Occurrence, Antibiotic Susceptibility, Biofilm Formation and Molecular Characterization of Staphylococcus aureus Isolated from Raw Shrimp in China
Abstract
1. Introduction
2. Materials and Methods
2.1. Sampling Sites
2.2. Isolation and Identification of S. aureus
2.3. Antimicrobial Susceptibility Testing for Shrimp-Related S. aureus
2.4. Biofilm Formation Assay
2.5. Whole-Genome Sequencing and Assembly
2.6. Determination of STs, Spa Types, SCCmec Types, Virulence Genes and ARG Genes
2.7. Nucleotide Sequence Accession Numbers
3. Results
3.1. Isolation and Identification of S. aureus from Shrimp in China
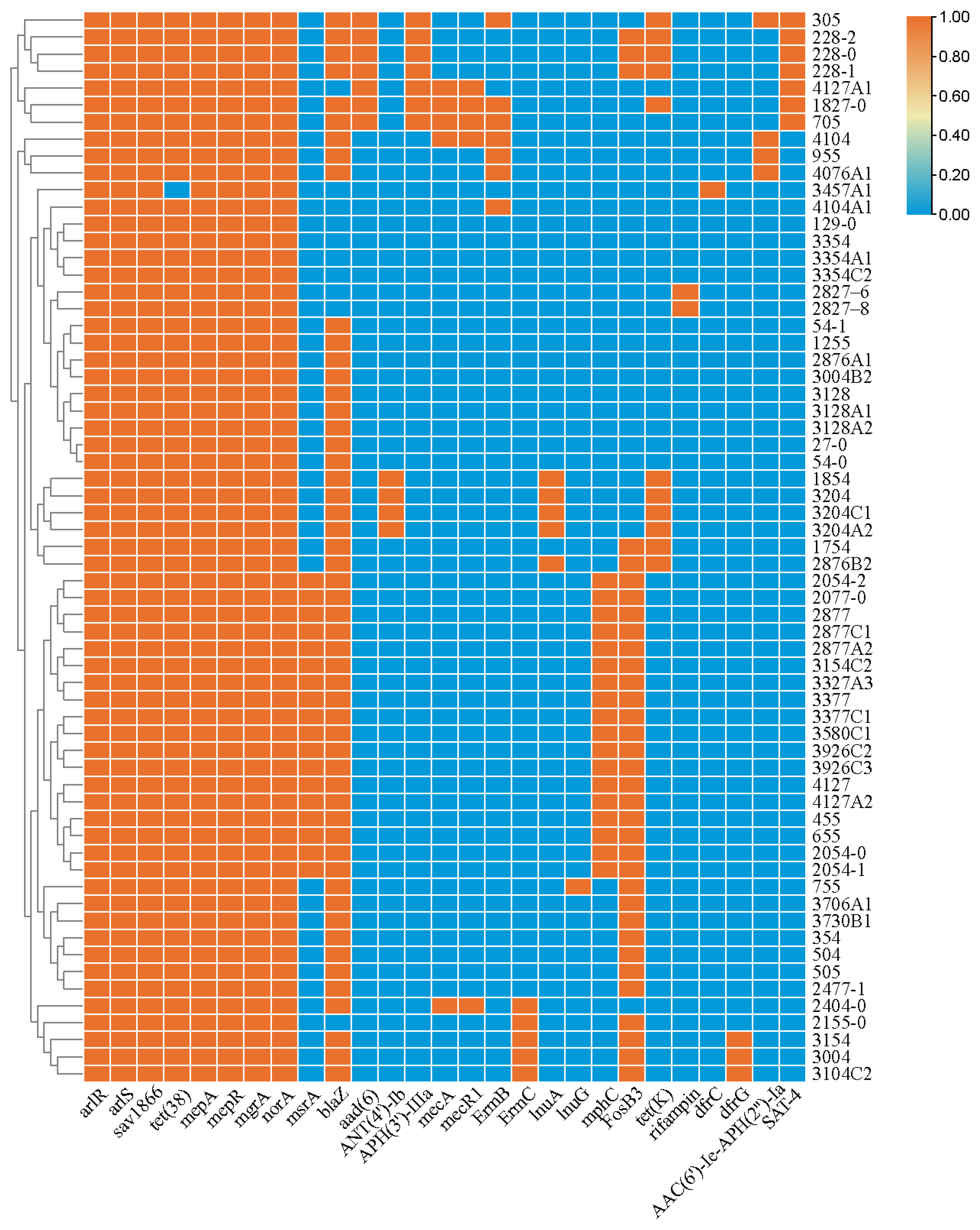
3.2. Antibiotic Susceptibility and Antibiotic-Resistant Genes (ARGs)
3.3. Biofilm Production and the Presence of Biofilm-Related Genes
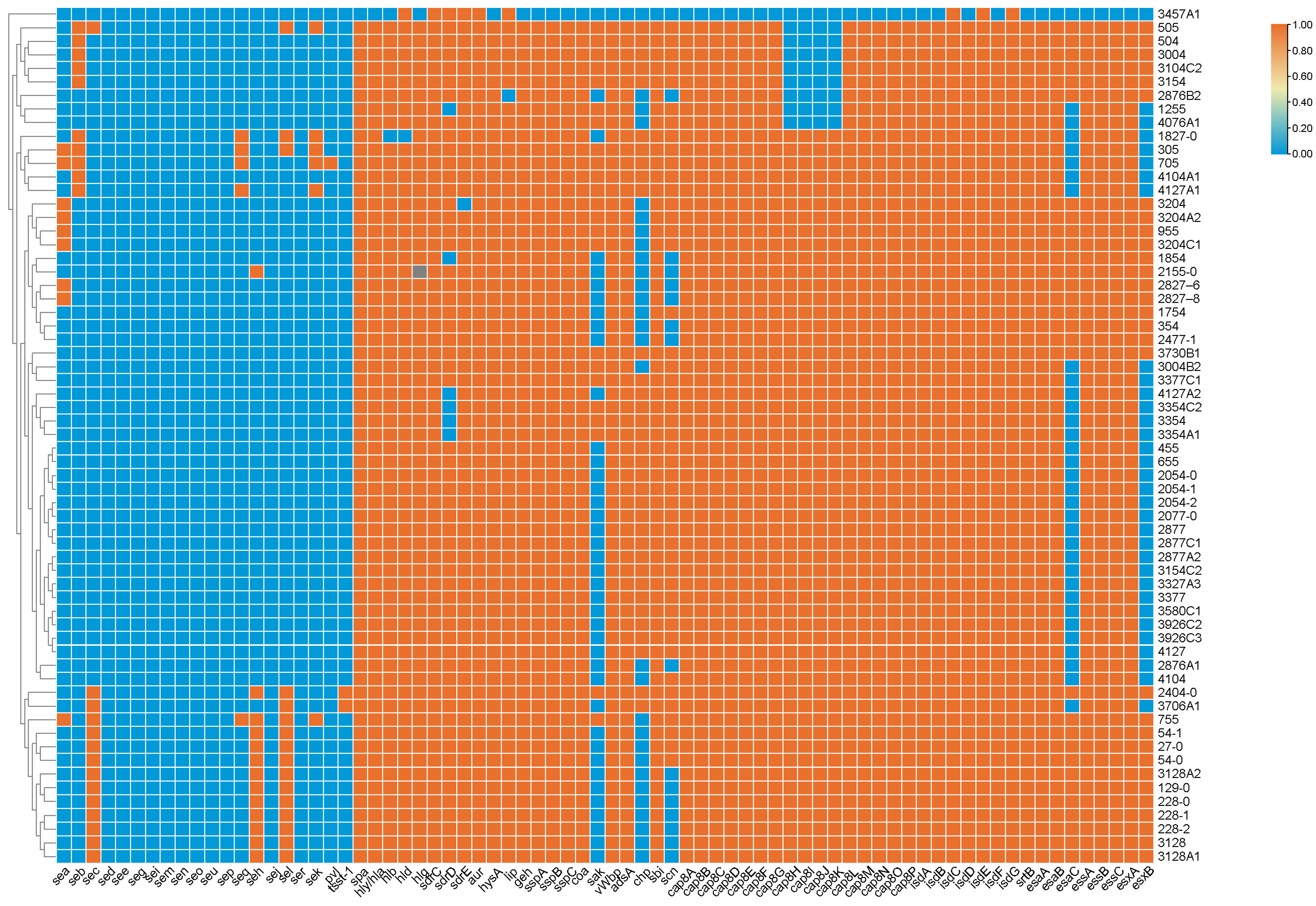
3.4. Prevalence of Virulence-Associated Genes
3.5. Molecular Typing of Shrimp-Related S. aureus Isolates
4. Discussion
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Norhana, M.; Poole, S.E.; Deeth, H.C.; Dykes, G.A. Prevalence, persistence and control of Salmonella and Listeria in shrimp and shrimp products: A review. Food Control 2010, 21, 343–361. [Google Scholar] [CrossRef]
- Letchumanan, V.; Yin, W.F.; Lee, L.H.; Chan, K.G. Prevalence and antimicrobial susceptibility of Vibrio parahaemolyticus isolated from retail shrimps in Malaysia. Front. Microbiol. 2015, 6, 1602–1608. [Google Scholar] [CrossRef] [PubMed]
- Ertas Onmaz, N.; Abay, S.; Karadal, F.; Hizlisoy, H.; Telli, N.; Al, S. Occurence and antimicrobial resistance of Staphylococcus aureus and Salmonella spp. in retail fish samples in Turkey. Mar. Pollut. Bull. 2015, 90, 242–246. [Google Scholar] [CrossRef]
- Lowy, F.; Lowy, F.D. Staphylococcus aureus infections. New Engl. J. Med. 1998, 339, 520–532. [Google Scholar] [CrossRef] [PubMed]
- David, M.Z.; Daum, R.S. Community-Associated Methicillin-Resistant Staphylococcus aureus: Epidemiology and Clinical Consequences of an Emerging Epidemic. Clin. Microbiol. Rev. 2010, 23, 616–687. [Google Scholar] [CrossRef] [PubMed]
- Spanu, V.; Spanu, C.; Virdis, S.; Cossu, F.; Scarano, C.; De Santis, E.P. Virulence factors and genetic variability of Staphylococcus aureus strains isolated from raw sheep’s milk cheese. Int. J. Food Microbiol. 2012, 153, 53–57. [Google Scholar] [CrossRef] [PubMed]
- Dai, J.; Wu, S.; Huang, J.; Wu, Q.; Zhang, F.; Zhang, J.; Wang, J.; Ding, Y.; Zhang, S.; Yang, X.; et al. Prevalence and Characterization of Staphylococcus aureus Isolated from Pasteurized Milk in China. Front. Microbiol. 2019, 10, 641. [Google Scholar] [CrossRef]
- Hennekinne, J.-A.; De Buyser, M.-L.; Dragacci, S. Staphylococcus aureus and its food poisoning toxins: Characterization and outbreak investigation. FEMS Microbiol. Rev. 2012, 36, 815–836. [Google Scholar] [CrossRef]
- Denayer, S.; Delbrassinne, L.; Nia, Y.; Botteldoorn, N. Food-Borne Outbreak Investigation and Molecular Typing: High Diversity of Staphylococcus aureus Strains and Importance of Toxin Detection. Toxins 2017, 9, 407. [Google Scholar] [CrossRef]
- Formosa-Dague, C.; Feuillie, C.; Beaussart, A.; Derclaye, S.; Kucharíková, S.; Lasa, I.; Van Dijck, P.; Dufrêne, Y.F. Sticky Matrix: Adhesion Mechanism of the Staphylococcal Polysaccharide Intercellular Adhesin. ACS Nano 2016, 10, 3443–3452. [Google Scholar] [CrossRef]
- Speziale, P.; Pietrocola, G. The Multivalent Role of Fibronectin-Binding Proteins A and B (FnBPA and FnBPB) of Staphylococcus aureus in Host Infections. Front. Microbiol. 2020, 11, 2054. [Google Scholar] [CrossRef]
- Farnsworth, C.W.; Schott, E.M.; Jensen, S.E.; Zukoski, J.; Benvie, A.M.; Refaai, M.A.; Kates, S.L.; Schwarz, E.M.; Zuscik, M.J.; Gill, S.R. Adaptive upregulation of clumping factor A (ClfA) by Staphylococcus aureus in the obese, type 2 diabetic host mediates increased virulence. Infect. Immun. 2017, 85, e01005–e01016. [Google Scholar] [CrossRef]
- Foster, T.J.; Geoghegan, J.A.; Ganesh, V.K.; Höök, M. Adhesion, invasion and evasion: The many functions of the surface proteins of Staphylococcus aureus. Nat. Rev. Microbiol. 2014, 12, 49–62. [Google Scholar] [CrossRef]
- Demir, C.; Demirci, M.; Yigin, A.; Tokman, H.B.; Cetik Yildiz, S. Presence of biofilm and adhesin genes in Staphylococcus aureus strains taken from chronic wound infections and their genotypic and phenotypic antimicrobial sensitivity patterns. Photodiagnosis Photodyn. Ther. 2020, 29, 101584. [Google Scholar] [CrossRef]
- Enright, M.C. The evolution of a resistant pathogen–The case of MRSA. Curr. Opin. Pharmacol. 2003, 3, 474–479. [Google Scholar] [CrossRef]
- Beleneva, I.A. Incidence and characteristics of Staphylococcus aureus and Listeria monocytogenes from the Japan and South China seas. Mar. Pollut. Bull. 2011, 62, 382–387. [Google Scholar] [CrossRef]
- Lee, J.H. Methicillin (Oxacillin)-Resistant Staphylococcus aureus Strains Isolated from Major Food Animals and Their Potential Transmission to Humans. Appl. Environ. Microbiol. 2003, 69, 6489–6494. [Google Scholar] [CrossRef] [PubMed]
- Normanno, G.; Firinu, A.; Virgilio, S.; Mula, G.; Dambrosio, A.; Poggiu, A.; Decastelli, L.; Mioni, R.; Scuota, S.; Bolzoni, G. Coagulase-positive Staphylococci and Staphylococcus aureus in food products marketed in Italy. Int. J. Food Microbiol. 2005, 98, 73–79. [Google Scholar] [CrossRef] [PubMed]
- Pereira, V.; Lopes, C.; Castro, A.; Silva, J.; Gibbs, P. Characterization for enterotoxin production, virulence factors, and antibiotic susceptibility of Staphylococcus aureus isolates from various foods in Portugal. Food Microbiol. 2009, 26, 278–282. [Google Scholar] [CrossRef] [PubMed]
- Zmantar, T.; Chaieb, K.; Makni, H.; Miladi, H.; Abdallah, F.B.; Mahdouani, K.; Bakhrouf, A. Detection by PCR of adhesins genes and slime production in clinical Staphylococcus aureus. J. Basic Microbiol. 2010, 48, 308–314. [Google Scholar] [CrossRef]
- Basanisi, M.G.; Nobili, G.; La Bella, G.; Russo, R.; Spano, G.; Normanno, G.; La Salandra, G. Molecular characterization of Staphylococcus aureus isolated from sheep and goat cheeses in southern Italy. Small Rumin. Res. 2016, 135, 17–19. [Google Scholar] [CrossRef]
- Vasudevan, P.; Nair, M.K.M.; Annamalai, T.; Venkitanarayanan, K.S. Phenotypic and genotypic characterization of bovine mastitis isolates of Staphylococcus aureus for biofilm formation. Vet. Microbiol. 2003, 92, 179–185. [Google Scholar] [CrossRef] [PubMed]
- Camacho, C.; Coulouris, G.; Avagyan, V.; Ma, N.; Papadopoulos, J.; Bealer, K.; Madden, T.L. BLAST+: Architecture and applications. BMC Bioinform. 2009, 10, 421. [Google Scholar] [CrossRef]
- Maiden, M.C.J.; van Rensburg, M.J.J.; Bray, J.E.; Earle, S.G.; Ford, S.A.; Jolley, K.A.; McCarthy, N.D. MLST revisited: The gene-by-gene approach to bacterial genomics. Nat. Rev. Microbiol. 2013, 11, 728–736. [Google Scholar] [CrossRef]
- Bartels, M.D.; Petersen, A.; Worning, P.; Nielsen, J.B.; Larner-Svensson, H.; Johansen, H.K.; Andersen, L.P.; Jarløv, J.O.; Boye, K.; Larsen, A.R.; et al. Comparing Whole-Genome Sequencing with Sanger Sequencing for spaTyping of Methicillin-Resistant Staphylococcus aureus. J. Clin. Microbiol. 2014, 52, 4305–4308. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Zheng, D.; Liu, B.; Yang, J.; Jin, Q. VFDB 2016: Hierarchical and refined dataset for big data analysis--10 years on. Nucleic Acids Res. 2016, 44, D694–D697. [Google Scholar] [CrossRef]
- Jia, B.; Raphenya, A.R.; Alcock, B.; Waglechner, N.; Guo, P.; Tsang, K.K.; Lago, B.A.; Dave, B.M.; Pereira, S.; Sharma, A.N.; et al. CARD 2017: Expansiosn and model-centric curation of the comprehensive antibiotic resistance database. Nucleic Acids Res. 2017, 45, D566–D573. [Google Scholar] [CrossRef]
- Zankari, E.; Hasman, H.; Cosentino, S.; Vestergaard, M.; Rasmussen, S.; Lund, O.; Aarestrup, F.M.; Larsen, M.V. Identification of acquired antimicrobial resistance genes. J. Antimicrob. Chemother. 2012, 67, 2640–2644. [Google Scholar] [CrossRef]
- Wu, S.; Huang, J.; Wu, Q.; Zhang, J.; Zhang, F.; Yang, X.; Wu, H.; Zeng, H.; Chen, M.; Ding, Y.; et al. Staphylococcus aureus Isolated From Retail Meat and Meat Products in China: Incidence, Antibiotic Resistance and Genetic Diversity. Front. Microbiol. 2018, 9, 2767. [Google Scholar] [CrossRef]
- Arfatahery, N.; Mirshafiey, A.; Abedimohtasab, T.P.; Zeinolabedinizamani, M. Study of the Prevalence of Staphylococcus Aureus in Marine and Farmed Shrimps in Iran Aiming the Future Development of a Prophylactic Vaccine. Procedia Vaccinol. 2015, 9, 44–49. [Google Scholar] [CrossRef]
- Beshiru, A.; Igbinosa, I.H.; Igbinosa, E.O. Characterization of enterotoxigenic Staphylococcus aureus from ready-to-eat seafood (RTES). LWT 2021, 135, 110042. [Google Scholar] [CrossRef]
- Simon, S.S.; Sanjeev, S. Prevalence of enterotoxigenic Staphylococcus aureus in fishery products and fish processing factory workers. Food Control 2007, 18, 1565–1568. [Google Scholar] [CrossRef]
- Argudín, M.Á.; Mendoza, M.C.; Rodicio, M.R. Food poisoning and Staphylococcus aureus enterotoxins. Toxins 2010, 2, 1751. [Google Scholar] [CrossRef] [PubMed]
- Chieffi, D.; Fanelli, F.; Cho, G.-S.; Schubert, J.; Blaiotta, G.; Franz, C.M.; Bania, J.; Fusco, V. Novel insights into the enterotoxigenic potential and genomic background of Staphylococcus aureus isolated from raw milk. Food Microbiol. 2020, 90, 103482. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Baloch, Z.; Jiang, T.; Zhang, C.; Peng, Z.; Li, F.; Fanning, S.; Ma, A.; Xu, J. Enterotoxigenicity and Antimicrobial Resistance of Staphylococcus aureus Isolated from Retail Food in China. Front. Microbiol. 2017, 8, 2256. [Google Scholar] [CrossRef]
- Jones, T.F.; Kellum, M.E.; Porter, S.S.; Bell, M.; Schaffner, W. An Outbreak of Community-Acquired Foodborne Illness Caused by Methicillin-Resistant Staphylococcus aureus. Emerg. Infect. Dis. 2002, 8, 82–84. [Google Scholar] [CrossRef]
- Su, Y.C.; Wong, A.C. Detection of staphylococcal enterotoxin H by an enzyme-linked immunosorbent assay. J. Food Prot. 1996, 59, 327–330. [Google Scholar] [CrossRef]
- Jørgensen, H.J.; Mathisen, T.; Løvseth, A.; Omoe, K.; Qvale, K.S.; Loncarevic, S. An outbreak of staphylococcal food poisoning caused by enterotoxin H in mashed potato made with raw milk. FEMS Microbiol. Lett. 2005, 252, 267–272. [Google Scholar] [CrossRef]
- Pereira, M.L.; Carmo, L.S.D.; Santos, E.J.D.; Pereira, J.L.; Bergdoll, M.S. Enterotoxin H in Staphylococcal Food Poisoning. J. Food Prot. 1996, 59, 559–561. [Google Scholar] [CrossRef]
- Mclauchlin, J.; Narayanan, G.L.; Mithani, V.; O’Neill, G. The detection of enterotoxins and toxic shock syndrome toxin genes in Staphylococcus aureus by polymerase chain reaction. J. Food Prot. 2000, 63, 479–488. [Google Scholar] [CrossRef]
- Olaniyi, R.; Pozzi, C.; Grimaldi, L.; Bagnoli, F. Staphylococcus aureus-associated skin and soft tissue infections: Anatomical localization, epidemiology, therapy and potential prophylaxis. In Staphylococcus aureus. Current Topics in Microbiology and Immunology; Springer: Cham, Switzerland, 2017; pp. 199–227. [Google Scholar]
- Zheng, Y.; Qin, C.; Zhang, X.; Zhu, Y.; Li, A.; Wang, M.; Tang, Y.; Kreiswirth, B.N.; Chen, L.; Zhang, H. The tst gene associated Staphylococcus aureus pathogenicity island facilitates its pathogenesis by promoting the secretion of inflammatory cytokines and inducing immune suppression. Microb. Pathog. 2020, 138, 103797. [Google Scholar] [CrossRef] [PubMed]
- Kulhankova, K.; King, J.; Salgado-Pabón, W. Staphylococcal toxic shock syndrome: Superantigen-mediated enhancement of endotoxin shock and adaptive immune suppression. Immunol. Res. 2014, 59, 182–187. [Google Scholar] [CrossRef] [PubMed]
- Wu, S.; Zhang, F.; Huang, J.; Wu, Q.; Zhang, J.; Dai, J.; Zeng, H.; Yang, X.; Chen, M.; Pang, R.; et al. Phenotypic and genotypic characterization of PVL-positive Staphylococcus aureus isolated from retail foods in China. Int. J. Food Microbiol. 2019, 304, 119–126. [Google Scholar] [CrossRef] [PubMed]
- Shallcross, L.J.; Ellen, F.; Johnson, A.M.; Hayward, A.C. The role of the Panton-Valentine leucocidin toxin in staphylococcal disease: A systematic review and meta-analysis. Lancet Infect. Dis. 2013, 13, 43–54. [Google Scholar] [CrossRef] [PubMed]
- Gillet, Y.; Issartel, B.; Vanhems, P.; Fournet, J.C.; Lina, G.; Bes, M.; Vandenesch, F.; Piémont, Y.; Brousse, N.; Floret, D. Association between Staphylococcus aureus strains carrying gene for Panton-Valentine leukocidin and highly lethal necrotising pneumonia in young immunocompetent patients. Lancet 2002, 359, 753–759. [Google Scholar] [CrossRef]
- Alibayov, B.; Baba-Moussa, L.; Sina, H.; Zdeňková, K.; Demnerová, K. Staphylococcus aureus mobile genetic elements. Mol. Biol. Rep. 2014, 41, 5005–5018. [Google Scholar] [CrossRef]
- Gibson, H.; Taylor, J.; Hall, K.; Holah, J. Effectiveness of cleaning techniques used in the food industry in terms of the removal of bacterial biofilms. J. Appl. Microbiol. 1999, 87, 41–48. [Google Scholar] [CrossRef]
- Balaban, N.; Rasooly, A. Staphylococcal enterotoxins. Int. J. Food Microbiol. 2000, 61, 1–10. [Google Scholar] [CrossRef]
- Braga, L.; Shupp, J.; Cummings, C.; Jett, M.; Takahashi, J.; Carmo, L.; Chartone-Souza, E.; Nascimento, A. Pomegranate extract inhibits Staphylococcus aureus growth and subsequent enterotoxin production. J. Ethnopharmacol. 2005, 96, 335–339. [Google Scholar] [CrossRef]
- Hamadi, F.; Latrache, H.; Mabrrouki, M.; Elghmari, A.; Outzourhit, A.; Ellouali, M.; Chtaini, A. Effect of pH on distribution and adhesion of Staphylococcus aureus to glass. J. Adhes. Sci. Technol. 2005, 19, 73–85. [Google Scholar] [CrossRef]
- Di Ciccio, P.; Vergara, A.; Festino, A.; Paludi, D.; Zanardi, E.; Ghidini, S.; Ianieri, A. Biofilm formation by Staphylococcus aureus on food contact surfaces: Relationship with temperature and cell surface hydrophobicity. Food Control 2015, 50, 930–936. [Google Scholar] [CrossRef]
- Chavhan, S.K.; Kalorey, D.R.; Nagdive, A.A.; Purohit, H.J.; Barbuddhe, S.B.; Kurkure, N.V. Molecular characterization of intercellular adhesion gene in Staphylococcus aureus isolated from bovine mastitic milk. Trop. Anim. Health Prod. 2012, 44, 247–252. [Google Scholar] [CrossRef]
- Darwish, S.F.; Asfour, H.A. Investigation of biofilm forming ability in Staphylococci causing bovine mastitis using phenotypic and genotypic assays. Sci. World J. 2013, 2013, 378492. [Google Scholar] [CrossRef]
- Dhanawade, N.B.; Kalorey, D.R.; Srinivasan, R.; Barbuddhe, S.B.; Kurkure, N.V. Detection of intercellular adhesion genes and biofilm production in Staphylococcus aureus isolated from bovine subclinical mastitis. Vet. Res. Commun. 2010, 34, 81–89. [Google Scholar] [CrossRef] [PubMed]
- Khoramrooz, S.S.; Mansouri, F.; Marashifard, M.; Hosseini, S.A.A.M.; Chenarestane-Olia, F.A.; Ganavehei, B.; Gharibpour, F.; Shahbazi, A.; Mirzaii, M.; Darban-Sarokhalil, D. Detection of biofilm related genes, classical enterotoxin genes and agr typing among Staphylococcus aureus isolated from bovine with subclinical mastitis in southwest of Iran. Microb. Pathog. 2016, 97, 45–51. [Google Scholar] [CrossRef] [PubMed]
- Greene, C.; McDevitt, D.; Francois, P.; Vaudaux, P.; Lew, D.P.; Poster, T. Adhesion properties of mutants of Staphylococcus aureus defective in fibronectin-binding proteins and studies on the expression of fnb genes. Mol. Microbiol. 1995, 17, 1143–1152. [Google Scholar] [CrossRef]
- Vaudaux, P.E.; Francois, P.; Proctor, R.A.; McDevitt, D.; Foster, T.J.; Albrecht, R.M.; Lew, D.P.; Wabers, H.; Cooper, S.L. Use of adhesion-defective mutants of Staphylococcus aureus to define the role of specific plasma proteins in promoting bacterial adhesion to canine arteriovenous shunts. Infect. Immun. 1995, 63, 585–590. [Google Scholar] [CrossRef]
- Vaudaux, P.; Pittet, D.; Haeberli, A.; Lerch, P.G.; Morgenthaler, J.-J.; Proctor, R.A.; Waldvogel, F.A.; Lew, D.P. Fibronectin is more active than fibrin or fibrinogen in promoting Staphylococcus aureus adherence to inserted intravascular catheters. J. Infect. Dis. 1993, 167, 633–641. [Google Scholar] [CrossRef]
- Rohde, H.; Burandt, E.C.; Siemssen, N.; Frommelt, L.; Burdelski, C.; Wurster, S.; Scherpe, S.; Davies, A.P.; Harris, L.G.; Horstkotte, M.A. Polysaccharide intercellular adhesin or protein factors in biofilm accumulation of Staphylococcus epidermidis and Staphylococcus aureus isolated from prosthetic hip and knee joint infections. Biomaterials 2007, 28, 1711–1720. [Google Scholar] [CrossRef]
- Felipe, V.; Morgante, C.A.; Somale, P.S.; Varroni, F.; Zingaretti, M.L.; Bachetti, R.A.; Correa, S.G.; Porporatto, C. Evaluation of the biofilm forming ability and its associated genes in Staphylococcus species isolates from bovine mastitis in Argentinean dairy farms. Microb. Pathog. 2017, 104, 278–286. [Google Scholar] [CrossRef]
- Normanno, G.; La, S.G.; Dambrosio, A.; Quaglia, N.C.; Corrente, M.; Parisi, A.; Santagada, G.; Firinu, A.; Crisetti, E.; Celano, G.V. Occurrence, characterization and antimicrobial resistance of enterotoxigenic Staphylococcus aureus isolated from meat and dairy products. Int. J. Food Microbiol. 2007, 115, 290–296. [Google Scholar] [CrossRef] [PubMed]
- Basanisi, M.G.; Bella, G.L.; Nobili, G.; Franconieri, I.; Salandra, G.L. Genotyping of Methicillin-resistant Staphylococcus aureus (MRSA) isolated from milk and dairy products in South Italy. Food Microbiol. 2017, 62, 141. [Google Scholar] [CrossRef] [PubMed]
- Rong, D.; Xu, M.; Wu, Q.; Zhang, J.; Yu, S. Prevalence, Virulence Genes, Antimicrobial Susceptibility, and Genetic Diversity of Staphylococcus aureus from Retail Aquatic Products in China. Front. Microbiol. 2017, 8, 714. [Google Scholar] [CrossRef] [PubMed]
- Purrello, S.; Garau, J.; Giamarellos, E.; Mazzei, T.; Pea, F.; Soriano, A.; Stefani, S. Methicillin-resistant Staphylococcus aureus infections: A review of the currently available treatment options. J. Glob. Antimicrob. Resist. 2016, 7, 178–186. [Google Scholar] [CrossRef]
- Fetsch, A.; Contzen, M.; Hartelt, K.; Kleiser, A.; Maassen, S.; Rau, J.; Kraushaar, B.; Layer, F.; Strommenger, B. Staphylococcus aureus food-poisoning outbreak associated with the consumption of ice-cream. Int. J. Food Microbiol. 2014, 187, 1–6. [Google Scholar] [CrossRef]
- Cha, J.O.; Lee, J.K.; Jung, Y.H.; Yoo, J.I.; Park, Y.K.; Kim, B.S.; Lee, Y.S. Molecular analysis of Staphylococcus aureus isolates associated with staphylococcal food poisoning in South Korea. J. Appl. Microbiol. 2006, 101, 864–871. [Google Scholar] [CrossRef]
- Yan, X.; Wang, B.; Tao, X.; Hu, Q.; Cui, Z.; Zhang, J.; Lin, Y.; You, Y.; Shi, X.; Grundmann, H. Characterization of Staphylococcus aureus Strains Associated with Food Poisoning in Shenzhen, China. Appl. Environ. Microbiol. 2012, 78, 6637–6642. [Google Scholar] [CrossRef]
- Lv, G.; Jiang, R.; Zhang, H.; Wang, L.; Li, L.; Gao, W.; Zhang, H.; Pei, Y.; Wei, X.; Dong, H.; et al. Molecular Characteristics of Staphylococcus aureus From Food Samples and Food Poisoning Outbreaks in Shijiazhuang, China. Front. Microbiol. 2021, 12, 652276. [Google Scholar] [CrossRef]
- Benito, D.; Lozano, C.; Gómez-Sanz, E.; Zarazaga, M.; Torres, C. Detection of methicillin-susceptible Staphylococcus aureus ST398 and ST133 strains in gut microbiota of healthy humans in Spain. Microb. Ecol. 2013, 66, 105–111. [Google Scholar] [CrossRef]
- Eibach, D.; Nagel, M.; Hogan, B.; Azuure, C.; Krumkamp, R.; Dekker, D.; Gajdiss, M.; Brunke, M.; Sarpong, N.; Owusu-Dabo, E. Nasal carriage of Staphylococcus aureus among children in the Ashanti Region of Ghana. PLoS ONE 2017, 12, e0170320. [Google Scholar] [CrossRef]
- Li, G.; Wu, S.; Luo, W.; Su, Y.; Luan, Y.; Wang, X. Staphylococcus aureus ST6-t701 isolates from food-poisoning outbreaks (2006–2013) in Xi’an, China. Foodborne Pathog. Dis. 2015, 12, 203–206. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Wang, D.; Zhan, S.; Zheng, J.; Tao, Y.; Wang, L.; Yan, X.; Tao, X.; Zhang, J.; Kan, B. Identification of Staphylococcus aureus and SpA polymorphisms in Ma’anshan City. J. Public Health Prev. Med. 2011, 22, 50–53. [Google Scholar]
- Huang, Y.C.; Ho, C.F.; Chen, C.J.; Su, L.H.; Lin, T.Y. Comparative molecular analysis of community-associated and healthcare-associated methicillin-resistant Staphylococcus aureus isolates from children in northern Taiwan. Clin. Microbiol. Infect. 2010, 14, 1167–1172. [Google Scholar] [CrossRef]
- Li, S.; Sun, S.; Yang, C.; Chen, H.; Yin, Y.; Li, H.; Zhao, C.; Wang, H. The Changing Pattern of Population Structure of Staphylococcus aureus from Bacteremia in China from 2013 to 2016: ST239-030-MRSA Replaced by ST59-t437. Front. Microbiol. 2018, 9, 332. [Google Scholar] [CrossRef] [PubMed]
- Liang, B.; Mai, J.; Liu, Y.; Huang, Y.; Zhong, H.; Xie, Y.; Deng, Q.; Huang, L.; Yao, S.; He, Y.; et al. Prevalence and Characterization of Staphylococcus aureus Isolated from Women and Children in Guangzhou, China. Front. Microbiol. 2018, 9, 2790. [Google Scholar] [CrossRef] [PubMed]
| Positive Number | Positive Sample | Sampling Site | Quantitative Methods | Qualitative Methods |
|---|---|---|---|---|
| MPN Values (MPN/g) | ||||
| 1 | YXJ27 | Guangzhou | 2.3 | + |
| 2 | ZCC54 | Guangzhou | 24 | + |
| 3 | CHJ129 | Guangzhou | 0.36 | + |
| 4 | PYJ228 | Guangzhou | >110 | + |
| 5 | SGC305 | Shaoguan | 0.74 | + |
| 6 | ZJC354 | Zhanjiang | 110 | + |
| 7 | HYC455 | Heyuan | 2.1 | + |
| 8 | ZJC1754 | Zhanjiang | 3.6 | + |
| 9 | SGJ1827 | Shaoguan | 1.1 | + |
| 10 | HYC1854 | Heyuan | 1.5 | + |
| 11 | HKC504 | Haikou | 0.36 | + |
| 12 | HKC505 | Haikou | 0.92 | + |
| 13 | NNC655 | Nanning | 4.3 | + |
| 14 | FZC705 | Fuzhou | 0.36 | + |
| 15 | XMC755 | Xiamen | 0.36 | + |
| 16 | HKC2404 | Haikou | 0.36 | + |
| 17 | SYJ2477 | Sanya | 0.62 | + |
| 18 | WHC955 | Wuhan | 0.36 | + |
| 19 | TYC1255 | Taiyuan | 0.36 | + |
| 20 | LZC2155 | Lanzhou | 0.36 | + |
| 21 | BJC2054 | Beijing | 2 | + |
| 22 | BJJ2077 | Beijing | 2.3 | + |
| 23 | CSJ2827 | Changsha | <0.3 | + |
| 24 | HZJ2876 | Hangzhou | 0.74 | + |
| 25 | HZJ2877 | Hangzhou | <0.3 | + |
| 26 | XNC3004 | Xining | 0.3 | + |
| 27 | HHHTC3104 | Huhehaote | 0.3 | + |
| 28 | HHHTJ3128 | Huhehaote | 24 | + |
| 29 | SYC3154 | Shenyang | 0.3 | + |
| 30 | NJC3204 | Nanjing | 24 | + |
| 31 | ZZC3327 | Zhengzhou | 0.92 | + |
| 32 | LSC3354 | Lasa | 4.3 | + |
| 33 | LSJ3377 | Lasa | 2.3 | + |
| 34 | AMC3457 | Macau | 0.36 | + |
| 35 | CSJ3580 | Changsha | 2.3 | + |
| 36 | NJS3706 | Nanjing | 2.3 | + |
| 37 | NJJ3730 | Nanjing | 0.36 | + |
| 38 | ZZJ3926 | Zhengzhou | 1.1 | + |
| 39 | CCJ4076 | Changchun | 0.36 | + |
| 40 | AMC4104 | Macau | 0.36 | + |
| 41 | AMJ4127 | Macau | 24 | + |
| Antibiotics | Zone Diameters (mm) | S. aureus (n = 64) | |||||
|---|---|---|---|---|---|---|---|
| R | I | S | NO. of Resistant Strains (%) | NO. of Intermediate-Resistance Strains (%) | NO. of Susceptible (%) | ||
| β-Lactams | Amoxycillin/clavulanic acid | ≤19 | - | ≥20 | 7 (11.1%) | 0 (0%) | 56 (88.9%) |
| Ampicillin | ≤28 | - | ≥29 | 54 (85.7%) | 0 (0%) | 9 (14.3%) | |
| Cefepime | ≤14 | 15–17 | ≥18 | 3 (4.8%) | 2 (3.2%) | 58 (92.0%) | |
| Cefoxitin | ≤21 | - | ≥22 | 5 (7.9%) | 0 (0%) | 58 (92.1%) | |
| Penicllin G | ≤28 | - | ≥29 | 54 (85.7%) | 0 (0%) | 9 (14.3%) | |
| Ceftazidime | ≤14 | 15–17 | ≥18 | 5 (7.9%) | 4 (6.4%) | 54 (85.7%) | |
| Aminoglycosides | Amikacin | ≤14 | 15–16 | ≥17 | 1 (1.6%) | 14 (22.2%) | 48 (76.2%) |
| Gentamicin | ≤12 | 13–14 | ≥15 | 4 (6.4%) | 0 (0%) | 59 (93.6%) | |
| Kanamycin | ≤13 | 14–17 | ≥18 | 14 (22.2%) | 10 (15.9%) | 39 (61.9%) | |
| Streptomycin | ≤11 | 12–14 | ≥15 | 6 (9.5%) | 45 (71.4%) | 12 (19.1%) | |
| Phenicols | Chloramphenicol | ≤17 | 18–20 | ≥21 | 0 (0%) | 17 (27.0%) | 46 (73.0%) |
| Lincosamides | Clindamycin | ≤14 | 15–20 | ≥21 | 8 (12.7%) | 7 (11.1%) | 48 (76.2%) |
| Macrolides | Erythromycin | ≤13 | 14–22 | ≥23 | 30 (47.6%) | 5 (7.9%) | 28 (44.5%) |
| Telithromycin | ≤18 | 19–21 | ≥22 | 8 (12.7%) | 10 (15.9%) | 45 (71.4%) | |
| Fluoroquinolones | Ciprofloxacin | ≤15 | 16–20 | ≥21 | 2 (3.2%) | 3 (4.8%) | 58 (92.0%) |
| Norfloxacin | ≤12 | 13–16 | ≥17 | 2 (3.2%) | 3 (4.8%) | 58 (92.0%) | |
| Tetracyclines | Tetracycline | ≤14 | 15–18 | ≥19 | 11 (17.5%) | 0 (0%) | 52 (82.5%) |
| Oxazolidinones | Linezolid | ≤20 | - | ≥21 | 0 (0%) | 0 (0%) | 63 (100%) |
| Ansamycins | Rifampicin | ≤16 | 17–19 | ≥20 | 3 (4.8%) | 1 (1.6%) | 59 (93.6%) |
| Sulfonamides | Trimethoprim/sulphamethoxazole 1:19 | ≤10 | 11–15 | ≥16 | 1 (1.6%) | 1 (1.6%) | 61 (96.8%) |
| Quinolones | Quinupristin/dalfopristin | ≤15 | 16–18 | ≥19 | 0 (0%) | 3 (4.8%) | 60 (95.2%) |
| Glycopeptides | Teicoplanin | ≤10 | 11–13 | ≥14 | 0 (0%) | 16 (25.4%) | 47 (74.6%) |
| Nitrofurantoins | Nitrofurantoin | ≤14 | 15–16 | ≥17 | 0 (0%) | 6 (9.5%) | 57 (90.5%) |
| Fusidic acid | ≤24 | - | ≥25 | 14 (22.2%) | 0 (0%) | 49 (77.8%) | |
| No. | S. aureus Isolates | Biofilm Production Assay * | Biofilm Production Ability * | Biofilm-Producing Genes | Adhesion Genes | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| icaA | icaB | icaC | icaD | icaR | clfA | clfB | fnbA | fnbB | cna | map | ebp | sdrC | sdrD | sdrE | ||||
| 1 | 27-0 | 3.5427 | +++ | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| 2 | 54-0 | 2.6298 | +++ | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| 3 | 54-1 | 2.9668 | +++ | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| 4 | 129-0 | 2.9715 | +++ | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| 5 | 228-0 | 2.8057 | +++ | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| 6 | 228-1 | 3.3775 | +++ | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| 7 | 228-2 | 2.1027 | +++ | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| 8 | 305 | 1.2128 | +++ | + | + | + | + | + | + | + | + | + | - | + | + | + | + | + |
| 9 | 354 | 1.8072 | +++ | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| 10 | 455 | 2.9610 | +++ | + | + | + | + | + | + | + | + | + | - | + | + | + | + | + |
| 11 | 1754 | 1.1967 | +++ | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| 12 | 1827-0 | 2.0557 | +++ | + | + | + | + | + | + | + | + | + | - | + | + | + | + | + |
| 13 | 1854 | 2.9648 | +++ | + | + | + | + | + | + | + | + | + | - | + | + | + | - | + |
| 14 | 504 | 2.5967 | +++ | + | + | + | + | + | + | + | + | + | - | + | + | + | + | + |
| 15 | 505 | 1.0083 | ++ | + | + | + | + | + | + | + | + | + | - | + | + | + | + | + |
| 16 | 655 | 2.4525 | +++ | + | + | + | + | + | + | + | + | + | - | + | + | + | + | + |
| 17 | 705 | 2.8222 | +++ | + | + | + | + | + | + | + | + | - | - | + | + | + | + | + |
| 18 | 755 | 3.0410 | +++ | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| 19 | 2404-0 | 2.2565 | +++ | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| 20 | 2477-1 | 1.7952 | +++ | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| 21 | 955 | 2.1990 | +++ | + | + | + | + | + | + | + | + | + | - | + | + | + | + | + |
| 22 | 1255 | 2.7460 | +++ | + | + | + | + | + | + | + | + | + | + | + | + | + | - | + |
| 23 | 2155-0 | 3.4222 | +++ | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| 24 | 2054-0 | 1.3443 | +++ | + | + | + | + | + | + | + | + | + | - | + | + | + | + | + |
| 25 | 2054-1 | 1.8343 | +++ | + | + | + | + | + | + | + | + | + | - | + | + | + | + | + |
| 26 | 2054-2 | 1.3802 | +++ | + | + | + | + | + | + | + | + | + | - | + | + | + | + | + |
| 27 | 2077-0 | 2.2180 | +++ | + | + | + | + | + | + | + | + | + | - | + | + | + | + | + |
| 28 | 2827–6 | 1.0738 | ++ | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| 29 | 2827–8 | 1.4532 | +++ | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| 30 | 2876A1 | 1.5003 | +++ | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| 31 | 2876B2 | 2.0088 | +++ | - | - | - | - | - | + | + | + | + | - | + | + | + | + | + |
| 32 | 2877 | 3.4300 | +++ | + | + | + | + | + | + | + | + | + | - | + | + | + | + | + |
| 33 | 2877C1 | 3.4535 | +++ | + | + | + | + | + | + | + | + | + | - | + | + | + | + | + |
| 34 | 2877A2 | 2.1013 | +++ | + | + | + | + | + | + | + | + | + | - | + | + | + | + | + |
| 35 | 3004 | 2.3895 | +++ | + | + | + | + | + | + | + | + | + | - | + | + | + | + | + |
| 36 | 3004B2 | 2.1682 | +++ | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| 37 | 3104C2 | 2.5065 | +++ | + | + | + | + | + | + | + | + | + | - | + | + | + | + | + |
| 38 | 3128 | 2.2482 | +++ | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| 39 | 3128A1 | 2.8780 | +++ | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| 40 | 3128A2 | 2.4757 | +++ | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| 41 | 3154 | 2.3555 | +++ | + | + | + | + | + | + | + | + | + | - | + | + | + | + | + |
| 42 | 3154C2 | 3.5033 | +++ | + | + | + | + | + | + | + | + | + | - | + | + | + | + | + |
| 43 | 3204 | 3.2423 | +++ | + | + | + | + | + | + | + | + | + | - | + | + | + | + | - |
| 44 | 3204C1 | 3.3025 | +++ | + | + | + | + | + | + | + | + | + | - | + | + | + | + | + |
| 45 | 3204A2 | 3.1310 | +++ | + | + | + | + | + | + | + | + | + | - | + | + | + | + | + |
| 46 | 3327A3 | 3.5470 | +++ | + | + | + | + | + | + | + | + | + | - | + | + | + | + | + |
| 47 | 3354 | 2.5655 | +++ | + | + | + | + | + | + | + | + | + | + | + | + | + | - | + |
| 48 | 3354A1 | 2.8357 | +++ | + | + | + | + | + | + | + | + | + | + | + | + | + | - | + |
| 49 | 3354C2 | 2.2680 | +++ | + | + | + | + | + | + | + | + | + | + | + | + | + | - | + |
| 50 | 3377 | 2.4502 | +++ | + | + | + | + | + | + | + | + | + | - | + | + | + | + | + |
| 51 | 3377C1 | 2.0077 | +++ | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| 52 | 3457A1 | 3.7317 | +++ | + | + | + | + | + | + | + | - | - | - | - | - | + | + | + |
| 53 | 3580C1 | 3.2920 | +++ | + | + | + | + | + | + | + | + | + | - | + | + | + | + | + |
| 54 | 3706A1 | 3.6107 | +++ | - | + | + | + | + | + | + | + | + | - | + | + | + | + | + |
| 55 | 3730B1 | 1.3638 | +++ | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| 56 | 3926C2 | 2.1268 | +++ | + | + | + | + | + | + | + | + | + | - | + | + | + | + | + |
| 57 | 3926C3 | 2.6023 | +++ | + | + | + | + | + | + | + | + | + | - | + | + | + | + | + |
| 58 | 4076A1 | 1.7465 | +++ | + | + | + | + | + | + | + | + | - | + | + | + | + | + | + |
| 59 | 4104 | 2.3777 | +++ | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| 60 | 4104A1 | 2.6428 | +++ | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| 61 | 4127 | 2.9718 | +++ | + | + | + | + | + | + | + | + | + | - | + | + | + | + | + |
| 62 | 4127A1 | 3.1565 | +++ | + | + | + | + | + | + | + | + | + | - | + | + | + | + | + |
| 63 | 4127A2 | 2.3085 | +++ | + | + | + | + | + | + | + | + | + | - | + | + | + | - | + |
| No. | Strains | ST | spa-Type | Sample Origin | Antibiotic Profiles | SEs Gene |
|---|---|---|---|---|---|---|
| 27-0 | 1 | t5837 | Guangzhou | AMP-PEN | sec-seh-sel | |
| 2 | 54-0 | 1 | t127 | Guangzhou | AMP-PEN | sec-seh-sel |
| 3 | 54-1 | 1 | t127 | Guangzhou | AMP-PEN | sec-seh-sel |
| 4 | 129-0 | 1 | t127 | Guangzhou | - | sec-seh-sel |
| 5 | 228-0 | 1 | t127 | Guangzhou | AMP-PEN--KAN-TET-RIF | sec-seh-sel |
| 6 | 228-1 | 1 | t127 | Guangzhou | AMP-PEN--KAN-TET-RIF | sec-seh-sel |
| 7 | 228-2 | 1 | t127 | Guangzhou | AMP-PEN--KAN-TET-RIF | sec-seh-sel |
| 8 | 305 | 59 | t437 | Shaoguan | AMP-PEN--KAN-STR-CLI-ERY-TEL-TET-FD | sea-seb-sek-seq |
| 9 | 354 | 6 | t701 | Zhanjiang | AMP-PEN | - |
| 10 | 455 | 15 | t4309 | Heyuan | AMP-PEN-ERY | - |
| 11 | 1754 | 6 | t701 | Zhanjiang | AMP-PEN | - |
| 12 | 1827-0 | 59 | t437 | Shaoguan | AMP-FEP-FOX-PEN-CAZ-KAN-STR-CLI-ERY-TEL-TET | seb-sek-seq |
| 13 | 1854 | 7 | t091 | Heyuan | AMP-PEN-KAN-TET | - |
| 14 | 504 | 25 | t17887 | Haikou | AMP-PEN | seb |
| 15 | 505 | 25 | t078 | Haikou | AMP-PEN | seb-sec-sel-sek |
| 16 | 655 | 15 | t085 | Nanning | AMP-PEN-ERY | - |
| 17 | 705 | 59 | t437 | Fuzhou | AMC-AMP-FOX-PEN-CAZ-KAN-STR-CLI-ERY-TEL | sea-seb-sek-seq |
| 18 | 755 | 1 | t5837 | Xiamen | AMP-PEN | sea-sec-seh-sel-sek-seq |
| 19 | 2404-0 | 1 | t114 | Haikou | AMC-AMP-FEP-FOX-PEN-CAZ-ERY | sec-seh-sel |
| 20 | 2477-1 | 6 | t701 | Sanya | AMP-PEN | - |
| 21 | 955 | 7 | t796 | Wuhan | AMP-PEN-GEN-KAN-CLI-ERY-TEL-FD | sea |
| 22 | 1255 | 398 | t571 | Taiyuan | AMP-PEN-FD | - |
| 23 | 2155-0 | 398 | t034 | Lanzhou | ERY | seh |
| 24 | 2054-0 | 15 | t085 | Beijing | AMP-PEN-ERY | - |
| 25 | 2054-1 | 15 | t085 | Beijing | AMP-PEN-ERY | - |
| 26 | 2054-2 | 15 | t085 | Beijing | AMP-PEN-ERY | - |
| 27 | 2077-0 | 15 | t085 | Beijing | AMP-PEN-ERY | - |
| 28 | 2827–6 | 4071 | t17886 | Changsha | —— | sea |
| 29 | 2827–8 | 4071 | t17886 | Changsha | —— | sea |
| 30 | 2876A1 | 188 | t189 | Hangzhou | AMC-AMP-PEN-FD | - |
| 31 | 2876B2 | 630 | t377 | Hangzhou | AMP-PEN-TET-FD | - |
| 32 | 2877 | 15 | t085 | Hangzhou | AMP-PEN-ERY | - |
| 33 | 2877C1 | 15 | t085 | Hangzhou | AMP-PEN-ERY | - |
| 34 | 2877A2 | 15 | t085 | Hangzhou | AMP-PEN-ERY | - |
| 35 | 3004 | 2205 | t377 | Xining | AMC-AMP-PEN-ERY-CIP-NOR-TET | seb |
| 36 | 3004B2 | 188 | t189 | Xining | AMP-PEN | - |
| 37 | 3104C2 | 25 | t3033 | Huhehaote | AMP-PEN-ERY | seb |
| 38 | 3128 | 1 | t127 | Huhehaote | AMP-PEN-FD | sec-seh-sel |
| 39 | 3128A1 | 1 | t127 | Huhehaote | AMP-PEN | sec-seh-sel |
| 40 | 3128A2 | 1 | t127 | Huhehaote | AMP-PEN | sec-seh-sel |
| 41 | 3154 | 25 | t258 | Shenyang | AMP-PEN-ERY | seb |
| 42 | 3154C2 | 15 | t085 | Shenyang | AMP-PEN-ERY | - |
| 43 | 3204 | 7 | t1689 | Nanjing | AMP-PEN-KAN-TET | sea |
| 44 | 3204C1 | 7 | t091 | Nanjing | AMP-PEN-KAN-TET | sea |
| 45 | 3204A2 | 7 | t091 | Nanjing | AMP-PEN-KAN-TET | sea |
| 46 | 3327A3 | 15 | t085 | Zhengzhou | AMP-PEN-CLI-ERY | - |
| 47 | 3354 | 188 | t189 | Lasa | —— | - |
| 48 | 3354A1 | 188 | t189 | Lasa | —— | - |
| 49 | 3354C2 | 188 | t189 | Lasa | —— | - |
| 50 | 3377 | 15 | t085 | Lasa | AMP-PEN-ERY-FD | - |
| 51 | 3377C1 | 15 | t085 | Lasa | AMP-PEN-ERY-FD | - |
| 52 | 3457A1 | 72 | t3092 | Macau | —— | - |
| 53 | 3580C1 | 15 | t085 | Changsha | AMP-PEN-ERY | - |
| 54 | 3706A1 | 4692 | t17888 | Nanjing | AMP-PEN-FD | sec-sel |
| 55 | 3730B1 | 6 | t701 | Nanjing | AMP-PEN | - |
| 56 | 3926C2 | 15 | t085 | Zhengzhou | AMP-PEN-ERY-FD | - |
| 57 | 3926C3 | 15 | t085 | Zhengzhou | AMP-PEN-ERY-FD | - |
| 58 | 4076A1 | 398 | t034 | Changchun | AMC-AMP-PEN-AMK-GEN-KAN-CLI-ERY-TEL-SXT | - |
| 59 | 4104 | 188 | t189 | Macau | AMC-AMP-FEP-FOX-PEN-CAZ-GEN-KAN-STR-CLI-ERY-TEL-CIP-NOR | - |
| 60 | 4104A1 | 188 | t189 | Macau | TEL-FD | seb |
| 61 | 4127 | 15 | t085 | Macau | AMP-PEN-STR-ERY | - |
| 62 | 4127A1 | 59 | t437 | Macau | AMC-AMP-FOX-PEN-CAZ-KAN-STR-CLI-ERY-TEL-FD | seb-sek-seq |
| 63 | 4127A2 | 15 | t085 | Macau | AMP-PEN-ERY-FD | - |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dai, J.; Huang, J.; Wu, S.; Zhang, F.; Li, Y.; Rong, D.; Zhao, M.; Ye, Q.; Gu, Q.; Zhang, Y.; et al. Occurrence, Antibiotic Susceptibility, Biofilm Formation and Molecular Characterization of Staphylococcus aureus Isolated from Raw Shrimp in China. Foods 2023, 12, 2651. https://doi.org/10.3390/foods12142651
Dai J, Huang J, Wu S, Zhang F, Li Y, Rong D, Zhao M, Ye Q, Gu Q, Zhang Y, et al. Occurrence, Antibiotic Susceptibility, Biofilm Formation and Molecular Characterization of Staphylococcus aureus Isolated from Raw Shrimp in China. Foods. 2023; 12(14):2651. https://doi.org/10.3390/foods12142651
Chicago/Turabian StyleDai, Jingsha, Jiahui Huang, Shi Wu, Feng Zhang, Yuanyu Li, Dongli Rong, Miao Zhao, Qinghua Ye, Qihui Gu, Youxiong Zhang, and et al. 2023. "Occurrence, Antibiotic Susceptibility, Biofilm Formation and Molecular Characterization of Staphylococcus aureus Isolated from Raw Shrimp in China" Foods 12, no. 14: 2651. https://doi.org/10.3390/foods12142651
APA StyleDai, J., Huang, J., Wu, S., Zhang, F., Li, Y., Rong, D., Zhao, M., Ye, Q., Gu, Q., Zhang, Y., Wei, X., Zhang, J., & Wu, Q. (2023). Occurrence, Antibiotic Susceptibility, Biofilm Formation and Molecular Characterization of Staphylococcus aureus Isolated from Raw Shrimp in China. Foods, 12(14), 2651. https://doi.org/10.3390/foods12142651





