Probiotic Properties and Proteomic Analysis of Pediococcus pentosaceus 1101
Abstract
1. Introduction
2. Materials and Methods
2.1. Strain Propagation
2.2. Bacterial Identification
2.3. Genome Sequencing Assembly
2.4. Evaluation of Probiotic Properties
2.4.1. Antagonistic Activity
2.4.2. Growth Kinetics at Different pH Values
2.4.3. Proteolytic Activity at Different pH Values
2.4.4. Inhibitory Activity at Different pH Values
2.4.5. P. pentosaceus 1101 Survival after Exposure to Low pH and Bile Salts
2.4.6. Antibiotic Resistance
2.4.7. Cell-Adhesion Properties
2.5. Proteomic Analysis of P. pentosaceus 1101 Exposed under Control and Gastrointestinal Conditions
2.5.1. Strain Exposure under Control and Gastrointestinal Conditions
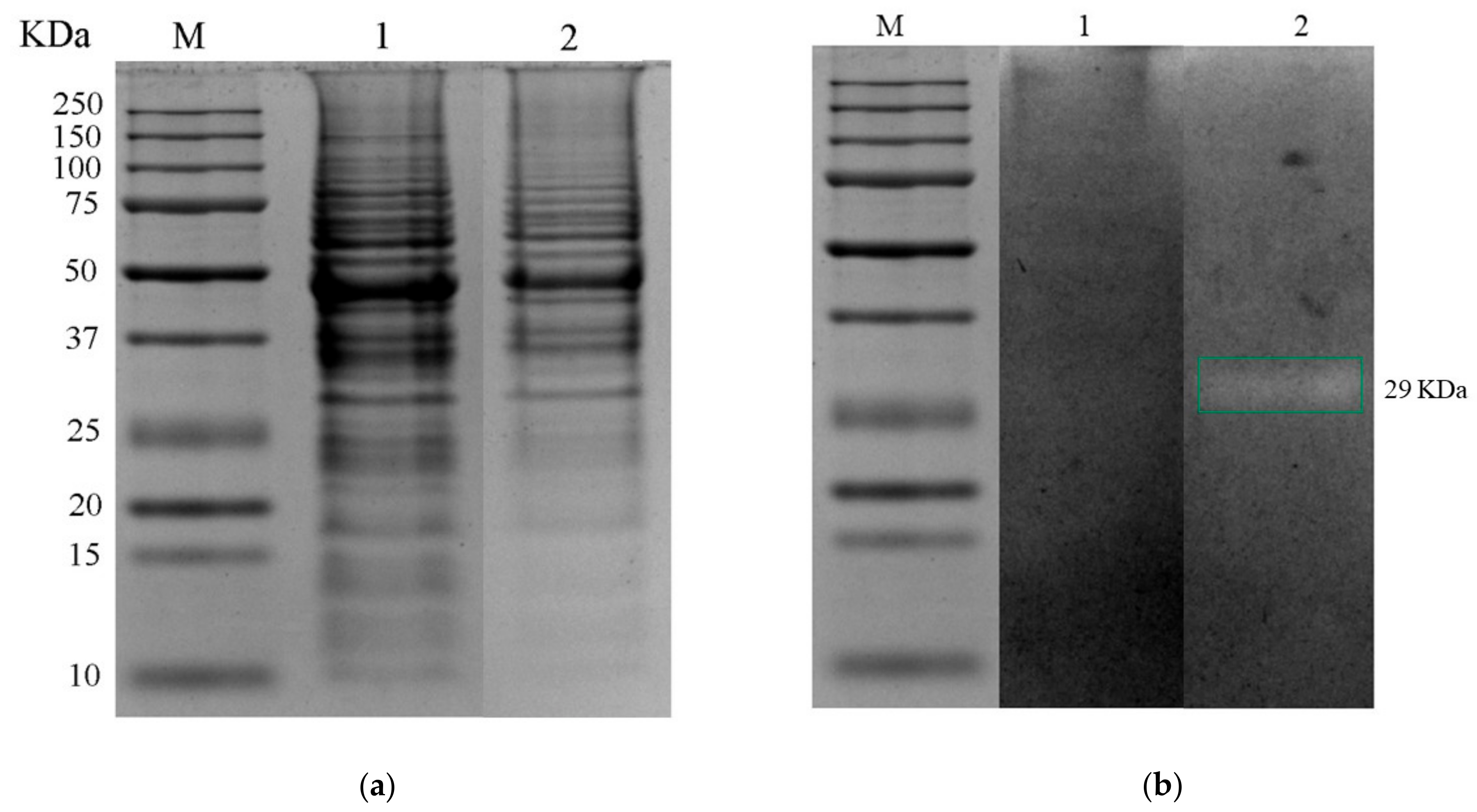
2.5.2. SDS–PAGE Electrophoretic Analysis and Lytic Activity by Zymograms
Protein Extraction
Electrophoretic Analysis
2.5.3. Protein Identification Using Liquid Chromatography Coupled to Tandem Mass Spectrometry (LC–MS/MS)
Extraction of Intracellular Protein Extracts from P. pentosaceus 1101
LC–MS/MS Analysis
Database Searching and Protein Identification Criteria
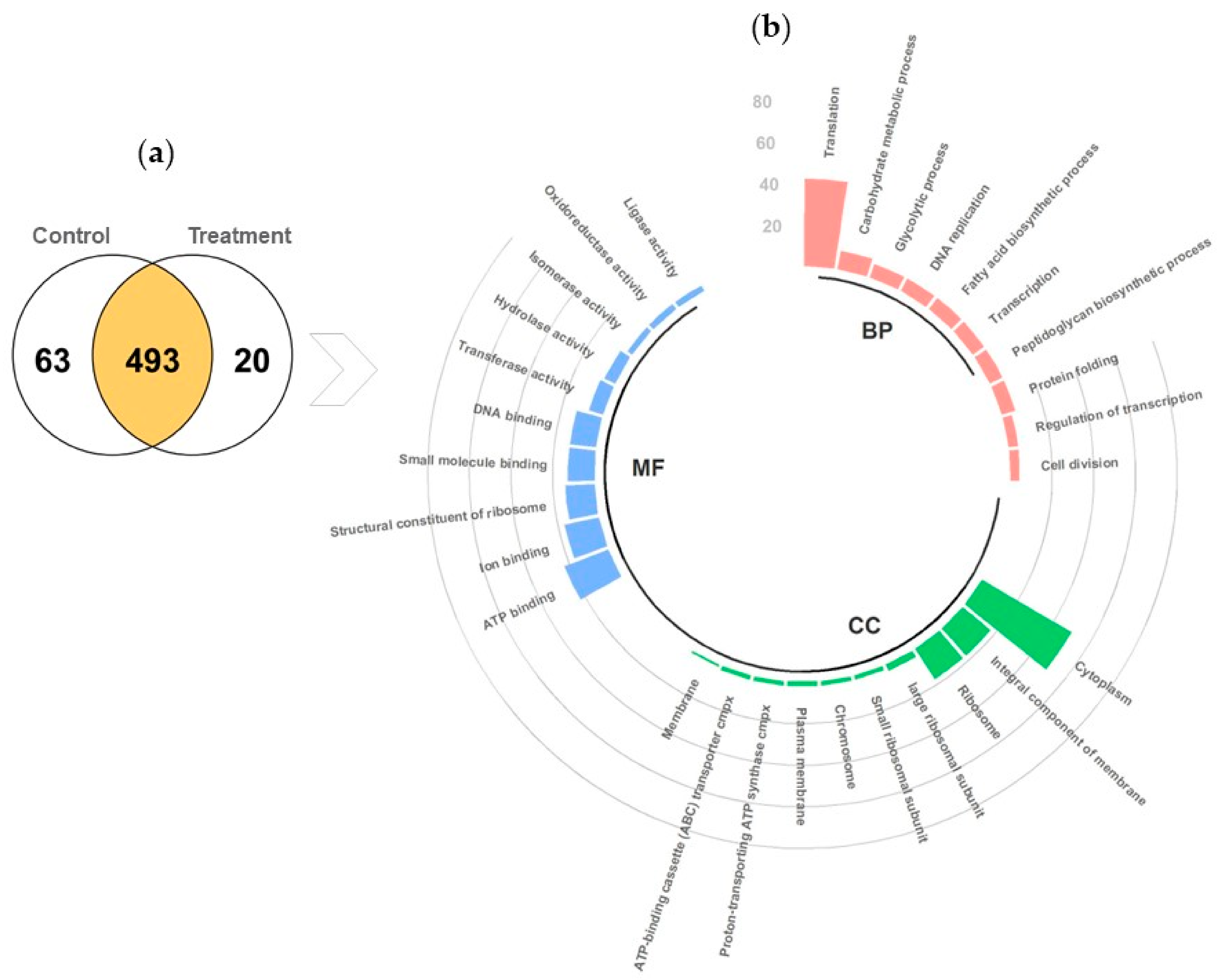
Bioinformatic Analysis
2.6. Statistical Analysis
3. Results and Discussion
3.1. Identification of the Strain
3.2. Genome Sequencing and Assembly
3.3. Evaluation of Probiotic Properties
3.3.1. Antagonistic Activity
3.3.2. Growth, Proteolytic and Inhibitory Activity at Different pH Values
3.3.3. P. pentosaceus 1101 Survival after Exposure to Low pH and Bile Salts
3.3.4. Antibiotic Resistance
3.3.5. Cell Adhesion Properties
3.4. Proteomic Analysis of P. pentosaceus 1101 Exposed under Control and Gastrointestinal Conditions
3.4.1. SDS–PAGE Analysis and Zymographic Antimicrobial Activity Determinations
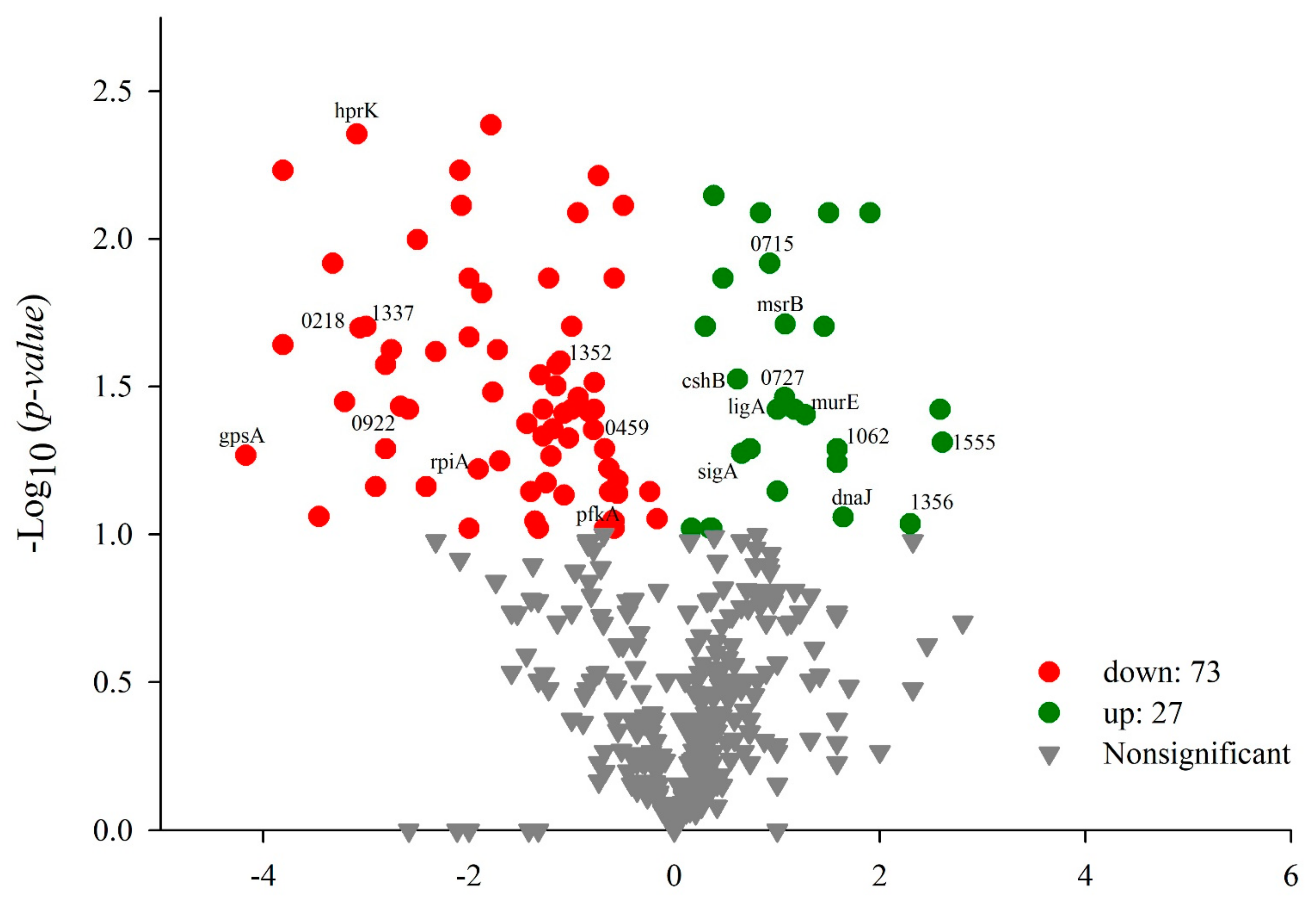
3.4.2. Protein Identification Using Liquid Chromatography Coupled to Tandem Mass Spectrometry (LC–MS/MS)
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Fan, S.; Xue, T.; Bai, B.; Bo, T.; Zhang, J. Probiotic Properties Including the Antioxidant and Hypoglycemic Ability of Lactic Acid Bacteria from Fermented Grains of Chinese Baijiu. Foods 2022, 11, 3476. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Xie, X.; Li, Y.; Chen, M.; Xue, L.; Wang, J.; Zhang, J.; Wu, S.; Ye, Q.; Zhang, S.; et al. Pediococcus pentosaceus IM96 Exerts Protective Effects against Enterohemorrhagic Escherichia coli O157: H7 Infection In Vivo. Foods 2021, 10, 2945. [Google Scholar] [CrossRef] [PubMed]
- Yang, S.J.; Kim, K.T.; Kim, T.Y.; Paik, H.D. Probiotic properties and antioxidant activities of Pediococcus pentosaceus SC28 and Levilactobacillus brevis KU15151 in fermented black gamju. Foods 2020, 9, 1154. [Google Scholar] [CrossRef]
- Ramirez-Chavarin, M.L.; Wacher, C.; Eslava-Campos, C.A.; Perez-Chabela, M.L. Probiotic potential of thermotolerant lactic acid bacteria strains isolated from cooked meat products. Int. Food Res. J. 2013, 20, 991–1000. [Google Scholar]
- Hernández-Alcántara, A.M.; Wacher, C.; Llamas, M.G.; López, P.; Pérez-Chabela, M.L. Probiotic properties and stress response of thermotolerant lactic acid bacteria isolated from cooked meat products. LWT Food Sci. Technol. 2018, 91, 249–257. [Google Scholar] [CrossRef]
- Śliżewska, K.; Chlebicz-Wójcik, A.; Nowak, A. Probiotic properties of new Lactobacillus strains intended to be used as feed additives for monogastric animals. Probiotics Antimicrob. Proteins 2021, 13, 146–162. [Google Scholar] [CrossRef]
- Bao, Y.; Zhang, Y.; Zhang, Y.; Liu, Y.; Wang, S.; Dong, X.; Wang, Y.; Zhang, H. Screening of potential probiotic properties of Lactobacillus fermentum isolated from traditional dairy products. Food Control 2010, 21, 695–701. [Google Scholar] [CrossRef]
- Gorjbach, S.L.; Nahas, L.; Lerner, P.I. Studies of intestinal microflora. I. Effects of diet, age, and periodic sampling on numbers of fecal microorganisms in man. Gastroenterology 1967, 53, 845–855. [Google Scholar]
- Erkkilä, S.; Petäjä, E. Screening of commercial meat starter cultures at low pH and in the presence of bile salts for potential probiotic use. Meat Sci. 2000, 55, 297–300. [Google Scholar] [CrossRef]
- Arief, I.I.; Jenie, B.S.L.; Astawan, M.; Fujiyama, K.; Witarto, A.B. Identification and probiotic characteristics of lactic acid bacteria isolated from Indonesian local beef. Asian-Australas. J. Anim. Sci. 2015, 9, 25–36. [Google Scholar]
- García-Cano, I.; Campos-Gómez, M.; Contreras-Cruz, M.; Serrano-Maldonado, C.E.; González-Canto, A.; Peña-Montes, C.; Rodríguez-Sanoja, R.; Sánchez, S.; Farrés, A. Expression, purification, and characterization of a bifunctional 99-kDa peptidoglycan hydrolase from Pediococcus acidilactici ATCC 8042. Appl. Microbiol. Biotechnol. 2015, 99, 8563–8573. [Google Scholar] [CrossRef] [PubMed]
- Eichelmann, C.G. Respuesta celular a estrés. Rev. Latinoam. Microbiol. 2006, 48, 162–172. [Google Scholar] [PubMed]
- Zheng, J.; Wittouck, S.; Salvetti, E.; Franz, C.M.; Harris, H.; Mattarelli, P.; O’Toole, P.W.; Pot, B.; Vandamme, P.; Walter, J.; et al. A taxonomic note on the genus Lactobacillus: Description of 23 novel genera, emended description of the genus Lactobacillus Beijerinck 1901, and union of Lactobacillaceae and Leuconostocaceae. Int. J. Syst. Evol. Microbiol. 2020, 70, 2782–2858. [Google Scholar] [CrossRef] [PubMed]
- Zommiti, M.; Bouffartigues, E.; Maillot, O.; Barreau, M.; Szunerits, S.; Sebei, K.; Feuilloley, M.; Connil, N.; Ferchichi, M. In vitro assessment of the probiotic properties and bacteriocinogenic potential of Pediococcus pentosaceus MZF16 isolated from artisanal Tunisian meat “dried ossban”. Front. Microbiol. 2018, 9, 2607. [Google Scholar] [CrossRef] [PubMed]
- Doyle, M.P.; Meng, J. Bacteria in food and beverage production. In The Prokaryotes, 3rd ed.; Dworkin, M., Falkow, S., Rosenberg, E., Schleifer, K.H., Stackebrandt, E., Eds.; Springer: New York, NY, USA, 2006; Volume 1, pp. 797–811. [Google Scholar]
- Abril, A.G.; Quintela-Baluja, M.; Villa, T.G.; Calo-Mata, P.; Barros-Velázquez, J.; Carrera, M. Proteomic Characterization of Virulence Factors and Related Proteins in Enterococcus Strains from Dairy and Fermented Food Products. Int. J. Mol. Sci. 2022, 23, 10971. [Google Scholar] [CrossRef]
- Baig, M.A.; Turner, M.S.; Liu, S.Q.; Al-Nabulsi, A.A.; Shah, N.P.; Ayyash, M.M. Potential Probiotic Pediococcus pentosaceus M41 Modulates Its Proteome Differentially for Tolerances Against Heat, Cold, Acid, and Bile Stresses. Front. Microbiol. 2021, 12, 2952. [Google Scholar] [CrossRef]
- Zhang, H.; Zhang, C.; Liu, H.; Chen, Q.; Kong, B. Proteomic response strategies of Pediococcus pentosaceus R1 isolated from Harbin dry sausages to oxidative stress. Food Biosci. 2021, 44, 101364. [Google Scholar] [CrossRef]
- Juárez-Castelán, C.; García-Cano, I.; Escobar-Zepeda, A.; Azaola-Espinosa, A.; Álvarez-Cisneros, Y.; Ponce-Alquicira, E. Evaluation of the bacterial diversity of Spanish-type chorizo during the ripening process using high-throughput sequencing and physicochemical characterization. Meat. Sci. 2019, 150, 7–13. [Google Scholar] [CrossRef]
- Najjari, A.; Ouzari, H.; Boudabous, A.; Zagorec, M. Method for reliable isolation of Lactobacillus sakei strains originating from Tunisian seafood and meat products. Int. J. Food Microbiol. 2008, 121, 342–351. [Google Scholar] [CrossRef]
- Mirás, V.I. Estudio de la Población de Bacterias Ácido Lácticas en un Embutido Cárnico Mediante MALDI TOF. Bachelor’s Thesis, Universidad de Valladolid, Valladolid, Spain, 2019. [Google Scholar]
- Bankevich, A.; Nurk, S.; Antipov, D.; Gurevich, A.A.; Dvorkin, M.; Kulikov, A.S.; Lesin, V.M.; Nikolenko, S.I.; Pham, S.; Prjibelski, A.D.; et al. SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 2012, 19, 455–477. [Google Scholar] [CrossRef]
- Seemann, T. Prokka: Rapid prokaryotic genome annotation. Bioinformatics 2014, 30, 2068–2069. [Google Scholar] [CrossRef] [PubMed]
- Aguilar-Zarate, P.; Cruz-Hernandez, M.A.; Montanez, J.C.; Belmares-Cerda, R.E.; Aguilar, C.N. Enhancement of tannase production by Lactobacillus plantarum CIR1: Validation in gas-lift bioreactor. Bioprocess Biosyst. Eng. 2014, 37, 2305–2316. [Google Scholar] [CrossRef] [PubMed]
- STATISTICA 2004 (Version 7). Available online: https://www.tibco.com/products/data-science (accessed on 22 May 2022).
- NCSS 2007 Update (Version 1). Available online: https://www.ncss.com/download/ncss/updates/ncss-2007-v1/ (accessed on 22 May 2022).
- Molin, G.; Ternström, A. Numerical taxonomy of psychrotrophic pseudomonads. J. Gen. Microbiol. 1982, 128, 1249–1264. [Google Scholar] [CrossRef] [PubMed]
- García-Cano, I.; Serrano-Maldonado, C.E.; Olvera-García, M.; Delgado-Arciniega, E.; Peña-Montes, C.; Mendoza-Hernández, G.; Quirasco, M. Antibacterial activity produced by Enterococcus spp. isolated from an artisanal Mexican dairy product, Cotija cheese. LWT-Food Sci. Technol. 2014, 59, 26–34. [Google Scholar] [CrossRef]
- Bradford, M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Khan, I.; Kang, S.C. Probiotic potential of nutritionally improved Lactobacillus plantarum DGK-17 isolated from Kimchi–A traditional Korean fermented food. Food Control 2016, 60, 88–94. [Google Scholar] [CrossRef]
- Savedboworn, W.; Riansa-ngawong, W.; Sinlapacharoen, W.; Pajakang, S.; Patcharajarukit, B.; Tipkanon, S.; Phattayakorn, K. Assessment of probiotic properties in lactic acid bacteria isolated from fermented vegetables. Int. J. Appl. Sci. Technol. 2014, 7, 53–65. [Google Scholar] [CrossRef][Green Version]
- Sánchez, L.; Tromps, J. Caracterización in vitro de bacterias ácido lácticas con potencial probiótico. Rev. Salud Anim. 2014, 36, 124–129. [Google Scholar]
- Collado, M.C.; Meriluoto, J.; Salminen, S. Adhesion and aggregation properties of probiotic and pathogen strains. Eur. Food Res. Technol. 2008, 226, 1065–1073. [Google Scholar] [CrossRef]
- Malik, A.; Sakamoto, M.; Hanazaki, S.; Osawa, M.; Suzuki, T.; Tochigi, M.; Kakii, K. Coaggregation among Nonflocculating Bacteria Isolated from Activated Sludge. Appl. Environ. Microbiol. 2003, 69, 6056–6063. [Google Scholar] [CrossRef]
- Kosin, B.; Rakshit, S.K. Induction of heat tolerance in autochthonous and allochthonous thermotolerant probiotics for application to white shrimp feed. Aquaculture 2010, 306, 302–309. [Google Scholar] [CrossRef]
- Navarro, U.C. Análisis Proteómico de las Vías de Señalización Mediadas por la Subunidad Gα Pga1 de una Proteína G Heterotrimérica de Penicillium chrysogenum. Ph.D. Thesis, Universidad Autónoma Metropolitana, Mexico City, Mexico, 2017. [Google Scholar]
- Laemmli, U.K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 1970, 227, 680–685. [Google Scholar] [CrossRef] [PubMed]
- Keller, A.; Nesvizhskii, A.I.; Kolker, E.; Aebersold, R. Empirical statistical model to estimate the accuracy of peptide identifications made by MS/MS and database search. Anal. Chem. 2002, 74, 5383–5392. [Google Scholar] [CrossRef] [PubMed]
- Nesvizhskii, A.I.; Keller, A.; Kolker, E.; Aebersold, R. A statistical model for identifying proteins by tandem mass spectrometry. Anal. Chem. 2003, 75, 4646–4658. [Google Scholar] [CrossRef] [PubMed]
- Zhu, W.H.; Smith, J.W.; Huang, C.-M. Mass spectrometry-based label-free quantitative proteomics. J. Biomed. Biotechnol. 2010, 2010, 840518. [Google Scholar] [CrossRef]
- Sherman, B.T.; Hao, M.; Qiu, J.; Jiao, X.; Baseler, M.W.; Lane, H.C.; Imamichi, T.; Chang, W. DAVID: A web server for functional enrichment analysis and functional annotation of gene lists (2021 update). Nucleic Acids Res. 2022, 10, 1–6. [Google Scholar] [CrossRef]
- Helander, I.M.; Von Wright, A.; Mattila-Sandholm, T.-M. Potential of lactic acid bacteria and novel antimicrobials against Gram-negative bacteria. Trends Food Sci. Technol. 1997, 8, 146–150. [Google Scholar] [CrossRef]
- Berardo, A.; Devreese, B.; De Maere, H.; Stavropoulou, D.A.; Van Royen, G.; Leroy, F.; De Smet, S. Actin proteolysis during ripening of dry fermented sausages at different pH values. Food Chem. 2017, 221, 1322–1332. [Google Scholar] [CrossRef]
- Sun, F.; Hu, Y.; Chen, Q.; Kong, B.; Liu, Q. Purification and biochemical characteristics of the extracellular protease from Pediococcus pentosaceus isolated from Harbin dry sausages. Meat Sci. 2019, 156, 156–165. [Google Scholar] [CrossRef]
- Engelhardt, T.; Albano, H.; Kiskó, G.; Mohácsi-Farkas, C.; Teixeira, P. Antilisterial activity of bacteriocinogenic Pediococcus acidilactici HA6111-2 and Lactobacillus plantarum ESB 202 grown under pH and osmotic stress conditions. Food Microbiol. 2015, 48, 109–115. [Google Scholar] [CrossRef]
- Juárez-Castelán, C. Evaluación de la Diversidad y Dinámica Bacteriana del Chorizo Tipo Español Durante su Proceso de Maduración, Mediante DGGE y Secuenciación Masiva. Ph.D. Thesis, Universidad Autónoma Metropolitana, Mexico City, Mexico, 2019. [Google Scholar]
- Montville, T.J.; Chen, Y. Mechanistic action of pediocin and nisin: Recent progress and unresolved questions. Appl. Microbiol. Biotechnol. 1998, 50, 511–519. [Google Scholar] [CrossRef] [PubMed]
- Havenaar, R.; Brink, B.T.; Huis, I.V.; Jos, H.J. Selection of Strains for Probiotic Use. In Probiotics; Springer: Dordrecht, Germany, 1992; pp. 209–224. [Google Scholar]
- Jacobsen, C.N.; Rosenfeldt, N.V.; Hayford, A.E.; Møller, P.L.; Michaelsen, K.F.; Paerregaard, A.; Sandström, B.; Tvede, M.; Jakobsen, M. Screening of probiotic activities of forty seven strains of Lactobacillus spp. by in vitro techniques and evaluation of colonization ability of five selected strains in human. Appl. Environ. Microbiol. 1999, 65, 4949–4956. [Google Scholar] [CrossRef] [PubMed]
- Martoni, C.; Bhathena, J.; Jones, M.L.; Urbanska, A.M.; Chen, H.; Prakash, S. Investigation of microencapsulated BSH active Lactobacillus in the simulated human GI tract. J. Biomed. Biotechnol. 2007, 2007, 13684. [Google Scholar] [CrossRef] [PubMed]
- Noriega, L.; Gueimonde, M.; Sánchez, B.; Margolles, A.; De Los Reyes-Gavilán, C.G. Effect of the adaptation to high bile salts concentrations on glycosidic activity, survival at low pH and cross-resistance to bile salts in Bifidobacterium. Int. J. Food Microbiol. 2004, 94, 79–86. [Google Scholar] [CrossRef] [PubMed]
- Ventura, M.; Turroni, F.; Van Sinderen, D. Probiogenomics as a tool to obtain genetic insights into adaptation of probiotic bacteria to the human gut. Bioeng. Bugs 2012, 3, 73–79. [Google Scholar] [CrossRef]
- Conway, P.L.; Gorbach, S.L.; Goldin, B.R. Survival of lactic acid bacteria in the human stomach and adhesion to intestinal cells. J. Dairy Sci. 1987, 70, 1–12. [Google Scholar] [CrossRef]
- Jaya, P.; Gill, H.; Smart, J.; Gopal, P.K. Selection and characterisation of Lactobacillus and Bifidobacterium strains for use as probiotics. Int. Dairy J. 1998, 8, 993–1002. [Google Scholar]
- Ruíz-Ramírez, Y.; Guadarrama-Mendoza, P.C.; Escalante, A.; Giles-Gómez, M.; Valadez-Blanco, R. Probiotic activity traits in vitro and production of antimicrobial peptides by Lactobacillaceae isolates from pulque using Lactobacillus acidophilus NCFM as control. Braz. J. Microbiol. 2022, 53, 921–933. [Google Scholar] [CrossRef]
- Colombo, M.; Nero, L.A.; Todorov, S.D. Safety profiles of beneficial lactic acid bacteria isolated from dairy systems. Braz. J. Microbiol. 2020, 51, 787–795. [Google Scholar] [CrossRef]
- Borges, S.; Barbosa, J.; Silva, J.; Teixeira, P. Evaluation of characteristics of Pediococcus spp. to be used as a vaginal probiotic. J. Appl. Microbiol. 2013, 115, 527–538. [Google Scholar] [CrossRef]
- Cebeci, A.; Gürakan, C. Properties of potential probiotic Lactobacillus plantarum strains. Food Microbiol. 2003, 20, 511–518. [Google Scholar] [CrossRef]
- Montassier, E.; Valdés-Mas, R.; Batard, E.; Zmora, N.; Dori-Bachash, M.; Suez, J.; Elinav, E. Probiotics impact the antibiotic resistance gene reservoir along the human GI tract in a person-specific and antibiotic-dependent manner. Nat. Microbiol. 2021, 6, 1043–1054. [Google Scholar] [CrossRef] [PubMed]
- Sharma, M.; Wasan, A.; Sharma, R.K. Recent developments in probiotics: An emphasis on Bifidobacterium. Food Biosci. 2021, 41, 100993. [Google Scholar] [CrossRef]
- Xu, H.; Jeong, H.S.; Lee, H.Y.; Ahn, J. Assessment of cell surface properties and adhesion potential of selected probiotic strains. Lett. Appl. Microbiol. 2009, 49, 434–442. [Google Scholar] [CrossRef]
- Papadimitriou, K.; Alegría, Á.; Bron, P.A.; De Angelis, M.; Gobbetti, M.; Kleerebezem, M.; Lemos, J.A.; Linares, D.M.; Ross, P.; Stanton, C.; et al. Stress physiology of lactic acid bacteria. Microbiol. Mol. 2016, 80, 837–890. [Google Scholar] [CrossRef]
- Lokesh, D.; Psn, S.; Kammara, R. PeSTK db a comprehensive data repository of Probiotic Serine Threonine kinases. Sci. Data 2022, 9, 282. [Google Scholar] [CrossRef]
- Párraga Solórzano, P.K.; Shupe, A.C.; Kehl-Fie, T.E. The sensor histidine kinase ArlS Is necessary for Staphylococcus aureus to activate ArlR in response to nutrient availability. J. Bacteriol. 2021, 203, e00422-21. [Google Scholar] [CrossRef]
- Fournier, B.; Hooper, D.C. A new two-component regulatory system involved in adhesion, autolysis, and extracellular proteolytic activity of Staphylococcus aureus. J. Bacteriol. 2000, 182, 3955–3964. [Google Scholar] [CrossRef]
- Shida, T.; Hattori, H.; Ise, F.; Sekiguchi, J. Mutational analysis of catalytic sites of the cell wall lytic N-acetylmuramoyl-L-alanine amidases CwlC and CwlV. JBC 2001, 276, 28140–28146. [Google Scholar] [CrossRef]
- Korndörfer, I.P.; Danzer, J.; Schmelcher, M.; Zimmer, M.; Skerra, A.; Loessner, M.J. The crystal structure of the bacteriophage PSA endolysin reveals a unique fold responsible for specific recognition of Listeria cell walls. J. Mol. Biol. 2006, 364, 678–689. [Google Scholar] [CrossRef]
- Gandhi, D.; Chanalia, P.; Bansal, P.; Dhanda, S. Peptidoglycan hydrolases of probiotic Pediococcus acidilactici NCDC 252: Isolation, physicochemical and in silico characterization. Int. J. Pept. Res. Ther. 2020, 26, 2119–2127. [Google Scholar] [CrossRef]
- Mora, D.; Musacchio, F.; Fortina, M.G.; Senini, L.; Manachini, P.L. Autolytic activity and pediocin-induced lysis in Pediococcus acidilactici and Pediococcus pentosaceus strains. J. Appl. Microbiol. 2003, 94, 561–570. [Google Scholar] [CrossRef] [PubMed]
- Govindasamy-Lucey, S.; Gopal, P.K.; Sullivan, P.A.; Pillidge, C.J. Varying influence of the autolysin, N-acetyl muramidase, and the cell envelope proteinase on the rate of autolysis of six commercial Lactococcus lactis cheese starter bacteria grown in milk. J. Dairy Res. 2000, 67, 585–596. [Google Scholar] [CrossRef] [PubMed]
- Llorente-Bousquets, A.; Pérez-Munguía, S.; Farrés, A. Novel extracellular proteolytic activity in Pediococcus acidilactici ATCC 8042. Can. J. Microbiol. 2008, 54, 694–699. [Google Scholar] [CrossRef] [PubMed]
- Jang, S.; Lee, J.; Jung, U.; Choi, H.S.; Suh, H.J. Identification of an anti-listerial domain from Pediococcus pentosaceus T1 derived from Kimchi, a traditional fermented vegetable. Food Control 2014, 43, 42–48. [Google Scholar] [CrossRef]
- Hamon, E.; Horvatovich, P.; Izquierdo, E.; Bringel, F.; Marchioni, E.; Aoudé-Werner, D.; Ennahar, S. Comparative proteomic analysis of Lactobacillus plantarumfor the identification of key proteins in bile tolerance. BMC Microbiol. 2011, 11, 63. [Google Scholar] [CrossRef]
- Álvarez-Ordóñez, A.; Begley, M.; Clifford, T.; Deasy, T.; Collins, B.; Hill, C. Transposon mutagenesis reveals genes involved in osmotic stress and drying in Cronobacter sakazakii. Food Res. Int. 2014, 55, 45–54. [Google Scholar] [CrossRef]
- Rafii, F.; Park, M. Detection and characterization of an ABC transporter in Clostridium hathewayi. Arch. Microbiol. 2008, 190, 417–426. [Google Scholar] [CrossRef]
- Pérez-Chabela, M.D.L.; Totosaus, A.; Guerrero, I. Evaluation of thermotolerant capacity of lactic acid bacteria isolated from commercial sausages and the effects of their addition on the quality of cooked sausages. Food. Sci. Technol. 2008, 28, 132–138. [Google Scholar] [CrossRef]
- Delgado, A.; Brito, D.; Peres, C.; Noe-Arroyo, F.; Garrido-Fernández, A. Bacteriocin production by Lactobacillus pentosus B96 can be expressed as a function of temperature and NaCl concentration. Food. Microbiol. 2005, 22, 521–528. [Google Scholar] [CrossRef]

| Strain | Antagonistic Activity |
|---|---|
| Escherichia coli DH5α GenBank:NZ_JRYM00000000.1 a | + |
| Enterococcus faecalis 1351 b | + |
| Listeria monocytogenes 1639 b | + |
| Salmonella enterica subsp. enterica serovar Typhimurium ATCC 14,028 a | + |
| Staphylococcus aureus ATCC 6538 a | + |
| Weisella viridescens GenBank: UAM-MG5: MT814884 b | + |
| Condition | O.D.max | µ (h−1) | Initial Count log10 CFU/mL | Final Count log10 CFU/mL | Logarithmic Survival Rate (%) |
|---|---|---|---|---|---|
| Resistance to bile salts | |||||
| Control | 1.30 ± 0.01 | 0.69 ± 0.01 b | |||
| 0.5% | 1.28 ± 0.01 | 1.08 ± 0.01 a | 9.448 ± 0.005 | 9.352 ± 0.002 | 99.1 |
| Resistance to acidity | |||||
| Control | 1.26 ± 0.01 a | 0.72 ± 0.01 a | |||
| pH 2 | 0.45 ± 0.13 b | 0.19 ± 0.08 b | 9.115 ± 0.012 | 7.914 ± 0.003 | 86.8 b |
| pH 3 | 0.55 ± 0.05 b | 0.19 ± 0.05 b | 9.115 ± 0.012 | 8.556 ± 0.024 | 93.9 a |
| Antibiotic | Resistance Profile |
|---|---|
| Cell-wall-biosynthesis inhibitors | |
| Amoxicillin/Clavulanic acid | S |
| Amoxicillin | S |
| Penicillin | S |
| Cephalexin | R |
| Cefazolin | S |
| Cefuroxime | S |
| Piperacillin | R |
| Protein-synthesis inhibitors | |
| Azithromycin | R |
| Erythromycin | R |
| Chloramphenicol | S |
| Tetracycline | R |
| DNA-synthesis inhibitors | |
| Ciprofloxacin | R |
| Ofloxacin | R |
| Metabolic products inhibitor | |
| Cotrimoxazole | R |
| LAB | Solvent | |
|---|---|---|
| Hexane (%) | Chloroform (%) | |
| P. pentosaceus 1101 | 91.5 ± 0.3 a | 19.6 ± 0.4 b |
| L. acidophilus NCFM® | 51.8 ± 0.3 b | 94.1 ± 0.5 a |
| LAB | 2 h | 4 h | 6 h | 20 h | 24 h |
|---|---|---|---|---|---|
| P. pentosaceus 1101 | 10.7 ± 0.3 b/51.2 ± 0.2 b | 14.7 ± 0.5 b/56.0 ± 0.2 b | 21.4 ± 0.4 b/60.1 ± 0.2 b | 62.9 ± 0.5 b/64.4 ± 0.1 b | 86.7± 0.2 b/74.3 ± 0.2 b |
| L. acidophilus NCFM | 14.3 ± 0.2 a/56.3 ± 0.1 a | 25.0 ± 0.1 a/58.7 ± 0.1 a | 32.2± 0.3 a/71.9 ± 0.2 a | 84.8 ± 0.3 a/80.1 ± 0.1 a | 96.4 ± 0.2 a/85.2 ± 0.2 a |
| Protein | Gene | Molecular Weight (kDa) | Function |
|---|---|---|---|
| Signal transduction histidine-protein kinase ArlS a | 0715 | 57 | Two-component regulatory system |
| Serine/threonine protein kinase b | 0832 | 57 | Phosphorylation |
| N-acetylmuramoyl-L-alanine amidase b | 1117 | 32 | Peptidoglycan catabolic process |
| Peptidoglycan transpeptidase, ErfK-YbiS-YhnG family a UDP-N-acetylmuramoyl-L-alanyl-D-glutamate--L-lysine ligase a | 1555 murE | 51 56 | Peptidoglycan biosynthetic process |
| LysM-domain-containing protein | 1356 | 23 | Hydrolase activity |
| Periplasmic protease a | 1062 | 52 | Proteolysis |
| ATPase component of ABC transporter with duplicated ATPase domains b | 1071 | 72 | ATP binding |
| RNA polymerase sigma factor SigA a | sigA | 43 | DNA binding |
| DNA ligase a | ligA | 75 | DNA repair |
| Single-stranded-DNA-specific exonuclease RecJ b | 1130 | 87 | |
| Transcription-repair-coupling factor, TRCF b | mfd | 132 | |
| Chaperone protein DnaJ a | dnaJ | 40 | Response to stress |
| Peptide methionine sulfoxide reductase MsrB a | msrB | 17 | |
| DEAD-box ATP-dependent RNA helicase CshB a | cshB | 52 | |
| Protein RadA b | radA | 50 | |
| Surface adhesin b | 0086 | 34 | Cell adhesion |
| Cell-elongation-specific peptidoglycan D,D-transpeptidase a | 0727 | 74 | Penicillin binding |
| Protein | Gene | Molecular Weight (kDa) | Function |
|---|---|---|---|
| Glycerol-3-phosphate dehydrogenase a | gpsA | 36 | Carbohydrate metabolic process |
| HPr kinase/phosphorylase a | hprK | 35 | |
| Fructose-bisphosphate aldolase a | 1352 | 31 | |
| Glyceraldehyde-3-phosphate dehydrogenase a | 0459 | 37 | |
| ATP-dependent 6-phosphofructokinase a | pfkA | 34 | |
| Ribose-5-phosphate isomerase a | rpiA | 25 | |
| L-lactate oxidase a | 0922 | 40 | Oxidoreductase |
| Malate dehydrogenase (NAD) a | 0218 | 33 | Carboxylic acid metabolic process |
| ABC-type multidrug transport system a | 1337 | 64 | Transporter activity |
| Energy-coupling factor transporter ATP-binding protein EcfA b | 1390 | 31 | |
| ABC-type antimicrobial peptide transport system b | 1650 | 71 | Antibiotic resistance |
| Predicted esterase of the alpha-beta hydrolase superfamily b | 0212 | 33 | Lipid catabolism |
| Aspartate racemase b | 0662 | 27 | Nitrogen compound metabolism |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Escobar-Sánchez, M.; Carrasco-Navarro, U.; Juárez-Castelán, C.; Lozano-Aguirre Beltrán, L.; Pérez-Chabela, M.L.; Ponce-Alquicira, E. Probiotic Properties and Proteomic Analysis of Pediococcus pentosaceus 1101. Foods 2023, 12, 46. https://doi.org/10.3390/foods12010046
Escobar-Sánchez M, Carrasco-Navarro U, Juárez-Castelán C, Lozano-Aguirre Beltrán L, Pérez-Chabela ML, Ponce-Alquicira E. Probiotic Properties and Proteomic Analysis of Pediococcus pentosaceus 1101. Foods. 2023; 12(1):46. https://doi.org/10.3390/foods12010046
Chicago/Turabian StyleEscobar-Sánchez, Monserrat, Ulises Carrasco-Navarro, Carmen Juárez-Castelán, Luis Lozano-Aguirre Beltrán, M. Lourdes Pérez-Chabela, and Edith Ponce-Alquicira. 2023. "Probiotic Properties and Proteomic Analysis of Pediococcus pentosaceus 1101" Foods 12, no. 1: 46. https://doi.org/10.3390/foods12010046
APA StyleEscobar-Sánchez, M., Carrasco-Navarro, U., Juárez-Castelán, C., Lozano-Aguirre Beltrán, L., Pérez-Chabela, M. L., & Ponce-Alquicira, E. (2023). Probiotic Properties and Proteomic Analysis of Pediococcus pentosaceus 1101. Foods, 12(1), 46. https://doi.org/10.3390/foods12010046





