GrainGenes: Tools and Content to Assist Breeders Improving Oat Quality
Abstract
:1. Introduction
2. How to Search and Reach Information on GrainGenes
- (A).
- The Quick Links section in the middle of GrainGenes’ home page provides an intuitive and visually informative landing area that links out to various pages containing useful tools and resources for users.
- (B).
- “Search and Browse GrainGenes” within the Quick Link collection, on the left menu, and in the GrainGenes Tools dropdown on the top menu, leads to a page that contains three different types of search capabilities for static and dynamic pages (https://wheat.pw.usda.gov/cgi-bin/GG3/browse.cgi, accessed on 17 March 2022), as well as external datasets related to wheat through the Wheat Information System (WheatIS) [12]. Since the third search type is specific to wheat, here we will only discuss the first two search types: (1) for static pages, users can search pre-indexed GrainGenes Web and database pages by using the search algorithm provided by Google™; (2) for dynamic pages within the GrainGenes database, which includes a wide range of data types such as genetic markers, genetic maps, QTL, traits, and phenotypes, users are able to deploy flexible search terms encompassing either all types of data or a specific data class. Users can browse a specific data type by clicking on the data class on the search page.
- (C).
- Within the GrainGenes Tools dropdown menu, “Advanced Queries” allows advanced users to access data programmatically using structured query language (SQL). Data models and database schema are provided here. For those who are interested in writing their own codes to access data in GrainGenes, we encourage them to visit the site at https://wheat.pw.usda.gov/GG3/advanced-queries, accessed on 17 March 2022 and follow the links there. Batch queries within the GrainGenes Advanced Query menu enable users to query more than one object; for example, to list all the QTL, significant markers, and maps for a given trait. As with the SQL Interface page, pre-made queries are also provided as an example.
- (D).
- A direct link into the collection of genome browsers. All individual browsers are grouped by crop, and colorful links take the user to the selected browser or open another group of browsers, in the case of pangenomes. The top collection is the most current or most popular browser for each crop. At the time of this writing, the most current genome browser for Avena is the PepsiCo OT3098 v2 Hexaploid oat (2021).
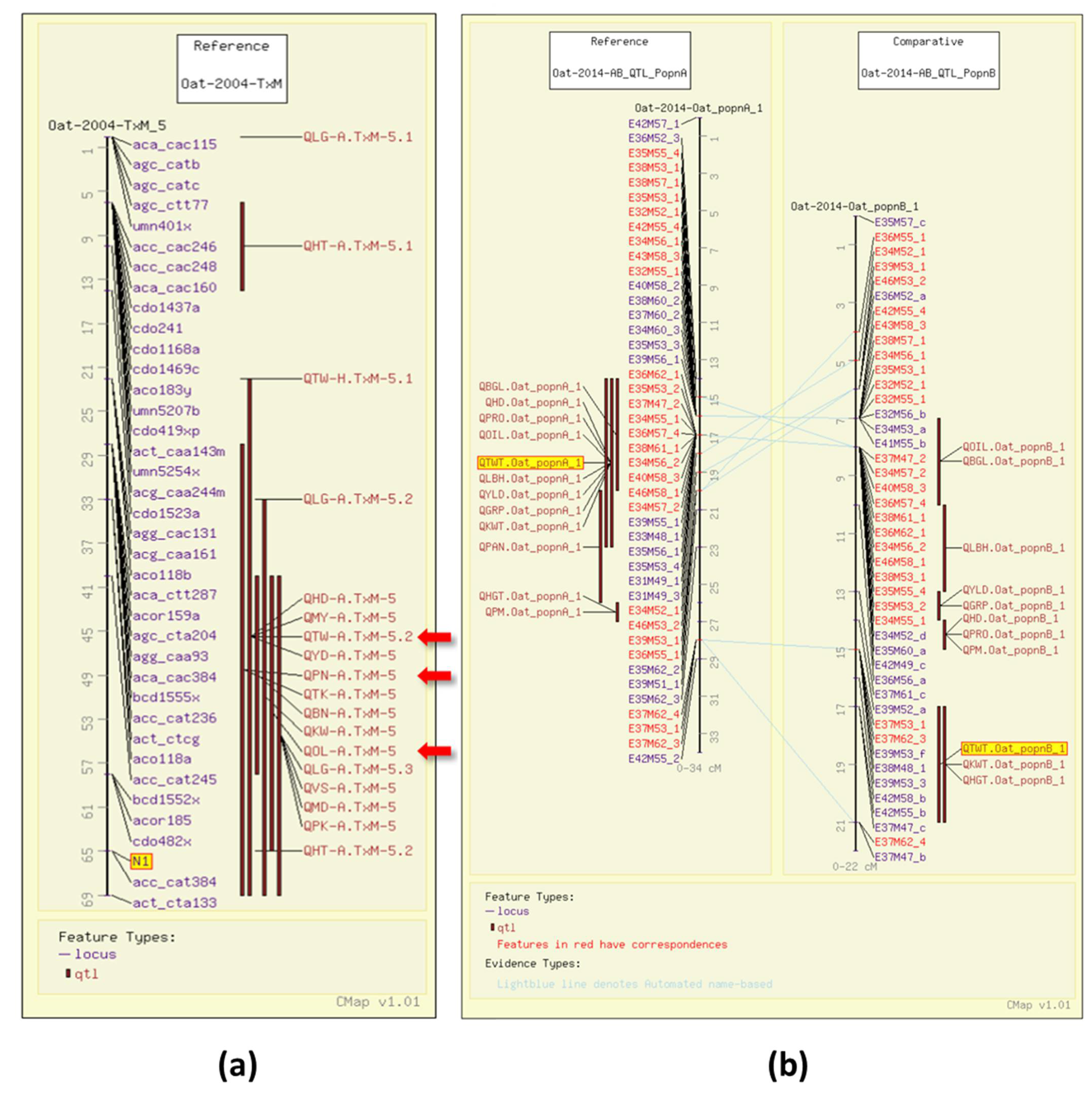
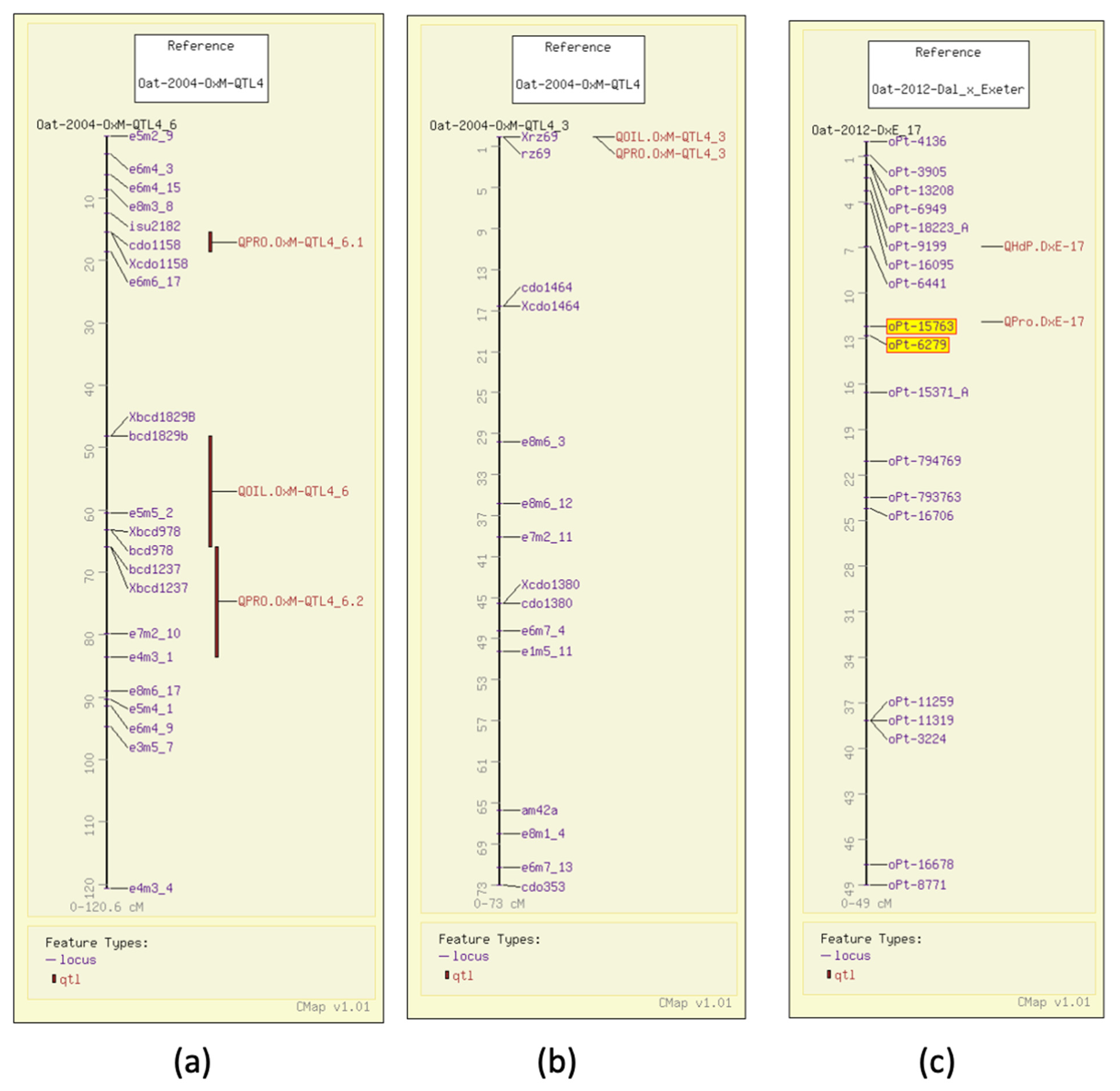
3. Oat Grain Quality Information in the GrainGenes Database
3.1. Test Weight
3.2. Groat Percentage
3.3. Grain Protein
3.4. Fat Content
3.5. Beta-Glucan Content
4. Oat Genome Browser Views
5. Reaching out to the GrainGenes Team
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Martínez-Villaluenga, C.; Peñas, E. Health benefits of oat: Current evidence and molecular mechanisms. Curr. Opin. Food Sci. 2017, 14, 26–31. [Google Scholar] [CrossRef] [Green Version]
- Blake, V.C.; Woodhouse, M.R.; Lazo, G.R.; Odell, S.G.; Wight, C.P.; Tinker, N.A.; Wang, Y.; Gu, Y.Q.; Birkett, C.L.; Jannink, J.-L.; et al. GrainGenes: Centralized small grain resources and digital platform for geneticists and breeders. Database 2019, 2019, 18. [Google Scholar] [CrossRef]
- Odell, S.G.; Lazo, G.R.; Woodhouse, M.R.; Hane, D.L.; Sen, T. The art of curation at a biological database: Principles and application. Curr. Plant Biol. 2017, 11–12, 2–11. [Google Scholar] [CrossRef]
- Maughan, P.J.; Lee, R.; Walstead, R.; Vickerstaff, R.J.; Fogarty, M.C.; Brouwer, C.R.; Reid, R.R.; Jay, J.J.; Bekele, W.A.; Jackson, E.W.; et al. Genomic insights from the first chromosome-scale assemblies of oat (Avena spp.) diploid species. BMC Biol. 2019, 17, 92. [Google Scholar] [CrossRef] [PubMed]
- Gorash, A.; Armonienė, R.; Fetch, J.M.; Liatukas, Ž.; Danytė, V. Aspects in oat breeding: Nutrition quality, nakedness and disease resistance, challenges and perspectives. Ann. Appl. Biol. 2017, 171, 281–302. [Google Scholar] [CrossRef]
- De Koeyer, D.L.; Tinker, N.A.; Wight, C.P.; Deyl, J.; Burrows, V.D.; O’Donoughue, L.S.; Lybaert, A.; Molnar, S.J.; Armstrong, K.C.; Fedak, G.; et al. A molecular linkage map with associated QTLs from a hulless × covered spring oat population. Theor. Appl. Genet. 2004, 108, 1285–1298. [Google Scholar] [CrossRef]
- Zhu, S.; Rossnagel, B.G.; Kaeppler, H.F. Genetic analysis of quantitative trait loci for groat protein and oil content in oat. Crop Sci. 2004, 44, 254–260. [Google Scholar] [CrossRef]
- Klos, K.E.; Yimer, B.A.; Howarth, C.J.; McMullen, M.S.; Sorrells, M.E.; Tinker, N.A.; Yan, W.; Beattie, A.D. The genetic architecture of milling quality in spring oat lines of the collaborative oat research enterprise. Foods 2021, 10, 2479. [Google Scholar] [CrossRef] [PubMed]
- Zimmer, C.M.; McNish, I.G.; Klos, K.E.; Oro, T.; Arruda, K.M.A.; Gutkoski, L.C.; Pacheco, M.T.; Smith, K.P.; Federizzi, L.C. Genome-wide association for β-glucan content, population structure, and linkage disequilibrium in elite oat germplasm adapted to subtropical environments. Mol. Breed. 2020, 40, 103. [Google Scholar] [CrossRef]
- Fogarty, M.C.; Smith, S.M.; Sheridan, J.L.; Hu, G.; Islamovic, E.; Reid, R.; Jackson, E.W.; Maughan, P.J.; Ames, N.P.; Jellen, E.N.; et al. Identification of mixed linkage β-glucan quantitative trait loci and evaluation of AsCslF6 homoeologs in hexaploid oat. Crop Sci. 2020, 60, 914–933. [Google Scholar] [CrossRef]
- Youens-Clark, K.; Faga, B.; Yap, I.V.; Stein, L.; Ware, D. CMap 1.01: A comparative mapping application for the Internet. Bioinformatics 2009, 25, 3040–3042. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sen, T.Z.; Caccamo, M.; Edwards, D.; Quesneville, H. Building a successful international research community through data sharing: The case of the Wheat Information System (WheatIS). F1000Research 2020, 9, 536. [Google Scholar] [CrossRef] [PubMed]
- Bekele, W.A.; Wight, C.P.; Chao, S.; Howarth, C.J.; Tinker, N.A. Haplotype-based genotyping-by-sequencing in oat genome research. Plant Biotechnol. J. 2018, 16, 1452–1463. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Doehlert, D.C.; McMullen, M.S.; Jannink, J.-L. Oat grain/groat size ratios: A physical basis for test weight. Cereal Chem. 2006, 83, 114–118. [Google Scholar] [CrossRef] [Green Version]
- Herrmann, M.H.; Yu, J.; Beuch, S.; Weber, W.E. Quantitative trait loci for quality and agronomic traits in two advanced backcross populations in oat (Avena sativa L.). Plant Breed. 2014, 133, 588–601. [Google Scholar] [CrossRef]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Priyam, A.; Woodcroft, B.; Rai, V.; Moghul, I.; Munagala, A.; Ter, F.; Chowdhary, H.; Pieniak, I.; Maynard, L.J.; Gibbins, M.A.; et al. Sequenceserver: A modern graphical user interface for custom BLAST databases. Mol. Biol. Evol. 2019, 36, 2922–2924. [Google Scholar] [CrossRef]
- Groh, S.; Kianian, S.F.; Phillips, R.L.; Rines, H.W.; Stuthman, D.D.; Wesenberg, D.M.; Fulcher, R.G. Analysis of factors influencing milling yield and their association to other traits by QTL analysis in two hexaploid oat populations. Theor. Appl. Genet. 2001, 103, 9–18. [Google Scholar] [CrossRef]
- Colombo, F.; Di Lorenzo, C.; Biella, S.; Bani, C.; Restani, P. Ancient and modern cereals as ingredients of the gluten-free diet: Are they safe enough for celiac consumers? Foods 2021, 10, 906. [Google Scholar] [CrossRef] [PubMed]
- Chesnut, R.S.; Shotwell, M.A.; Boyer, S.K.; Larkins, B.A. Analysis of avenin proteins and the expression of their mRNAs in developing oat seeds. Plant Cell 1989, 1, 913–924. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brückner-Gühmann, M.; Kratzsch, A.; Sozer, N.; Drusch, S. Oat protein as plant-derived gelling agent: Properties and potential of modification. Future Foods 2021, 4, 100053. [Google Scholar] [CrossRef]
- Gangopadhyay, N.; Hossain, M.B.; Rai, D.K.; Brunton, N.P. A Review of extraction and analysis of bioactives in oat and barley and scope for use of novel food processing technologies. Molecules 2015, 20, 10884–10909. [Google Scholar] [CrossRef] [PubMed]
- Peterson, D.M. Protein concentration, concentration of protein fractions, and amino acid balance in oats. Crop Sci. 1976, 16, 663–666. [Google Scholar] [CrossRef]
- Kim, S.; Charbonnier, L.; Mossé, J. Heterogeneity of avenin, the oat prolamin: Fractionation, molecular weight and amino acid composition. Biochim. Biophys. Acta 1978, 537, 22–30. [Google Scholar] [CrossRef]
- Portyanko, V.A.; Portmortsev, A.A.; Kalashnik, N.A.; Bogachkov, V.I.; Sozinov, A.A. Genetic control of avenins and the principles of their classification. Sov. Genet. 1987, 23, 584–590. [Google Scholar]
- Shotwell, M.A.; Boyer, S.K.; Chesnut, R.S.; Larkins, B.A. Analysis of seed storage protein genes of oats. J. Biol. Chem. 1990, 265, 9652–9658. [Google Scholar] [CrossRef]
- Potier, B. Oat Seed Storage Protein Genes: Promoter Studies in Transgenic Tobacco Plants. Ph.D. Thesis, University of Ottawa, Ottawa, ON, Canada, 1994. [Google Scholar]
- Tanchak, M.A.; Giband, M.; Potier, B.; Schernthaner, J.P.; Dukiandjiev, S.; Altosaar, I. Genomic clones encoding 11S globulins in oats (Avena sativa L.). Genome 1995, 38, 627–634. [Google Scholar] [CrossRef] [PubMed]
- Wight, C.P.; Yan, W.; Fetch, J.M.; Deyl, J.; Tinker, N.A. A set of new simple sequence repeat and avenin DNA markers suitable for mapping and fingerprinting studies in oat (Avena spp.). Crop Sci. 2010, 50, 1207–1218. [Google Scholar] [CrossRef]
- Howes, N.K.; Chong, J.; Brown, P.D. Oat endosperm proteins associated with resistance to stem rust of oats. Genome 1992, 35, 120–125. [Google Scholar] [CrossRef]
- Hizbai, B.T.; Gardner, K.M.; Wight, C.P.; Dhanda, R.K.; Molnar, S.J.; Johnson, D.; Frégeau-Reid, J.; Yan, W.; Rossnagel, B.G.; Holland, J.; et al. Quantitative trait loci affecting oil content, oil composition, and other agronomically important traits in oat. Plant Genome 2012, 5, 164–175. [Google Scholar] [CrossRef] [Green Version]
- Kianian, S.F.; Egli, M.A.; Phillips, R.L.; Rines, H.W.; Somers, D.A.; Gengenbach, B.G.; Webster, F.H.; Livingston, S.M.; Groh, S.; O’Donoughue, L.S.; et al. Association of a major groat oil content QTL and an acetyl-CoA carboxylase gene in oat. Theor. Appl. Genet. 1999, 98, 884–894. [Google Scholar] [CrossRef]
- Mathews, R.; Kamil, A.; Chu, Y. Global review of heart health claims for oat beta-glucan products. Nutr. Rev. 2020, 78, 78–97. [Google Scholar] [CrossRef]
- Kianian, S.F.; Phillips, R.L.; Rines, H.W.; Fulcher, R.G.; Webster, F.H.; Stuthman, D.D. Quantitative trait loci influencing β-glucan content in oat (Avena sativa, 2n=6x=42). Theor. Appl. Genet. 2000, 101, 1039–1048. [Google Scholar] [CrossRef]
- Skinner, M.E.; Uzilov, A.V.; Stein, L.D.; Mungall, C.J.; Holmes, I.H. JBrowse: A next-generation genome browser. Genome Res. 2009, 19, 1630–1638. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lipps, H.J.; Rhodes, D. G-quadruplex structures: In vivo evidence and function. Trends Cell Biol. 2009, 19, 414–422. [Google Scholar] [CrossRef] [PubMed]
- Cagirici, H.B.; Budak, H.; Sen, T.Z. Genome-wide discovery of G-quadruplexes in barley. Sci. Rep. 2021, 11, 7876. [Google Scholar] [CrossRef]
- Cagirici, H.B.; Sen, T.Z. Genome-wide discovery of g-quadruplexes in wheat: Distribution and putative functional roles. G3 Genes Genomes Genet. 2020, 10, 2021–2032. [Google Scholar] [CrossRef] [PubMed] [Green Version]




| Acetyl Co-A Carboxylase (1) | Ligule Development (4) |
| Acid phosphatase (2) | Malate dehydrogenase (2) |
| alpha-Amylase (2) | Male sterility (2) |
| alpha-Esterase activity (2) | Maturity (1) |
| Avenin (7) | Multiple glumes (1) |
| Awn pubescence (1) | NADH dehydrogenase (1) |
| Awnedness (6) | Naked karyopsis (4) |
| Basal articulation (9) | Nodal pubescence (2) |
| beta-Esterase activity (1) | Nonheading (1) |
| beta-Galactosidase (1) | Panicle type (7) |
| beta-glucanase (1) | Pathogenesis-related protein (1) |
| beta-n-Acetylglucosaminidase (1) | Peduncle development (1) |
| Blast (1) | Peduncle length (1) |
| Blunt spikelet (1) | Peroxidase (7) |
| Chlorophyll deficiency-albino (6) | Phosphatase (1) |
| Chlorophyll deficiency—albovirescens (1) | Phosphoglucomutase (1) |
| Chlorophyll deficiency—chlorina (4) | Phosphogluconate dehydrogenase (2) |
| Chlorophyll deficiency—lutescens (3) | PHYA (1) |
| Chlorophyll deficiency—netting (2) | Plumule color (1) |
| Chlorophyll deficiency—stripe (3) | Rachilla pubescence (5) |
| Coleoptile color (1) | Reaction to Ditylenchus dipsaci (1) |
| Daylength insensitivity (1) | Reaction to Erysiphe graminis DC. (5) |
| Diaphorase activity (1) | Reaction to Helminthosporium victoriae (1) |
| Dwarf (1) | Reaction to Heterodera avenae (3) |
| Dwarfness (9) | Reaction to Pseudomonas coronafaciens (3) |
| Esterase (13) | Reaction to Puccinia coronata f. sp. avenae (119) |
| Fatuoid (1) | Reaction to Puccinia graminis Pers. (17) |
| Floret development (1) | Reaction to Puccinia recondita Rob. ex Desm. (1) |
| Floret disjunction (2) | Reaction to Schizaphis graminum Rond. (1) |
| Gametophyte (1) | Reaction to Ustilago avenae (11) |
| Giantism (2) | Reaction to Ustilago avenae and Ustilago kolleri (9) |
| Growth habit (1) | Reaction to Ustilago kolleri (10) |
| Isocitrate dehydrogenase (1) | Red seedling pigmentation (3) |
| Kernel pubescence (11) | Response to vernalization (1) |
| Leaf blade pubescence (2) | Seed pigmentation (1) |
| Leaf margin pubescence (2) | Seed proteins (1) |
| Leaf necrosis (1) | Semiglumeless (1) |
| Leaf sheath pubescence (4) | Shikimate dehydrogenase (1) |
| Lemma color (16) | Straw color (1) |
| Lemma fluorescence (2) | Synapsis (5) |
| Lemma pubescence (11) | Tertiary seedset (2) |
| Lemma waxiness (1) | Time to flowering (1) |
| Lethal (4) | Vivipary (3) |
| Leucine aminopeptidase activity (1) |
| Awn Frequency (2) | Milling Difficulty (oat) (1) |
|---|---|
| beta-glucan (FIA) (13) | Milling yield (oat) (2) |
| beta-glucan (NIR) (3) | Naked (2) |
| beta-glucan (oat) (88) | Oat winter hardiness (crown meristem freeze tolerance) (1) |
| Biomass (3) | Oat winter hardiness (field survival) (1) |
| BYDV (58) | Oleic acid content (8) |
| Days to heading (136) | Palmitic acid content (4) |
| Days to maturity (2) | Panicle number (2) |
| Flowering time (56) | Panicle type (5) |
| Grain filling period (2) | Phenology (1) |
| Grain yield (oat) (15) | Physiological maturity (2) |
| Groat oil (29) | Protein (1) |
| Groat percentage (46) | Reaction to barley yellow dwarf virus (2) |
| Groat protein (24) | Reaction to crown rust (oat, adult) (62) |
| Growth phase index (3) | Reaction to crown rust (oat, seedling) (11) |
| Harvest index (1) | Reaction to powdery mildew (8) |
| Height (155) | Root growth rate (30) |
| Kernel area (oat) (5) | Root surface area (6) |
| Kernel length (8) | Severity of crown rust (oat, adult) (56) |
| Kernel percent hull (3) | Shoot growth rate (14) |
| Kernel plumpness (4) | Shoot length (3) |
| Kernel weight (20) | Stearic acid content (4) |
| Kernel width (oat) (5) | Straw stiffness index (6) |
| Kernels per spikelet (1) | Straw yield (23) |
| Lemma pigment (2) | Test weight (63) |
| Linoleic acid content (7) | Thin kernels (5) |
| Linolenic acid content (4) | Tillers per plot (9) |
| Lodging (9) | Visual score (1) |
| Lodging at flowering (2) | Yield (79) |
| Lodging at harvest (4) | Aluminum tolerance (A. strigosa) (4) |
| Map Name | Date | Parent | Markers | Type | Maps |
|---|---|---|---|---|---|
| Oat-2020-CORE-SV | 2020 | Oat Core Collection | 80 | 53 Seed Vigor QTL, SNP, SSR | 22 linkage groups |
| Oat-2020-UFRGS-Bg | 2020 | UFRGS Panel | 27 | 27 b-glucan QTL, SNP, STS | 6 linkage groups |
| Oat-2020-UMNF-Pc | 2020 | UMN Founder Population | 42 | 43 Pc, HD QTL, SNP, RFLP | 7 linkage groups |
| Oat-2020-Celx9210_Pc39 | 2020 | Celer x STH9210 | 66 | DArT, SNP, SCAR, Pc39 | 1 linkage group |
| Oat-2020-CORE-BG | 2020 | Oat Core Collection | 219 | 56 QTL, SNPs | 19 linkage groups |
| Oat-2020-HxS-BG | 2020 | HiFi x Sol-Fi | 4 | AsCslF6_D, SNPs | 1 linkage group |
| Oat-2020-Pc98 | 2020 | Pc98, Bingo, Kasztan | 21 | KASPs, Pc98 | Mrg20 in 2 popns |
| Oat-2019-TX07CSxHidalgo | 2019 | TX07CS-1948 x Hidalgo | 6721 | SNPs, 31 QTL for seedling crown rust, height, heading date | 21 Linkage Groups |
| Oat-2019-CrownRust | 2019 | 9 parents (see mapdata) | 36 | SNPs Pc45, PcKM | 1 Linkage group from 5 popns |
| Oat-2018-Consensus | 2018 | Oat Diversity Panel | 99,846 | SNP, RFLP, Genes | 21 Merged Groups |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Blake, V.C.; Wight, C.P.; Yao, E.; Sen, T.Z. GrainGenes: Tools and Content to Assist Breeders Improving Oat Quality. Foods 2022, 11, 914. https://doi.org/10.3390/foods11070914
Blake VC, Wight CP, Yao E, Sen TZ. GrainGenes: Tools and Content to Assist Breeders Improving Oat Quality. Foods. 2022; 11(7):914. https://doi.org/10.3390/foods11070914
Chicago/Turabian StyleBlake, Victoria C., Charlene P. Wight, Eric Yao, and Taner Z. Sen. 2022. "GrainGenes: Tools and Content to Assist Breeders Improving Oat Quality" Foods 11, no. 7: 914. https://doi.org/10.3390/foods11070914
APA StyleBlake, V. C., Wight, C. P., Yao, E., & Sen, T. Z. (2022). GrainGenes: Tools and Content to Assist Breeders Improving Oat Quality. Foods, 11(7), 914. https://doi.org/10.3390/foods11070914






