Dynamic Modelling to Describe the Effect of Plant Extracts and Customised Starter Culture on Staphylococcus aureus Survival in Goat’s Raw Milk Soft Cheese
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Materials’ and Extracts’ Preparation
2.2. Bacterial Strains
2.3. Inoculation of Bacterial Strains in Milk and Cheese Production: Incorporation of Starter Culture or Plant Extract
2.4. Microbiological and Physicochemical Analysis throughout Cheese Ripening
2.5. Modelling of S. aureus and LAB Behaviour during Cheese Ripening
2.5.1. S. aureus Behaviour during Cheese Ripening
2.5.2. LAB Behaviour during Cheese Ripening
2.5.3. Estimation of Parameters
3. Results and Discussion
3.1. pH Decay during Cheese Ripening
3.2. S. aureus Behaviour during Cheese Ripening
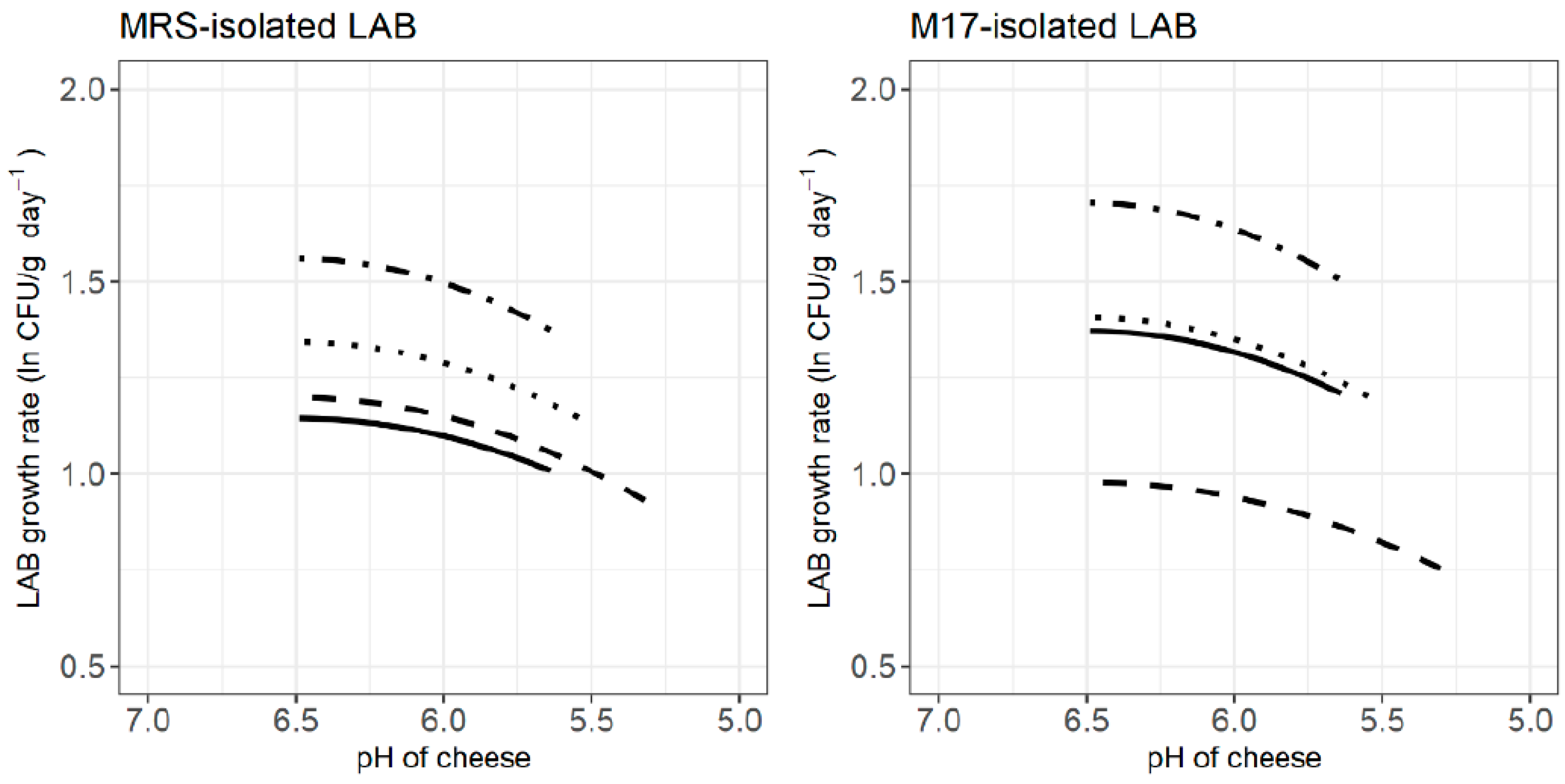
3.3. LAB Behaviour during Cheese Ripening
4. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Jamali, H.; Paydar, M.; Radmehr, B.; Ismail, S.; Dadrasnia, A. Prevalence and antimicrobial resistance of Staphylococcus aureus isolated from raw milk and dairy products. Food Control 2015, 54, 383–388. [Google Scholar] [CrossRef]
- Cremonesi, P.; Perez, G.; Pisoni, G.; Moroni, P.; Morandi, S.; Luzzana, M.; Brasca, M.; Castiglioni, B. Detection of enterotoxigenic Staphylococcus aureus isolates in raw milk cheese. Lett. Appl. Microbiol. 2007, 45, 586–591. [Google Scholar] [CrossRef] [PubMed]
- Gücükoğlu, A.; Kevenk, T.O.; Uyanik, T.; Çadirci, Ö.; Terzi, G.; Alişarli, M. Detection of Enterotoxigenic Staphylococcus aureus in Raw Milk and Dairy Products by Multiplex PCR. J. Food Sci. 2012, 77, M620–M623. [Google Scholar] [CrossRef] [PubMed]
- Rosengren, Å.; Fabricius, A.; Guss, B.; Sylvén, S.; Lindqvist, R. Occurrence of foodborne pathogens and characterization of Staphylococcus aureus in cheese produced on farm-dairies. Int. J. Food Microbiol. 2010, 144, 263–269. [Google Scholar] [CrossRef]
- Rola, J.G.; Czubkowska, A.; Korpysa-Dzirba, W.; Osek, J. Occurrence of Staphylococcus aureus on farms with small scale production of raw milk cheeses in Poland. Toxins 2016, 8, 62. [Google Scholar] [CrossRef]
- Prabakusuma, A.S.; Zhu, J.; Shi, Y.; Ma, Q.; Zhao, Q.; Yang, Z.; Xu, Y.; Huang, A. Prevalence and antimicrobial resistance profiling of Staphylococcus aureus isolated from traditional cheese in Yunnan, China. 3 Biotech 2022, 12, 1. [Google Scholar] [CrossRef]
- Armani, M.; Macori, G.; Gallina, S.; Tavella, A.; Giusti, M.; Paolazzi, G.; Trentini, L.; Rabini, M.; Decastelli, L.; Lombardo, D. Coagulase positive staphylococci and food poisoning toxins—A case study of an outbreak investigation occurred in a sheperd hut. Int. J. Infect. Dis. 2016, 45, 464. [Google Scholar] [CrossRef]
- Jørgensen, H.J.; Mørk, T.; Rørvik, L.M. The occurrence of Staphylococcus aureus on a farm with small-scale production of raw milk cheese. J. Dairy Sci. 2005, 88, 3810–3817. [Google Scholar] [CrossRef]
- Johler, S.; Weder, D.; Bridy, C.; Huguenin, M.-C.; Robert, L.; Hummerjohann, J.; Stephan, R. Outbreak of staphylococcal food poisoning among children and staff at a Swiss boarding school due to soft cheese made from raw milk. J. Dairy Sci. 2015, 98, 2944–2948. [Google Scholar] [CrossRef]
- Filipello, V.; Bonometti, E.; Campagnani, M.; Bertoletti, I.; Romano, A.; Zuccon, F.; Campanella, C.; Losio, M.N.; Finazzi, G. Investigation and Follow-Up of a Staphylococcal Food Poisoning Outbreak Linked to the Consumption of Traditional Hand-Crafted Alm Cheese. Pathogens 2020, 9, 1064. [Google Scholar] [CrossRef]
- Gajewska, J.; Zakrzewski, A.; Chajęcka-Wierzchowska, W.; Zadernowska, A. Meta-analysis of the global occurrence of S. aureus in raw cattle milk and artisanal cheeses. Food Control 2023, 147, 109603. [Google Scholar] [CrossRef]
- Medveďová, A.; Valík, L.; Sirotná, Z.; Liptáková, D. Growth characterisation of Staphylococcus aureus in milk: A quantitative approach. Czech J. Food Sci. 2009, 27, 443–453. [Google Scholar] [CrossRef]
- Mohamed, F.A.E.F.; Salama, H.H.; El-Sayed, S.M.; El-Sayed, H.S.; Zahran, H.A.H. Utilization of Natural Antimicrobial and Antioxidant of Moringa oleifera Leaves Extract in Manufacture of Cream Cheese. J. Biol. Sci. 2018, 18, 92–106. [Google Scholar] [CrossRef]
- Carvalho, F.; Rodrigues, A.; Gomes, D.M.G.S.; Ferreira, F.M.L.; Dias, S.P.; Pereira, C.J.D.; Henriques, M.H.F. Improvement of Ripened Cheese Quality and Safety with Thymus mastichina L. Bioactive Extracts. Adv. Biotechnol. Food Ind. 2018, 14, 197–211. [Google Scholar] [CrossRef]
- Shan, B.; Cai, Y.-Z.; Brooks, J.D.; Corke, H. Potential application of spice and herb extracts as natural preservatives in cheese. J. Med. Food 2011, 14, 284–290. [Google Scholar] [CrossRef]
- Gonzales-Barron, U.; Campagnollo, F.B.; Schaffner, D.W.; Sant’Ana, A.S.; Cadavez, V.A.P. Behavior of Listeria monocytogenes in the presence or not of intentionally-added lactic acid bacteria during ripening of artisanal Minas semi-hard cheese. Food Microbiol. 2020, 91, 103545. [Google Scholar] [CrossRef]
- Le Marc, Y.; Valík, Ľ.; Medveďová, A. Modelling the effect of the starter culture on the growth of Staphylococcus aureus in milk. Int. J. Food Microbiol. 2009, 129, 306–311. [Google Scholar] [CrossRef]
- Alomar, J.; Loubiere, P.; Delbes, C.; Nouaille, S.; Montel, M.C. Effect of Lactococcus garvieae, Lactococcus lactis and Enterococcus faecalis on the behaviour of Staphylococcus aureus in microfiltered milk. Food Microbiol. 2008, 25, 502–508. [Google Scholar] [CrossRef]
- European Food and Safety Authority. Introduction of a Qualified Presumption of Safety (QPS) approach for assessment of selected microorganisms referred to EFSA—Opinion of the Scientific Committee. EFSA J. 2007, 5, 587. [Google Scholar] [CrossRef]
- Silva, B.N.; Cadavez, V.; Ferreira-Santos, P.; Alves, M.J.; Ferreira, I.C.F.R.; Barros, L.; Teixeira, J.A.; Gonzales-Barron, U. Chemical Profile and Bioactivities of Extracts from Edible Plants Readily Available in Portugal. Foods 2021, 10, 673. [Google Scholar] [CrossRef]
- Silva, B.N.; Cadavez, V.; Caleja, C.; Pereira, E.; Calhelha, R.C.; Añibarro-Ortega, M.; Finimundy, T.; Kostić, M.; Soković, M.; Teixeira, J.A.; et al. Phytochemical Composition and Bioactive Potential of Melissa officinalis L., Salvia officinalis L. and Mentha spicata L. Extracts. Foods 2023, 12, 947. [Google Scholar] [CrossRef] [PubMed]
- Silva, B.N.; Faria, A.S.; Cadavez, V.; Teixeira, J.A.; Gonzales-Barron, U. Technological Potential of Lactic Acid Bacteria Isolated from Portuguese Goat’s Raw Milk Cheeses. Biol. Life Sci. Forum 2021, 6, 9. [Google Scholar] [CrossRef]
- Cadavez, V.A.P.; Campagnollo, F.B.; Silva, R.A.; Duffner, C.M.; Schaffner, D.W.; Sant’ana, A.S.; Gonzales-Barron, U. A comparison of dynamic tertiary and competition models for describing the fate of Listeria monocytogenes in Minas fresh cheese during refrigerated storage. Food Microbiol. 2019, 79, 48–60. [Google Scholar] [CrossRef]
- ISO 6888-1:2021; Microbiology of the Food Chain—Horizontal Method for the Enumeration of Coagulase-Positive Staphylococci (Staphylococcus aureus and Other Species)—Part 1: Method Using Baird-Parker Agar Medium. International Organization of Standardization: Geneva, Switzerland, 2021.
- ISO 15214:1998; Microbiology of Food and Animal Feeding Stuffs—Horizontal Method for the Enumeration of Mesophilic Lactic Acid Bacteria—Colony-Count Technique at 30 Degrees C. International Organization for Standardization: Geneva, Switzerland, 1998.
- Geeraerd, A.H.; Herremans, C.H.; Van Impe, J.F. Structural model requirements to describe microbial inactivation during a mild heat treatment. Int. J. Food Microbiol. 2000, 59, 185–209. [Google Scholar] [CrossRef] [PubMed]
- Geeraerd, A.H.; Valdramidis, V.P.; Van Impe, J.F. GInaFiT, a freeware tool to assess non-log-linear microbial survivor curves. Int. J. Food Microbiol. 2005, 102, 95–105. [Google Scholar] [CrossRef] [PubMed]
- Huang, L. Mathematical modeling and numerical analysis of the growth of non-O157 Shiga toxin-producing Escherichia coli in spinach leaves. Int. J. Food Microbiol. 2012, 160, 32–41. [Google Scholar] [CrossRef]
- Huang, L. Optimization of a new mathematical model for bacterial growth. Food Control 2013, 32, 283–288. [Google Scholar] [CrossRef]
- Rosso, L.; Lobry, J.R.; Bajard, S.; Flandrois, J.P. Convenient model to describe the combined effects of temperature and pH on microbial growth. Appl. Environ. Microbiol. 1995, 61, 610–616. [Google Scholar] [CrossRef]
- Wolf, B.F.; Fogler, H.S. Growth of Leuconostoc mesenteroides NRRL-B523 in an alkaline medium: Suboptimal pH growth inhibition of a lactic acid bacterium. Biotechnol. Bioeng. 2005, 89, 96–101. [Google Scholar] [CrossRef]
- Drosinos, E.H.; Mataragas, M.; Nasis, P.; Galiotou, M.; Metaxopoulos, J. Growth and bacteriocin production kinetics of Leuconostoc mesenteroides E131. J. Appl. Microbiol. 2005, 99, 1314–1323. [Google Scholar] [CrossRef]
- Krier, F.; Revol-Junelles, A.M.; Germain, P. Influence of temperature and pH on production of two bacteriocins by Leuconostoc mesenteroides subsp. mesenteroides FR52 during batch fermentation. Appl. Microbiol. Biotechnol. 1998, 50, 359–363. [Google Scholar] [CrossRef] [PubMed]
- Kim, W.S.; Ren, J.; Dunn, N.W. Differentiation of Lactococcus lactis subspecies lactis and subspecies cremoris strains by their adaptive response to stresses. FEMS Microbiol. Lett. 1999, 171, 57–65. [Google Scholar] [CrossRef] [PubMed]
- Di Biase, M.; Le Marc, Y.; Bavaro, A.R.; De Bellis, P.; Lonigro, S.L.; Lavermicocca, P.; Postollec, F.; Valerio, F. A Predictive Growth Model for Pro-technological and Probiotic Lacticaseibacillus paracasei Strains Fermenting White Cabbage. Front. Microbiol. 2022, 13, 907393. [Google Scholar] [CrossRef] [PubMed]
- M’hamed, A.C.; Ncib, K.; Merghni, A.; Migaou, M.; Lazreg, H.; Mejdi, S.; Noumi, E.; Mansour, M.B.; Maaroufi, R.M. Characterization of Probiotic Properties of Lacticaseibacillus paracasei L2 Isolated from a Traditional Fermented Food “Lben”. Life 2023, 13, 21. [Google Scholar] [CrossRef]
- Mataragas, M.; Metaxopoulos, J.; Galiotou, M.; Drosinos, E.H. Influence of pH and temperature on growth and bacteriocin production by Leuconostoc mesenteroides L124 and Lactobacillus curvatus L442. Meat Sci. 2003, 64, 265–271. [Google Scholar] [CrossRef] [PubMed]
- Holzapfel, W.H.; Björkroth, J.A.; Dicks, L.M.T. Leuconostoc. In Bergey’s Manual of Systematics of Archaea and Bacteria; Wiley: Hoboken, NJ, USA, 2015; pp. 1–23. [Google Scholar] [CrossRef]
- Sánchez, C.; Neves, A.R.; Cavalheiro, J.; dos Santos, M.M.; García-Quintáns, N.; López, P.; Santos, H. Contribution of citrate metabolism to the growth of Lactococcus lactis CRL264 at low pH. Appl. Environ. Microbiol. 2008, 74, 1136–1144. [Google Scholar] [CrossRef]
- Hutkins, R.W.; Nannen, N.L. pH Homeostasis in Lactic Acid Bacteria. J. Dairy Sci. 1993, 76, 2354–2365. [Google Scholar] [CrossRef]
- Sawatari, Y.; Yokota, A. Diversity and Mechanisms of Alkali Tolerance in Lactobacilli. Appl. Environ. Microbiol. 2007, 73, 3909–3915. [Google Scholar] [CrossRef]
- Ricciardi, A.; Storti, L.V.; Giavalisco, M.; Parente, E.; Zotta, T. The Effect of Respiration, pH, and Citrate Co-Metabolism on the Growth, Metabolite Production and Enzymatic Activities of Leuconostoc mesenteroides subsp. cremoris E30. Foods 2022, 11, 535. [Google Scholar] [CrossRef]
- Rallu, F.; Gruss, A.; Ehrlich, S.D.; Maguin, E. Acid- and multistress-resistant mutants of Lactococcus lactis: Identification of intracellular stress signals. Mol. Microbiol. 2000, 35, 517–528. [Google Scholar] [CrossRef]
- Mercade, M.; Lindley, N.D.; Loubière, P. Metabolism of Lactococcus lactis subsp. cremoris MG 1363 in acid stress conditions. Int. J. Food Microbiol. 2000, 55, 161–165. [Google Scholar] [CrossRef]
- Reale, A.; Di Renzo, T.; Rossi, F.; Zotta, T.; Iacumin, L.; Preziuso, M.; Parente, E.; Sorrentino, E.; Coppola, R. Tolerance of Lactobacillus casei, Lactobacillus paracasei and Lactobacillus rhamnosus strains to stress factors encountered in food processing and in the gastro-intestinal tract. LWT—Food Sci. Technol. 2015, 60, 721–728. [Google Scholar] [CrossRef]
- Possas, A.; Bonilla-Luque, O.M.; Valero, A. From Cheese-Making to Consumption: Exploring the Microbial Safety of Cheeses through Predictive Microbiology Models. Foods 2021, 10, 355. [Google Scholar] [CrossRef] [PubMed]
- Campagnollo, F.; Margalho, L.; Kamimura, B.; Feliciano, M.; Freire, L.; Lopes, L.; Alvarenga, V.; Cadavez, V.; Gonzales-Barron, U.; Schaffner, D.; et al. Selection of indigenous lactic acid bacteria presenting anti-listerial activity, and their role in reducing the maturation period and assuring the safety of traditional Brazilian cheeses. Food Microbiol. 2018, 73, 288–297. [Google Scholar] [CrossRef] [PubMed]
- Martinez, R.C.; Staliano, C.D.; Vieira, A.D.S.; Villarreal, M.L.M.; Todorov, S.D.; Saad, S.M.I.; Franco, B.D.G.M. Bacteriocin production and inhibition of Listeria monocytogenes by Lactobacillus sakei subsp. sakei 2a in a potentially synbiotic cheese spread. Food Microbiol. 2015, 48, 143–152. [Google Scholar] [CrossRef] [PubMed]
- Pingitore, E.V.; Todorov, S.D.; Sesma, F.; Franco, B.D.G.M. Application of bacteriocinogenic Enterococcus mundtii CRL35 and Enterococcus faecium ST88Ch in the control of Listeria monocytogenes in fresh Minas cheese. Food Microbiol. 2012, 32, 38–47. [Google Scholar] [CrossRef] [PubMed]
- Nascimento, M.S.; Moreno, I.; Kuaye, A.Y. Applicability of bacteriocin-producing Lactobacillus plantarum, Enterococcus faecium and Lactococcus lactis ssp. lactis as adjunct starter in Minas Frescal cheesemaking. Int. J. Dairy Technol. 2008, 61, 352–357. [Google Scholar] [CrossRef]
- Østergaard, N.B.; Eklöw, A.; Dalgaard, P. Modelling the effect of lactic acid bacteria from starter- and aroma culture on growth of Listeria monocytogenes in cottage cheese. Int. J. Food Microbiol. 2014, 188, 15–25. [Google Scholar] [CrossRef]
- Mahajan, D.; Bhat, Z.F.; Kumar, S. Pine needles (Cedrus deodara (Roxb.) Loud.) extract as a novel preservative in cheese. Food Pack. Shelf Life 2016, 7, 20–25. [Google Scholar] [CrossRef]
- Abd El-Aziz, M.; Mohamed, S.H.S.; Seleet, F.L. Production and Evaluation of Soft Cheese Fortified with Ginger Extract as a Functional Dairy Food. Polish J. Food Nutr. Sci. 2012, 62, 77–83. [Google Scholar] [CrossRef]
- Ritota, M.; Manzi, P. Natural Preservatives from Plant in Cheese Making. Animals 2020, 10, 749. [Google Scholar] [CrossRef] [PubMed]
- Li, H.-Z.; Ren, Z.; Reddy, N.V.; Hou, T.; Zhang, Z.J. In silico evaluation of antimicrobial, antihyaluronidase and bioavailability parameters of rosmarinic acid in Perilla frutescens leaf extracts. SN Appl. Sci. 2020, 2, 1547. [Google Scholar] [CrossRef]
- Abedini, A.; Roumy, V.; Mahieux, S.; Biabiany, M.; Standaert-Vitse, A.; Rivière, C.; Sahpaz, S.; Bailleul, F.; Neut, C.; Hennebelle, T. Rosmarinic Acid and Its Methyl Ester as Antimicrobial Components of the Hydromethanolic Extract of Hyptis atrorubens Poit. (Lamiaceae). Evid. Based Complement. Altern. Med. 2013, 2013, 604536. [Google Scholar] [CrossRef] [PubMed]
- Honório, V.G.; Bezerra, J.; Souza, G.T.; Carvalho, R.J.; Gomes-Neto, N.J.; Figueiredo, R.C.B.Q.; Melo, J.V.; Souza, E.L.; Magnani, M. Inhibition of Staphylococcus aureus cocktail using the synergies of oregano and rosemary essential oils or carvacrol and 1,8-cineole. Front. Microbiol. 2015, 6, 1223. [Google Scholar] [CrossRef] [PubMed]
- Bais, H.P.; Walker, T.S.; Schweizer, H.P.; Vivanco, J.M. Root specific elicitation and antimicrobial activity of rosmarinic acid in hairy root cultures of Ocimum basilicum. Plant Physiol. Biochem. 2002, 40, 983–995. [Google Scholar] [CrossRef]
- Borges, A.; Ferreira, C.; Saavedra, M.J.; Simões, M. Antibacterial activity and mode of action of ferulic and gallic acids against pathogenic bacteria. Microb. Drug Resist. 2013, 19, 256–265. [Google Scholar] [CrossRef]
- Rúa, J.; Fernández-Álvarez, L.; Gutiérrez-Larraínzar, M.; Del Valle, P.; De Arriaga, D.; García-Armesto, M.R. Screening of Phenolic Antioxidants for Their Inhibitory Activity Against Foodborne Staphylococcus aureus Strains. Foodborne Pathog. Dis. 2010, 7, 695–705. [Google Scholar] [CrossRef]
- Tavares, W.S.; Martin-Pastor, M.; Tavares, A.G.; Sousa, F.F.O. Biopharmaceutical Activities Related to Ellagic Acid, Chitosan, and Zein and Their Improvement by Association. J. Food Sci. 2018, 83, 2970–2975. [Google Scholar] [CrossRef]
- Alves, M.J.; Ferreira, I.C.F.R.; Froufe, H.J.C.; Abreu, R.M.V.; Martins, A.; Pintado, M. Antimicrobial activity of phenolic compounds identified in wild mushrooms, SAR analysis and docking studies. J. Appl. Microbiol. 2013, 115, 346–357. [Google Scholar] [CrossRef]
- Yang, Y.; Tao, B.; Gong, Y.; Chen, R.; Yang, W.; Lin, C.; Chen, M.; Qin, L.; Jia, Y.; Cai, K. Functionalization of Ti substrate with pH-responsive naringin-ZnO nanoparticles for the reconstruction of large bony after osteosarcoma resection. J. Biomed. Mater. Res. A 2020, 108, 2190–2205. [Google Scholar] [CrossRef]
- Attia, G.H.; Marrez, D.A.; Mohammed, M.A.; Albarqi, H.A.; Ibrahim, A.M.; El Raey, M.A. Synergistic Effect of Mandarin Peels and Hesperidin with Sodium Nitrite against Some Food Pathogen Microbes. Molecules 2021, 26, 3186. [Google Scholar] [CrossRef] [PubMed]
- Wemmenhove, E.; van Valenberg, H.J.F.; van Hooijdonk, A.C.M.; Wells-Bennik, M.H.J.; Zwietering, M.H. Factors that inhibit growth of Listeria monocytogenes in nature-ripened Gouda cheese: A major role for undissociated lactic acid. Food Control 2018, 84, 413–418. [Google Scholar] [CrossRef]
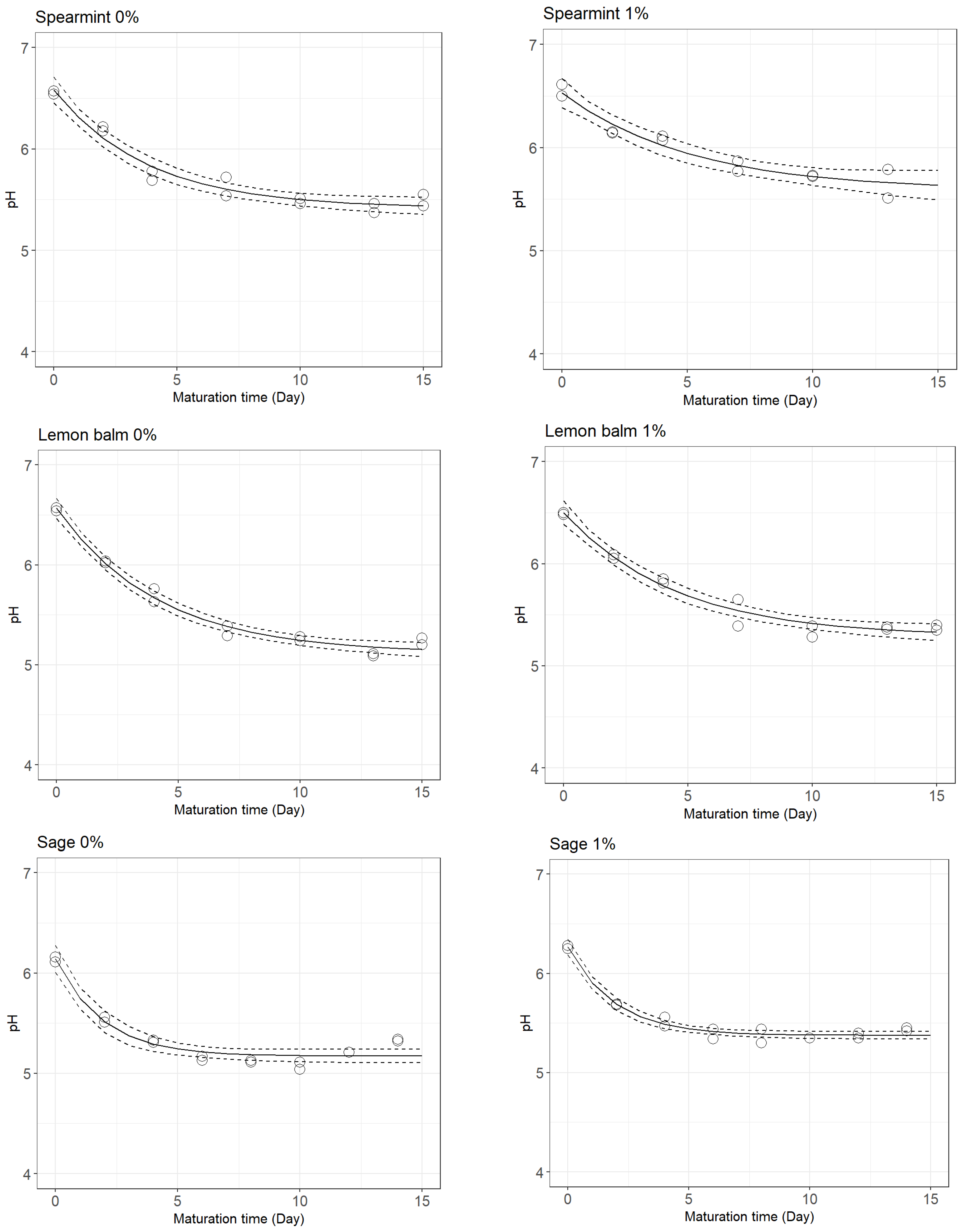
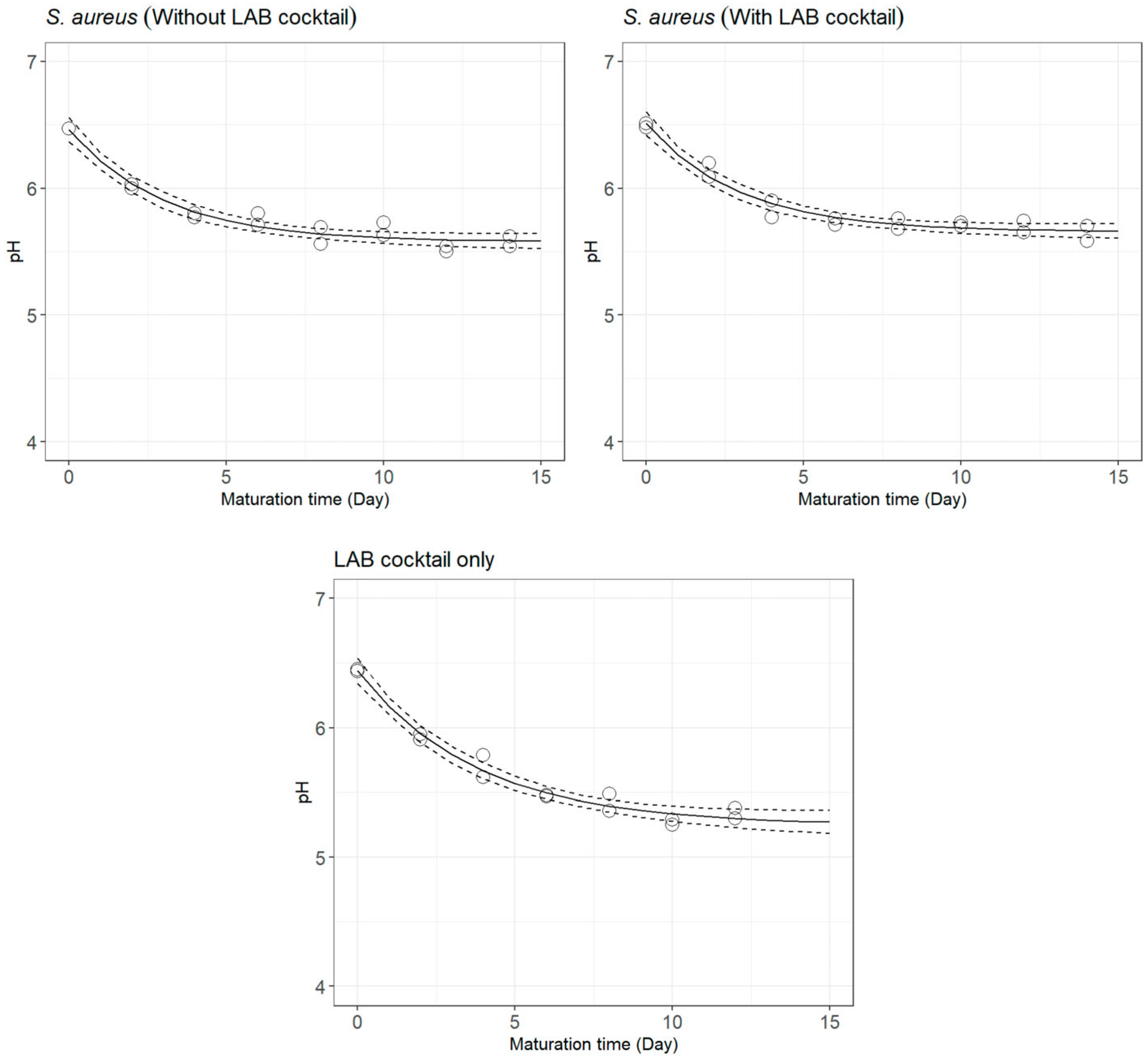
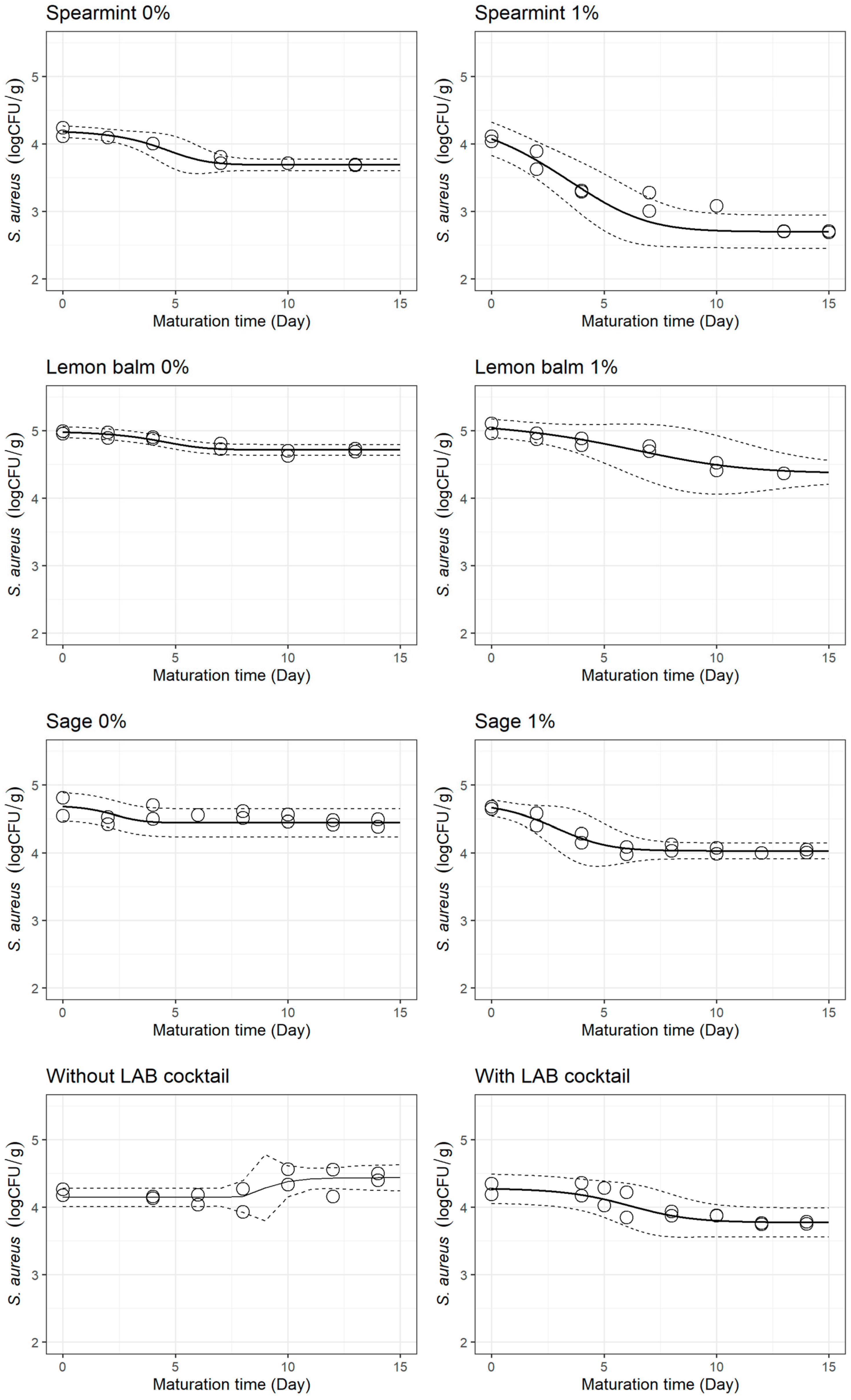
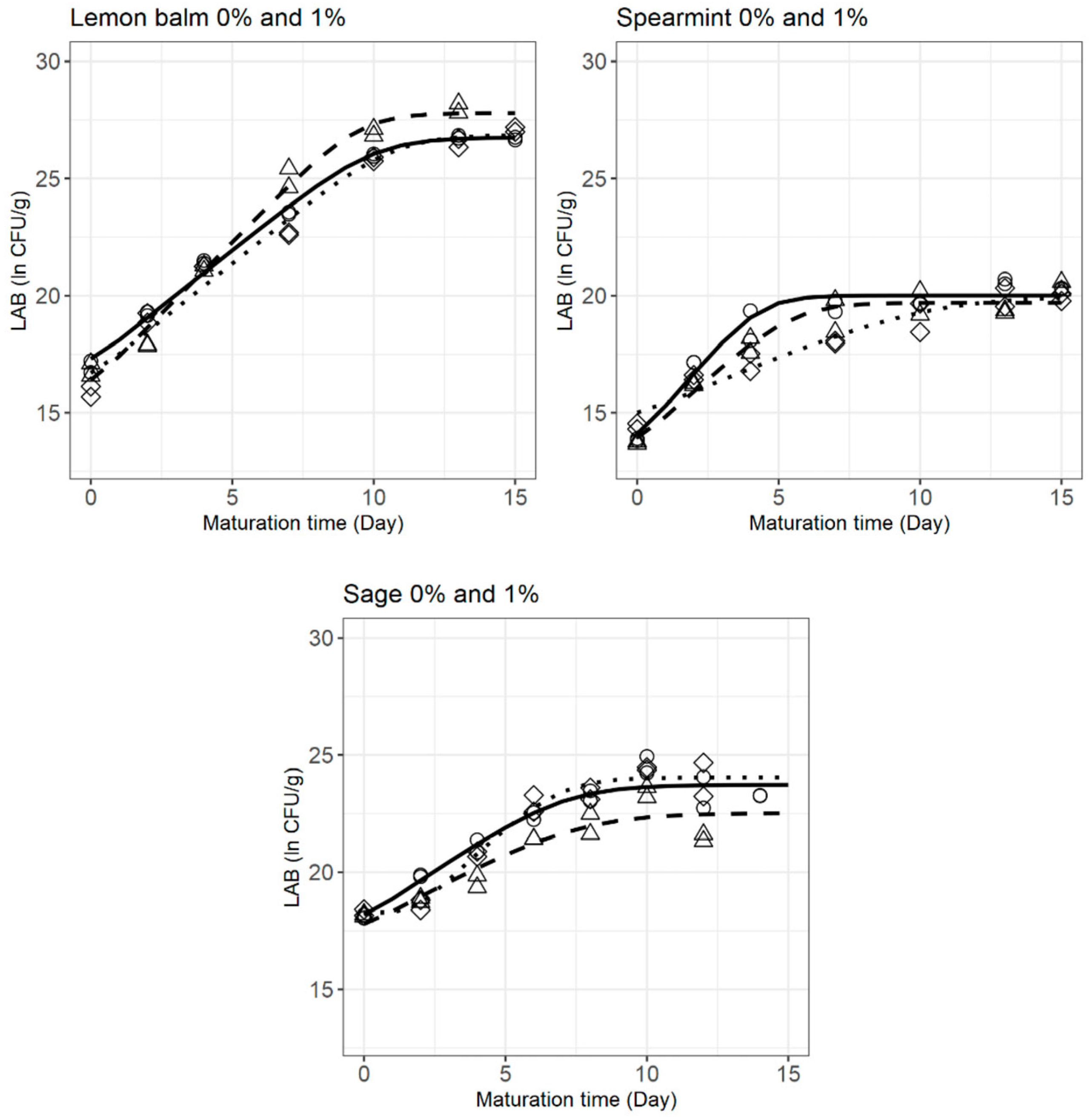
| Treatment | Parameters | Mean ± SE | Pr (>|t|) | Goodness-of-Fit Measures | |
|---|---|---|---|---|---|
| S. aureus + Spearmint 0% | 6.581 ± 0.058 | <0.0001 | S2 = 0.006 RMSE = 0.075 MAE = 0.061 | 1.163 | |
| 5.418 ± 0.050 | <0.0001 | ||||
| 0.262 ± 0.041 | <0.0001 | ||||
| S. aureus + Spearmint 1% | 6.530 ± 0.062 | <0.0001 | S2 = 0.007 RMSE = 0.079 MAE = 0.067 | 0.946 | |
| 5.584 ± 0.107 | <0.0001 | ||||
| 0.194 ± 0.058 | 0.008 | ||||
| S. aureus + Lemon balm 0% | 6.567 ± 0.046 | <0.0001 | S2 = 0.004 RMSE = 0.059 MAE = 0.047 | 1.452 | |
| 5.115 ± 0.043 | <0.0001 | ||||
| 0.240 ± 0.025 | <0.0001 | ||||
| S. aureus + Lemon balm 1% | 6.502 ± 0.053 | <0.0001 | S2 = 0.005 RMSE = 0.069 MAE = 0.050 | 1.216 | |
| 5.286 ± 0.055 | <0.0001 | ||||
| 0.223 ± 0.034 | <0.0001 | ||||
| S. aureus + Sage 0% | 6.142 ± 0.061 | <0.0001 | S2 = 0.007 RMSE = 0.079 MAE = 0.063 | 0.970 | |
| 5.172 ± 0.031 | <0.0001 | ||||
| 0.522 ± 0.092 | <0.0001 | ||||
| S. aureus + Sage 1% | 6.265 ± 0.036 | <0.0001 | S2 = 0.002 RMSE = 0.046 MAE = 0.037 | 0.888 | |
| 5.377 ± 0.018 | <0.0001 | ||||
| 0.521 ± 0.058 | <0.0001 | ||||
| S. aureus without LAB cocktail | 6.461 ± 0.044 | <0.0001 | S2 = 0.003 RMSE = 0.057 MAE = 0.045 | 0.885 | |
| 5.576 ± 0.031 | <0.0001 | ||||
| 0.330 ± 0.047 | <0.0001 | ||||
| S. aureus with LAB cocktail | 6.509 ± 0.042 | <0.0001 | S2 = 0.003 RMSE = 0.054 MAE = 0.043 | 0.853 | |
| 5.656 ± 0.029 | <0.0001 | ||||
| 0.337 ± 0.047 | <0.0001 | ||||
| LAB cocktail only | 6.440 ± 0.044 | <0.0001 | S2 = 0.004 RMSE = 0.058 MAE = 0.044 | 1.190 | |
| 5.250 ± 0.051 | <0.0001 | ||||
| 0.263 ± 0.035 | <0.0001 |
| Treatment | Parameters | Mean ± SE | Pr (>|t|) | Goodness-of-Fit Measures | △Y0–12 (log CFU/g) |
|---|---|---|---|---|---|
| Spearmint 0% ((0) = 1.5) | log Dref | 0.993 ± 0.190 | 0.001 | S2 = 0.002 RMSE = 0.040 MAE = 0.035 | 0.491 |
| zpH | 1.599 ± 0.358 | <0.0001 | |||
| Spearmint 1% ((0) = 0.01) | log Dref | 0.621 ± 0.061 | <0.0001 | S2 = 0.015 RMSE = 0.116 MAE = 0.098 | 1.373 |
| zpH | 3.172 ± 0.655 | <0.0001 | |||
| Lemon balm 0% ((0) = 1.5) | log Dref | 0.996 ± 0.056 | <0.0001 | S2 = 0.002 RMSE = 0.037 MAE = 0.033 | 0.262 |
| zpH | 1.851 ± 0.066 | <0.0001 | |||
| Lemon balm 1% ((0) = 0.01) | log Dref | 1.190 ± 0.200 | <0.0001 | S2 = 0.004 RMSE = 0.063 MAE = 0.056 | 0.611 |
| zpH | 2.340 ± 0.835 | 0.019 | |||
| Sage 0% ((0) = 1.5) | log Dref | 0.796 ± 0.068 | <0.0001 | S2 = 0.010 RMSE = 0.098 MAE = 0.077 | 0.238 |
| zpH | 2.054 ± 0.131 | <0.0001 | |||
| Sage 1% ((0) = 0.01) | log Dref | 0.996 ± 0.278 | 0.003 | S2 = 0.003 RMSE = 0.057 MAE = 0.047 | 0.634 |
| zpH | 2.006 ± 0.677 | 0.010 | |||
| With LAB cocktail ((0) = 3) | log Dref | 0.756 ± 0.067 | <0.0001 | S2 = 0.011 RMSE = 0.103 MAE = 0.078 | 0.493 |
| zpH | 2.490 ± 0.487 | <0.0001 |
| Treatment | Parameters | Mean ± SE | Pr (>|t|) | Goodness-of-Fit Measures |
|---|---|---|---|---|
| Spearmint 1% | 13.94 ± 0.366 | <0.0001 | S2 = 0.277 RMSE = 0.508 MAE = 0.445 | |
| 19.70 ± 0.216 | <0.0001 | |||
| 1.088 ± 0.160 | <0.0001 | |||
| S. aureus + Spearmint 0% | 14.11 ± 0.360 | <0.0001 | S2 = 0.254 RMSE = 0.486 MAE = 0.438 | |
| 20.01 ± 0.195 | <0.0001 | |||
| 1.421 ± 0.189 | <0.0001 | |||
| S. aureus + Spearmint 1% | 15.00 ± 0.299 | <0.0001 | S2 = 0.249 RMSE = 0.481 MAE = 0.421 | |
| 19.99 ± 0.358 | <0.0001 | |||
| 0.503 ± 0.076 | <0.0001 | |||
| Lemon balm 1% | 16.40 ± 0.307 | <0.0001 | S2 = 0.252 RMSE = 0.275 MAE = 0.208 | |
| 27.79 ± 0.349 | <0.0001 | |||
| 1.219 ± 0.074 | <0.0001 | |||
| S. aureus + Lemon balm 0% | 17.31 ± 0.168 | <0.0001 | S2 = 0.081 RMSE = 0.275 MAE = 0.208 | |
| 26.75 ± 0.157 | <0.0001 | |||
| 0.960 ± 0.038 | <0.0001 | |||
| S. aureus + Lemon balm 1% | 16.71 ± 0.346 | <0.0001 | S2 = 0.361 RMSE = 0.579 MAE = 0.490 | |
| 26.87 ± 0.350 | <0.0001 | |||
| 0.967 ± 0.073 | <0.0001 | |||
| Sage 1% | 17.80 ± 0.435 | <0.0001 | S2 = 0.461 RMSE = 0.654 MAE = 0.539 | |
| 22.52 ± 0.457 | <0.0001 | |||
| 0.643 ± 0.138 | <0.0001 | |||
| S. aureus + Sage 0% | 18.19 ± 0.325 | <0.0001 | S2 = 0.262 RMSE = 0.496 MAE = 0.369 | |
| 23.73 ± 0.232 | <0.0001 | |||
| 0.806 ± 0.102 | <0.0001 | |||
| S. aureus + Sage 1% | 18.27 ± 0.338 | <0.0001 | S2 = 0.179 RMSE = 0.408 MAE = 0.336 | |
| 24.05 ± 0.229 | <0.0001 | |||
| 1.133 ± 0.174 | <0.0001 | |||
| λ | 1.749 ± 0.565 | 0.011 |
| Treatment | Parameters | Mean ± SE | Pr (>|t|) | Goodness-of-Fit Measures |
|---|---|---|---|---|
| Without LAB cocktail | 15.01 ± 0.258 | <0.0001 | S2 = 0.040 RMSE = 0.183 MAE = 0.135 | |
| 18.54 ± 0.137 | <0.0001 | |||
| 1.560 ± 0.260 | 0.009 | |||
| With LAB cocktail | 16.65 ± 0.340 | <0.0001 | S2 = 0.074 RMSE = 0.248 MAE = 0.198 | |
| 20.22 ± 0.199 | <0.0001 | |||
| 1.198 ± 0.260 | 0.019 | |||
| S. aureus without LAB cocktail | 15.16 ± 0.174 | <0.0001 | S2 = 0.021 RMSE = 0.134 MAE = 0.120 | |
| 18.83 ± 0.085 | <0.0001 | |||
| 1.343 ± 0.145 | 0.0007 | |||
| S. aureus with LAB cocktail | 16.37 ± 0.144 | <0.0001 | S2 = 0.016 RMSE = 0.120 MAE = 0.096 | |
| 20.40 ± 0.071 | <0.0001 | |||
| 1.144 ± 0.091 | <0.0001 |
| Treatment | Parameters | Mean ± SE | Pr (>|t|) | Goodness-of-Fit Measures |
|---|---|---|---|---|
| Without LAB cocktail | 15.27 ± 0.531 | <0.0001 | S2 = 0.190 RMSE = 0.404 MAE = 0.349 | |
| 19.57 ± 0.249 | <0.0001 | |||
| 1.705 ± 0.475 | 0.023 | |||
| With LAB cocktail | 16.72 ± 0.464 | <0.0001 | S2 = 0.168 RMSE = 0.3791 MAE = 0.342 | |
| 21.20 ± 0.265 | <0.0001 | |||
| 0.979 ± 0.236 | 0.014 | |||
| S. aureus without LAB cocktail | 15.35 ± 0.321 | <0.0001 | S2 = 0.074 RMSE = 0.253 MAE = 0.232 | |
| 19.96 ± 0.164 | <0.0001 | |||
| 1.407 ± 0.221 | 0.003 | |||
| S. aureus with LAB cocktail | 15.99 ± 0.392 | <0.0001 | S2 = 0.102 RMSE = 0.292 MAE = 0.261 | |
| 20.88 ± 0.236 | <0.0001 | |||
| 1.372 ± 0.246 | 0.011 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Silva, B.N.; Coelho-Fernandes, S.; Teixeira, J.A.; Cadavez, V.; Gonzales-Barron, U. Dynamic Modelling to Describe the Effect of Plant Extracts and Customised Starter Culture on Staphylococcus aureus Survival in Goat’s Raw Milk Soft Cheese. Foods 2023, 12, 2683. https://doi.org/10.3390/foods12142683
Silva BN, Coelho-Fernandes S, Teixeira JA, Cadavez V, Gonzales-Barron U. Dynamic Modelling to Describe the Effect of Plant Extracts and Customised Starter Culture on Staphylococcus aureus Survival in Goat’s Raw Milk Soft Cheese. Foods. 2023; 12(14):2683. https://doi.org/10.3390/foods12142683
Chicago/Turabian StyleSilva, Beatriz Nunes, Sara Coelho-Fernandes, José António Teixeira, Vasco Cadavez, and Ursula Gonzales-Barron. 2023. "Dynamic Modelling to Describe the Effect of Plant Extracts and Customised Starter Culture on Staphylococcus aureus Survival in Goat’s Raw Milk Soft Cheese" Foods 12, no. 14: 2683. https://doi.org/10.3390/foods12142683
APA StyleSilva, B. N., Coelho-Fernandes, S., Teixeira, J. A., Cadavez, V., & Gonzales-Barron, U. (2023). Dynamic Modelling to Describe the Effect of Plant Extracts and Customised Starter Culture on Staphylococcus aureus Survival in Goat’s Raw Milk Soft Cheese. Foods, 12(14), 2683. https://doi.org/10.3390/foods12142683








