Antifungal Effect of Metabolites from a New Strain Lactiplantibacillus Plantarum LPP703 Isolated from Naturally Fermented Yak Yogurt
Abstract
1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Strains
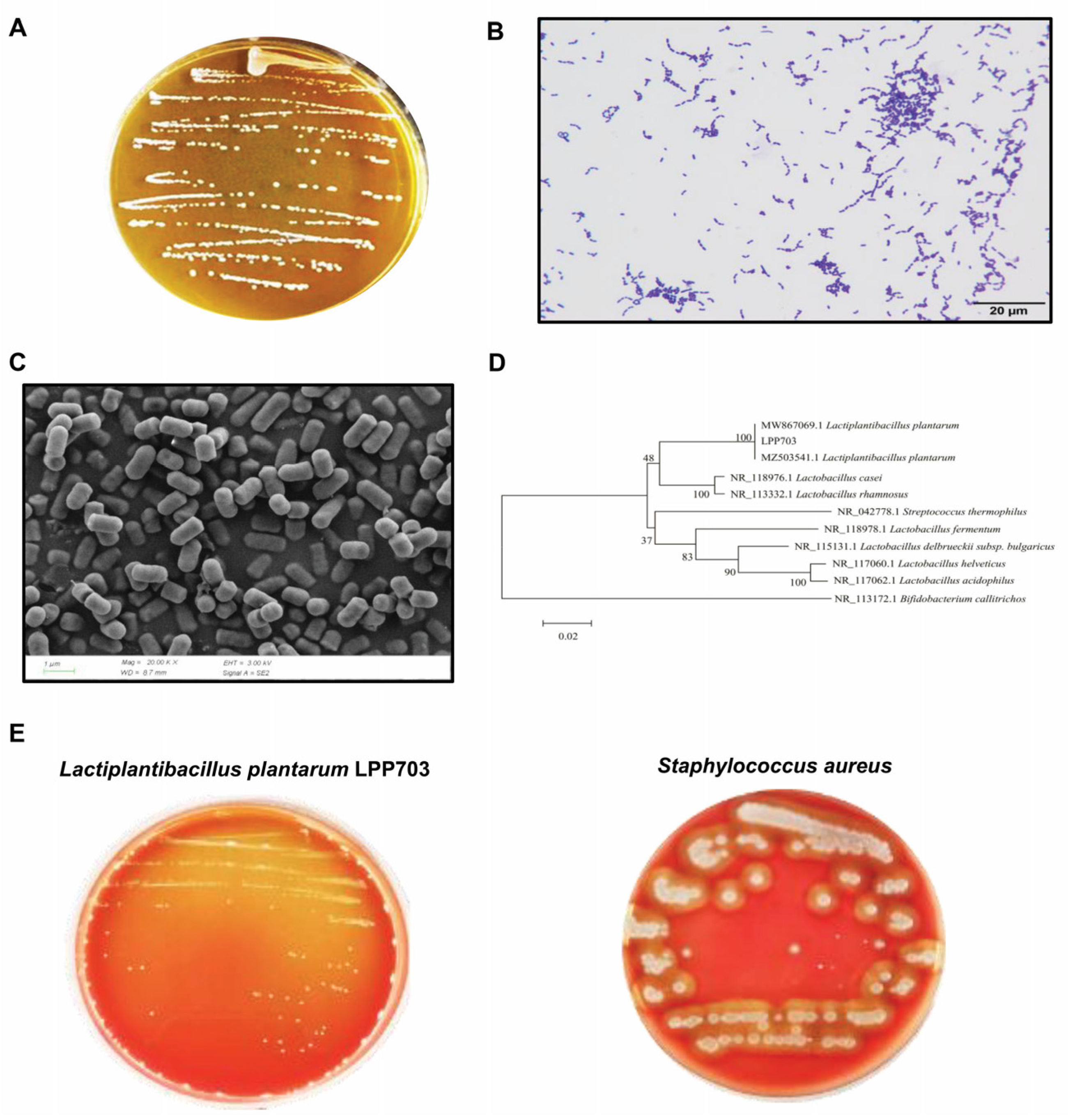
2.3. Morphological Analysis, Identification and Safety Evaluation of Lpb. Plantarum LPP703
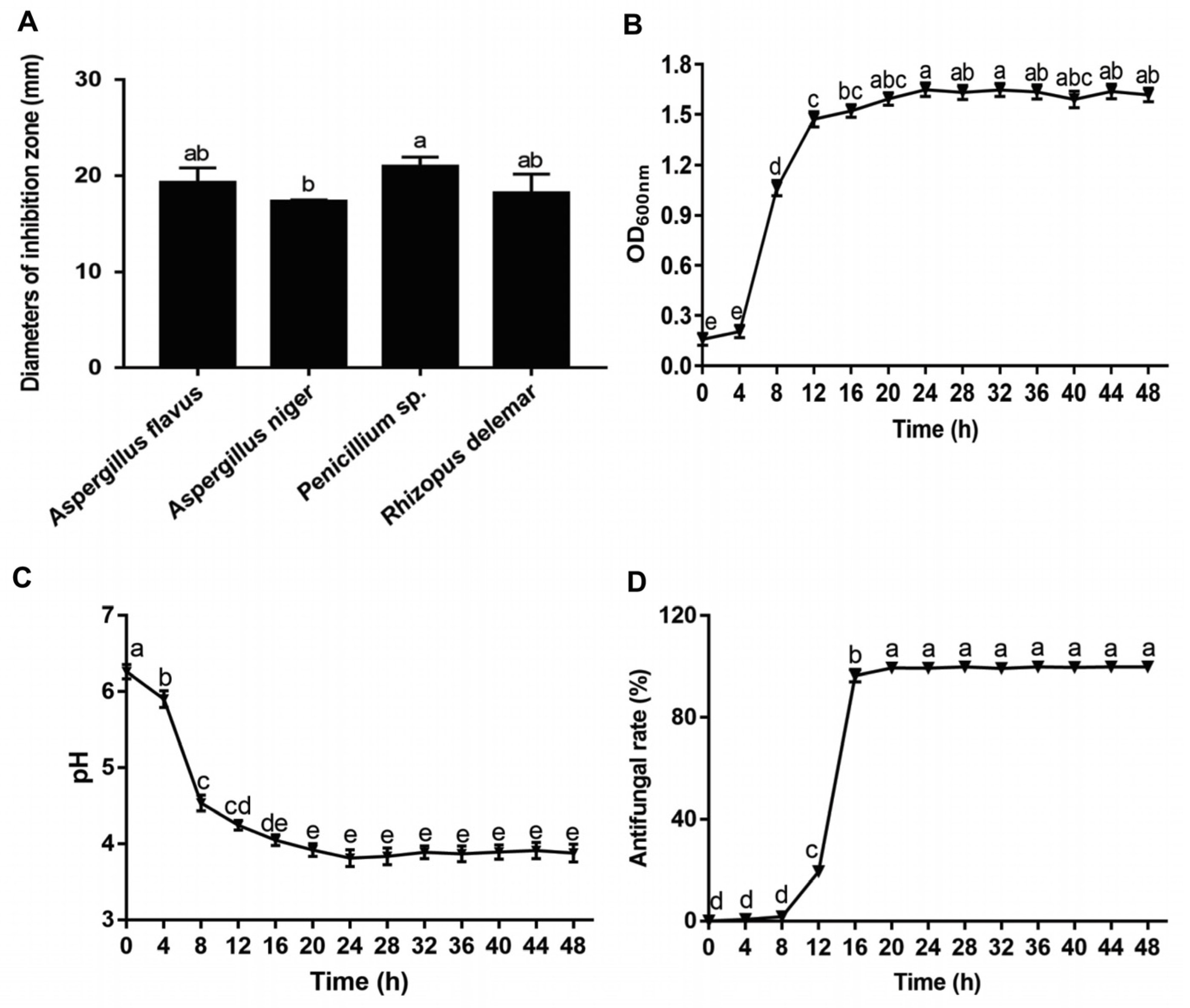
2.4. Determination of Antifungal Activity of Lpb. Plantarum LPP703 and Its Metabolites
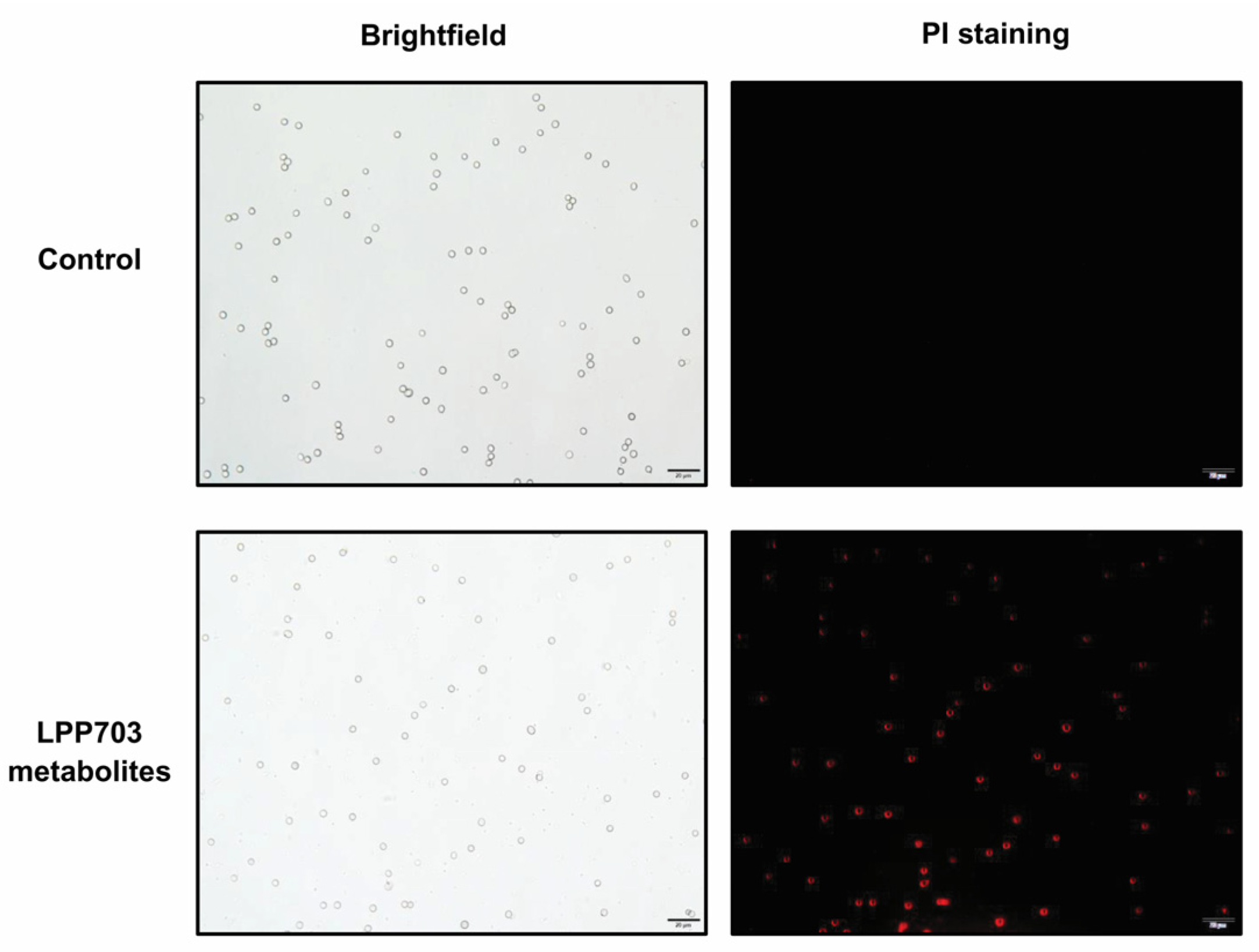
2.5. Determination of the Permeability of Cell Membrane
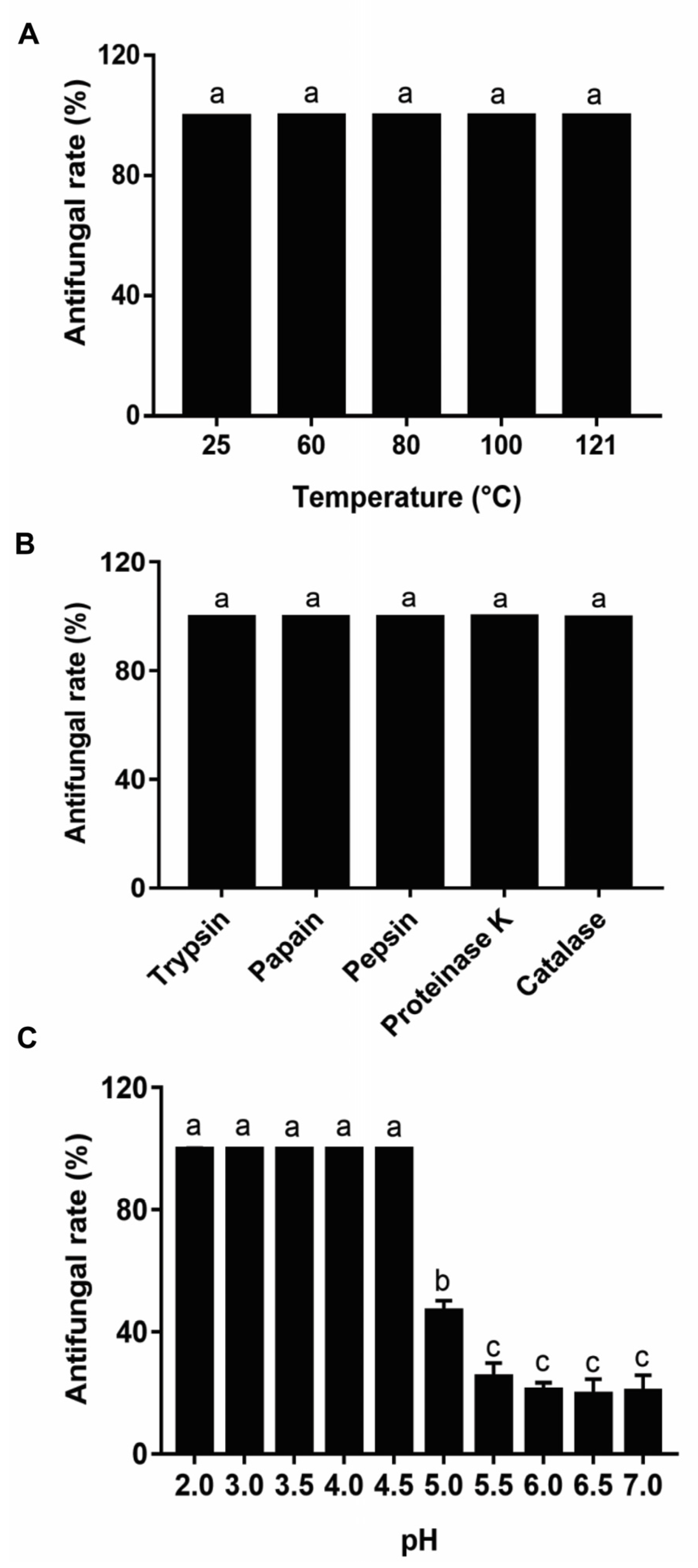
2.6. Antifungal Activity Stability of Metabolites from Lpb. Plantarum LPP703
2.7. Component Analysis of Metabolites from Lpb. Plantarum LPP703
2.8. Molecular Docking Prediction
2.9. Statistical Analysis
3. Results
3.1. Identification and Hemolytic Activity of Lpb. Plantarum LPP703 Strain
3.2. Antifungal Activity of Lpb. Plantarum LPP703 and Its Metabolites
3.3. Effect of Metabolites from Lpb. Plantarum LPP703 on Cell Membrane Integrity
3.4. Effect of Heat, Enzymatic Hydrolysis, and pH on the Antifungal Activity of Metabolites from Lpb. Plantarum LPP703
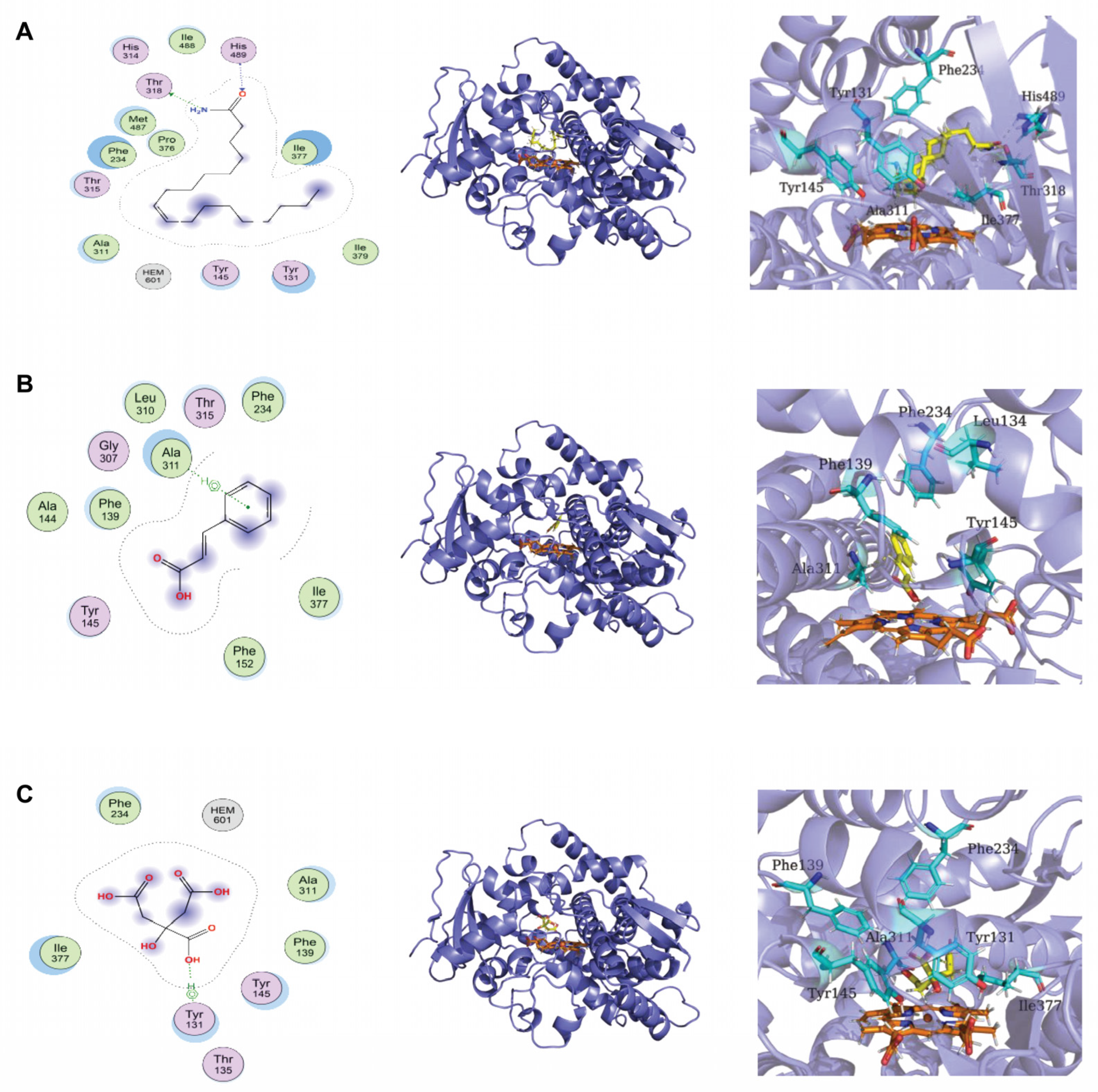
3.5. Chemical Composition of Lpb. Plantarum LPP703 Metabolites and Molecular Docking against CYP51
4. Discussion
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Alizadeh, A.M.; Hashempour-Baltork, F.; Khaneghah, A.M.; Hosseini, H. New perspective approaches in controlling fungi and mycotoxins in food using emerging and green technologies. Curr. Opin. Food Sci. 2021, 39, 7–15. [Google Scholar] [CrossRef]
- Das, S.; Singh, V.K.; Dwivedy, A.K.; Chaudhari, A.K.; Dubey, N.K. Nanostructured Pimpinella anisum essential oil as novel green food preservative against fungal infestation, aflatoxin B1 contamination and deterioration of nutritional qualities. Food Chem. 2021, 344, 128574. [Google Scholar] [CrossRef] [PubMed]
- Milićević, D.R.; Škrinjar, M.; Baltić, T. Real and perceived risks for mycotoxin contamination in foods and feeds: Challenges for food safety control. Toxins 2010, 2, 572–592. [Google Scholar] [CrossRef] [PubMed]
- Muhialdin, B.J.; Algboory, H.L.; Kadum, H.; Mohammed, N.K.; Saari, N.; Hassan, Z.; Hussin, A.S.M. Antifungal activity determination for the peptides generated by Lactobacillus plantarum TE10 against Aspergillus flavus in maize seeds. Food Control 2020, 109, 106898. [Google Scholar] [CrossRef]
- Pradhan, S.; Ananthanarayan, L.; Prasad, K.; Bhatnagar-Mathur, P. Anti-fungal activity of lactic acid bacterial isolates against aflatoxigenic fungi inoculated on peanut kernels. LWT 2021, 143, 111104. [Google Scholar] [CrossRef]
- Ramos-Pereira, J.; Mareze, J.; Fernández, D.; Rios, E.A.; Santos, J.A.; López-Díaz, T.-M. Antifungal activity of lactic acid bacteria isolated from milk against Penicillium commune, P. nordicum, and P. verrucosum. Int. J. Food Microbiol. 2021, 355, 109331. [Google Scholar] [CrossRef]
- Zhao, S.; Hao, X.; Yang, F.; Wang, Y.; Fan, X.; Wang, Y. Antifungal activity of Lactobacillus plantarum ZZUA493 and its application to extend the shelf life of Chinese steamed buns. Foods 2022, 11, 195. [Google Scholar] [CrossRef]
- Bukhari, S.A.; Salman, M.; Numan, M.; Javed, M.R.; Zubair, M.; Mustafa, G. Characterization of antifungal metabolites produced by Lactobacillus plantarum and Lactobacillus coryniformis isolated from rice rinsed water. Mol. Biol. Rep. 2020, 47, 1871–1881. [Google Scholar] [CrossRef]
- Kwak, M.-K.; Liu, R.; Kang, S.-O. Antimicrobial activity of cyclic dipeptides produced by Lactobacillus plantarum LBP-K10 against multidrug-resistant bacteria, pathogenic fungi, and influenza A virus. Food Control 2018, 85, 223–234. [Google Scholar] [CrossRef]
- Guimarães, A.; Venancio, A.; Abrunhosa, L. Antifungal effect of organic acids from lactic acid bacteria on Penicillium nordicum. Food Addit. Contam. 2018, 35, 1803–1818. [Google Scholar] [CrossRef]
- Schmidt, M.; Lynch, K.M.; Zannini, E.; Arendt, E.K. Fundamental study on the improvement of the antifungal activity of Lactobacillus reuteri R29 through increased production of phenyllactic acid and reuterin. Food Control 2018, 88, 139–148. [Google Scholar] [CrossRef]
- Abouloifa, H.; Gaamouche, S.; Rokni, Y.; Hasnaoui, I.; Bellaouchi, R.; Ghabbour, N.; Karboune, S.; Brasca, M.; D’hallewin, G.; Salah, R.B. Antifungal activity of probiotic Lactobacillus strains isolated from natural fermented green olives and their application as food bio-preservative. Biol. Control 2021, 152, 104450. [Google Scholar] [CrossRef]
- Song, J.; Peng, S.; Yang, J.; Zhou, F.; Suo, H. Isolation and identification of novel antibacterial peptides produced by Lactobacillus fermentum SHY10 in Chinese pickles. Food Chem. 2021, 348, 129097. [Google Scholar] [CrossRef] [PubMed]
- Mohammed, S.; Orukotan, A.; Shittu, A.; Onuoha, E.; Moriki, A.; Mukhtar, A. Characterization and identification of lactic acid bacteria isolated from fermented milk sold at central market, Kaduna. KASU J. Biochem. Biotechnol. 2019, 3, 1–8. [Google Scholar]
- Zhu, Y.; Xu, Y.; Yang, Q. Antifungal properties and AFB1 detoxification activity of a new strain of Lactobacillus plantarum. J. Hazard Mater. 2021, 414, 125569. [Google Scholar] [CrossRef]
- Kumar, S.; Nei, M.; Dudley, J.; Tamura, K. MEGA: A biologist-centric software for evolutionary analysis of DNA and protein sequences. Brief Bioinform. 2008, 9, 299–306. [Google Scholar] [CrossRef]
- Mount, D.W. Using the basic local alignment search tool (BLAST). Cold Spring Harb. Protoc. 2007, 2007, pdb.top17. [Google Scholar] [CrossRef]
- Russo, P.; Arena, M.P.; Fiocco, D.; Capozzi, V.; Drider, D.; Spano, G. Lactobacillus plantarum with broad antifungal activity: A promising approach to increase safety and shelf-life of cereal-based products. Int. J. Food Microbiol. 2017, 247, 48–54. [Google Scholar] [CrossRef]
- Li, Q.; Zhu, X. Vanillin and its derivatives, potential promising antifungal agents, inhibit Aspergillus flavus spores via destroying the integrity of cell membrane rather than cell wall. Oil Gas Sci. Technol. 2021, 4, 54–61. [Google Scholar] [CrossRef]
- Sadiq, F.A.; Yan, B.; Tian, F.; Zhao, J.; Zhang, H.; Chen, W. Lactic acid bacteria as antifungal and anti-mycotoxigenic agents: A comprehensive review. Compr. Rev. Food Sci. Food Saf. 2019, 18, 1403–1436. [Google Scholar] [CrossRef]
- Abouloifa, H.; Rokni, Y.; Bellaouchi, R.; Ghabbour, N.; Karboune, S.; Brasca, M.; Ben Salah, R.; Chihib, N.E.; Saalaoui, E.; Asehraou, A. Characterization of probiotic properties of antifungal Lactobacillus strains isolated from traditional fermenting green olives. Probiotics Antimicrob. Proteins 2020, 12, 683–696. [Google Scholar] [CrossRef] [PubMed]
- El Oirdi, S.; Lakhlifi, T.; Bahar, A.A.; Yatim, M.; Rachid, Z.; Belhaj, A. Isolation and identification of Lactobacillus plantarum 4F, a strain with high antifungal activity, fungicidal effect, and biopreservation properties of food. J. Food Process. Preserv. 2021, 45, e15517. [Google Scholar] [CrossRef]
- Bulgasem, B.Y.; Lani, M.N.; Hassan, Z.; Yusoff, W.M.W.; Fnaish, S.G. Antifungal activity of lactic acid bacteria strains isolated from natural honey against pathogenic Candida species. Mycobiology 2016, 44, 302–309. [Google Scholar] [CrossRef]
- Shehata, M.G.; Badr, A.N.; El Sohaimy, S.A.; Asker, D.; Awad, T.S. Characterization of antifungal metabolites produced by novel lactic acid bacterium and their potential application as food biopreservatives. Ann. Agric. Sci. 2019, 64, 71–78. [Google Scholar] [CrossRef]
- Zhao, S.; Guo, Y.; Wang, Q.; An, B. Antifungal effects of lycorine on Botrytis cinerea and possible mechanisms. Biotechnol. Lett. 2021, 43, 1503–1512. [Google Scholar] [CrossRef]
- Unal Cevik, I.; Dalkara, T. Intravenously administered propidium iodide labels necrotic cells in the intact mouse brain after injury. Cell Death Differ. 2003, 10, 928–929. [Google Scholar] [CrossRef]
- Li, K.; Zhang, W.; Kwok, L.-Y.; Menghe, B. Screening of Lactobacillus plantarum with broad-spectrum antifungal activity and its application in preservation of golden-red apples. Czech J. Food Sci. 2020, 38, 315–322. [Google Scholar] [CrossRef]
- GüLck, T.; Booth, J.; Carvalho, Â.; Khakimov, B.; Crocoll, C.; Motawia, M.; Møller, B.; Bohlmann, J.; Gallage, N. Synthetic biology of cannabinoids and cannabinoid glucosides in Nicotiana benthamiana and Saccharomyces cerevisiae. J. Nat. Prod. 2020, 83, 2877–2893. [Google Scholar] [CrossRef]
- Cao, H.; Meng, D.; Zhang, W.; Ye, T.; Yuan, M.; Yu, J.; Wu, X.; Li, Y.; Yin, F.; Fu, C. Growth inhibition of Fusarium graminearum and deoxynivalenol detoxification by lactic acid bacteria and their application in sourdough bread. Int. J. Food Sci. Tech. 2021, 56, 2304–2314. [Google Scholar] [CrossRef]
- Nora, L.; Bettache, G. Antifungal activity of newly isolates of lactic acid bacteria. Innov. Rom. Food Biotechnol. 2013, 13, 80–88. [Google Scholar]
- Li, H.; Liu, L.; Zhang, S.; Cui, W.; Lv, J. Identification of antifungal compounds produced by Lactobacillus casei AST18. Curr. Microbiol. 2012, 65, 156–161. [Google Scholar] [CrossRef] [PubMed]
- Le Lay, C.; Coton, E.; Le Blay, G.; Chobert, J.-M.; Haertlé, T.; Choiset, Y.; Van Long, N.N.; Meslet-Cladière, L.; Mounier, J. Identification and quantification of antifungal compounds produced by lactic acid bacteria and propionibacteria. Int. J. Food Microbiol. 2016, 239, 79–85. [Google Scholar] [CrossRef]
- Dagnas, S.; Gauvry, E.; Onno, B.; Membre, J.-M. Quantifying effect of lactic, acetic, and propionic acids on growth of molds isolated from spoiled bakery products. J Food Prot. 2015, 78, 1689–1698. [Google Scholar] [CrossRef] [PubMed]
- Chen, P.-T.; Hong, Z.-S.; Cheng, C.-L.; Ng, I.-S.; Lo, Y.-C.; Nagarajan, D.; Chang, J.-S. Exploring fermentation strategies for enhanced lactic acid production with polyvinyl alcohol-immobilized Lactobacillus plantarum 23 using microalgae as feedstock. Bioresour Technol. 2020, 308, 123266. [Google Scholar] [CrossRef] [PubMed]
- Chen, G.Q.; Eschbach, F.I.; Weeks, M.; Gras, S.L.; Kentish, S.E. Removal of lactic acid from acid whey using electrodialysis. Sep. Purif. Technol. 2016, 158, 230–237. [Google Scholar] [CrossRef]
- Nasrollahzadeh, A.; Mokhtari, S.; Khomeiri, M.; Saris, P.E. Antifungal preservation of food by lactic acid bacteria. Foods 2022, 11, 395. [Google Scholar] [CrossRef]
- Scano, P.; Pisano, M.B.; Murgia, A.; Cosentino, S.; Caboni, P. GC-MS metabolomics and antifungal characteristics of autochthonous Lactobacillus strains. Dairy 2021, 2, 326–335. [Google Scholar] [CrossRef]
- Nesci, A.V.; Etcheverry, M.G. Control of Aspergillus growth and aflatoxin production using natural maize phytochemicals under different conditions of water activity. Pest Manag. Sci. 2006, 62, 775–784. [Google Scholar] [CrossRef]
- Roy, S.; Nuckles, E.; Archbold, D.D. Effects of phenolic compounds on growth of Colletotrichum spp. in vitro. Curr. Microbiol. 2018, 75, 550–556. [Google Scholar] [CrossRef]
- Cai, J.; Yan, R.; Shi, J.; Chen, J.; Long, M.; Wu, W.; Kuca, K. Antifungal and mycotoxin detoxification ability of essential oils: A review. Phytother. Res. 2022, 36, 62–72. [Google Scholar] [CrossRef]
- Shokri, H. Evaluation of inhibitory effects of citric and tartaric acids and their combination on the growth of Trichophyton mentagrophytes, Aspergillus fumigatus, Candida albicans, and Malassezia furfur. Comp. Clin. Path. 2011, 20, 543–545. [Google Scholar] [CrossRef]
- Song, J.; Zhang, S.; Lu, L. Fungal cytochrome P450 protein Cyp51: What we can learn from its evolution, regulons and Cyp51-based azole resistance. Fungal Biol. Rev. 2018, 32, 131–142. [Google Scholar] [CrossRef]
- Liu, N.; Abramyan, E.D.; Cheng, W.; Perlatti, B.; Harvey, C.J.; Bills, G.F.; Tang, Y. Targeted genome mining reveals the biosynthetic gene clusters of natural product CYP51 inhibitors. J. Am. Chem. Soc. 2021, 143, 6043–6047. [Google Scholar] [CrossRef] [PubMed]
- Sun, B.; Dong, Y.; An, Y.; Liu, M.; Han, J.; Zhao, L.; Liu, X. Design, synthesis and bioactivity evaluation of novel arylalkene-amide derivatives as dual-target antifungal inhibitors. Eur. J. Med. Chem. 2020, 205, 112645. [Google Scholar] [CrossRef] [PubMed]
| Compound | Chemical Formula | Retention Time (min) | Proportion (%) |
|---|---|---|---|
| Oleamide | C18H35NO | 9.18 | 40.90 ± 1.73 |
| Trans-Cinnamic acid | C9H8O2 | 2.07 | 5.31 ± 0.19 |
| Citric acid | C6H8O7 | 1.04 | 4.18 ± 0.23 |
| Linoleamide | C18H33NO | 8.86 | 2.72 ± 0.83 |
| 2-Hydroxyphenethylamine | C8H11NO | 2.07 | 2.72 ± 0.12 |
| Omphalotin C | C76H125N13O19 | 3.97 | 2.29 ± 0.08 |
| Desglucocheirotoxol | C29H44O10 | 2.97 | 2.16 ± 0.10 |
| PS(MonoMe(11,5)/MonoMe(13,5)) | C50H86NO12P | 8.99 | 1.85 ± 0.11 |
| CL(16:1(9Z)/22:6(4Z,7Z,10Z,13Z,16Z,19Z)/20:4(5Z,8Z,11Z,14Z)/20:4(5Z,8Z,11Z,14Z)) | C87H140O17P2 | 3.97 | 1.59 ± 0.09 |
| Neocasomorphin (1–5) | C29H41N5O9 | 2.92 | 1.28 ± 0.07 |
| PE-NMe2(20:2(11Z,14Z)/22:4(7Z,10Z,13Z,16Z)) | C49H86NO8P | 8.79 | 1.22 ± 0.07 |
| Ethyl 7-epi-12-hydroxyjasmonate glucoside | C20H32O9 | 2.38 | 1.15 ± 0.05 |
| PE(22:2(13Z,16Z)/22:5(4Z,7Z,10Z,13Z,16Z)) | C49H84NO8P | 9.05 | 1.10 ± 0.07 |
| PE(DiMe(13,5)/DiMe(13,5)) | C53H94NO10P | 8.72 | 1.06 ± 0.05 |
| Receptor | Ligand | Docking Score (Kcal/mol) |
|---|---|---|
| CYP51 | Oleamide | −7.78 |
| CYP51 | Trans-Cinnamic acid | −4.66 |
| CYP51 | Citric acid | −5.10 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Peng, Q.; Yang, J.; Wang, Q.; Suo, H.; Hamdy, A.M.; Song, J. Antifungal Effect of Metabolites from a New Strain Lactiplantibacillus Plantarum LPP703 Isolated from Naturally Fermented Yak Yogurt. Foods 2023, 12, 181. https://doi.org/10.3390/foods12010181
Peng Q, Yang J, Wang Q, Suo H, Hamdy AM, Song J. Antifungal Effect of Metabolites from a New Strain Lactiplantibacillus Plantarum LPP703 Isolated from Naturally Fermented Yak Yogurt. Foods. 2023; 12(1):181. https://doi.org/10.3390/foods12010181
Chicago/Turabian StylePeng, Qian, Jing Yang, Qiang Wang, Huayi Suo, Ahmed Mahmoud Hamdy, and Jiajia Song. 2023. "Antifungal Effect of Metabolites from a New Strain Lactiplantibacillus Plantarum LPP703 Isolated from Naturally Fermented Yak Yogurt" Foods 12, no. 1: 181. https://doi.org/10.3390/foods12010181
APA StylePeng, Q., Yang, J., Wang, Q., Suo, H., Hamdy, A. M., & Song, J. (2023). Antifungal Effect of Metabolites from a New Strain Lactiplantibacillus Plantarum LPP703 Isolated from Naturally Fermented Yak Yogurt. Foods, 12(1), 181. https://doi.org/10.3390/foods12010181






