | 60054 | 1689 | NAD-dependent histone deacetylase SIR2 | - | NAD-dependent histone deacetylase SIR2 OS = Saccharomyces cerevisiae (strain ATCC 204,508/S288c) | GO:0005488; GO:0003824; GO:0032991; GO:0110165; GO:0065007; GO:0008152; GO:0009987; GO:0051179; GO:0050896 | COG0846 | O | K11121 | PF02146; PF04574 |
| gene2215 | Scaffold21 | 115232 | 116167 | 936 | PNP1p Purine nucleoside phosphorylase | punA | Purine nucleoside phosphorylase OS = Saccharomyces cerevisiae (strain ATCC 204,508/S288c) | GO:0003824; GO:0110165; GO:0008152; GO:0009987 | COG0005 | F | K03783 | PF01048 |
| gene3938 | Scaffold61 | 37137 | 38489 | 1353 | IMP 5′-nucleotidase | - | IMP-specific 5′-nucleotidase 1 OS = Saccharomyces cerevisiae (strain ATCC 204,508/S288c) | GO:0005488; GO:0003824; GO:0008152; GO:0009987 | - | - | K18550 | PF06437 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Song, M.; Yin, C.; Xu, Q.; Liu, Y.; Zhang, H.; Liu, X.; Yan, H. Enhanced Production of β-Nicotinamide Mononucleotide with Exogenous Nicotinamide Addition in Saccharomyces boulardii-YS01. Foods 2023, 12, 2897. https://doi.org/10.3390/foods12152897
Song M, Yin C, Xu Q, Liu Y, Zhang H, Liu X, Yan H. Enhanced Production of β-Nicotinamide Mononucleotide with Exogenous Nicotinamide Addition in Saccharomyces boulardii-YS01. Foods. 2023; 12(15):2897. https://doi.org/10.3390/foods12152897
Chicago/Turabian StyleSong, Meijie, Chunhua Yin, Qianqian Xu, Yang Liu, Haiyang Zhang, Xiaolu Liu, and Hai Yan. 2023. "Enhanced Production of β-Nicotinamide Mononucleotide with Exogenous Nicotinamide Addition in Saccharomyces boulardii-YS01" Foods 12, no. 15: 2897. https://doi.org/10.3390/foods12152897
APA StyleSong, M., Yin, C., Xu, Q., Liu, Y., Zhang, H., Liu, X., & Yan, H. (2023). Enhanced Production of β-Nicotinamide Mononucleotide with Exogenous Nicotinamide Addition in Saccharomyces boulardii-YS01. Foods, 12(15), 2897. https://doi.org/10.3390/foods12152897






