DNA-Based Tools to Certify Authenticity of Rice Varieties—An Overview
Abstract
1. Introduction
1.1. Rice Diversity
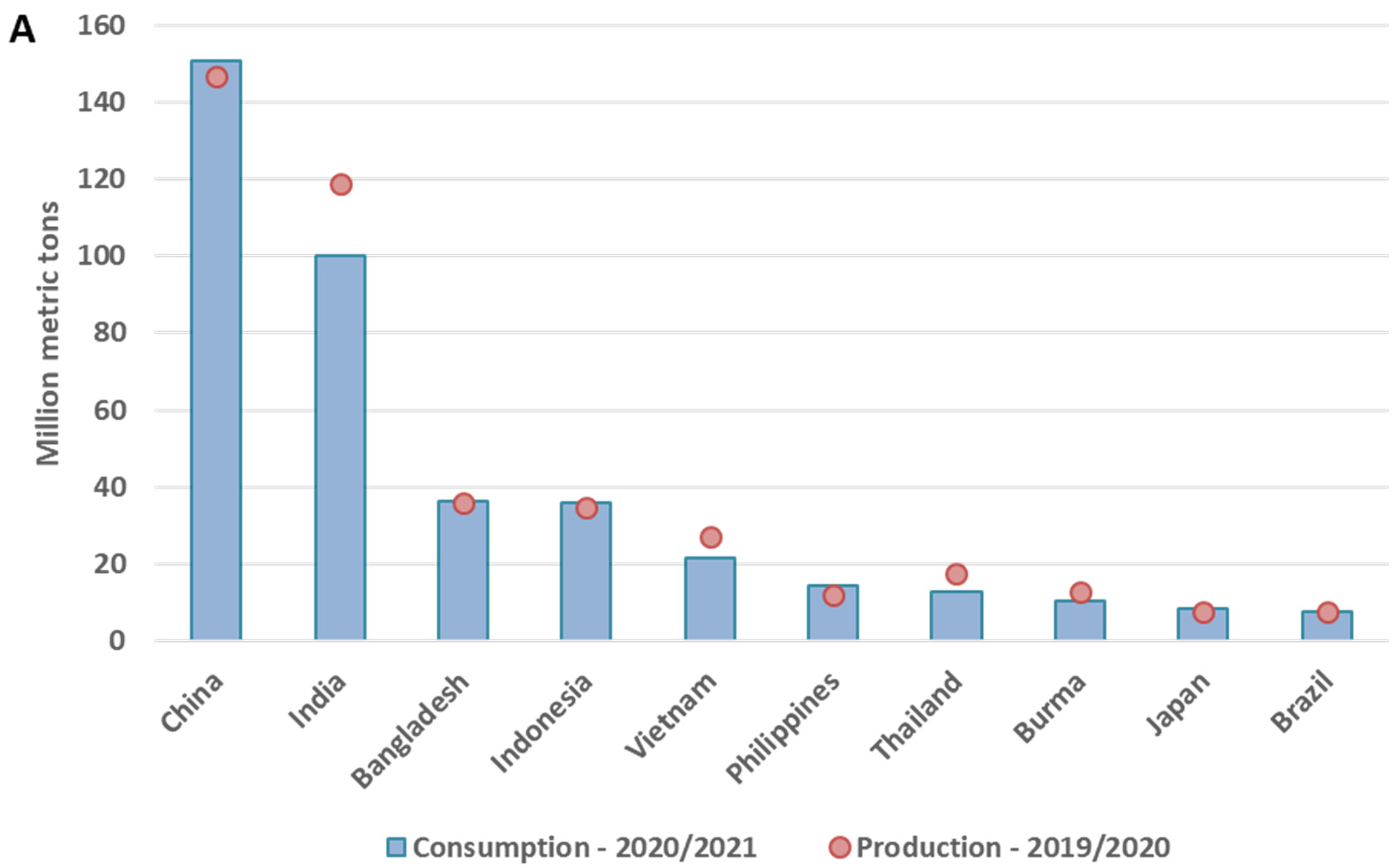
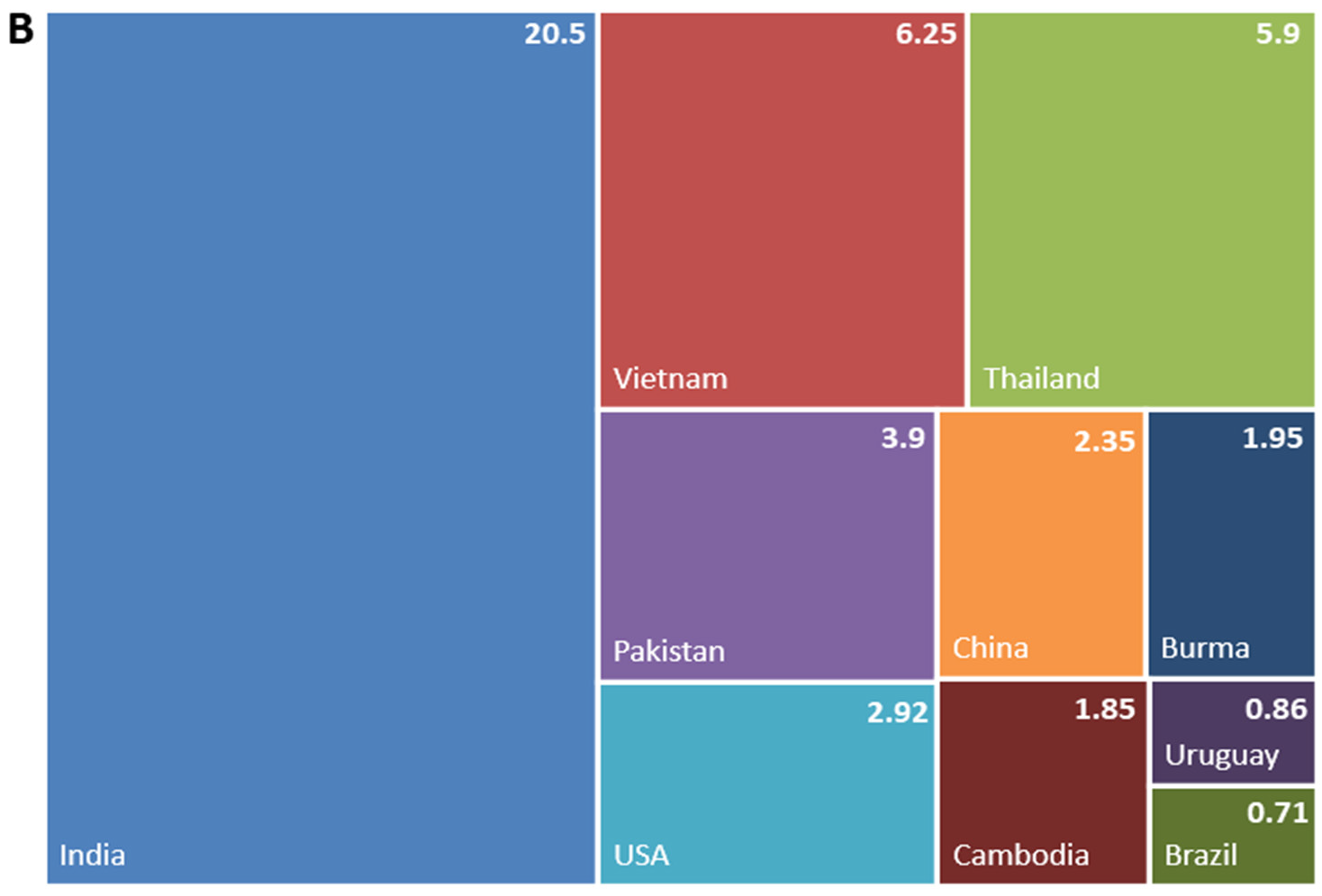
1.2. Rice in Europe and the Mediterranean Region—Production and Market
1.3. The Problem: Fraud in Trade Varieties
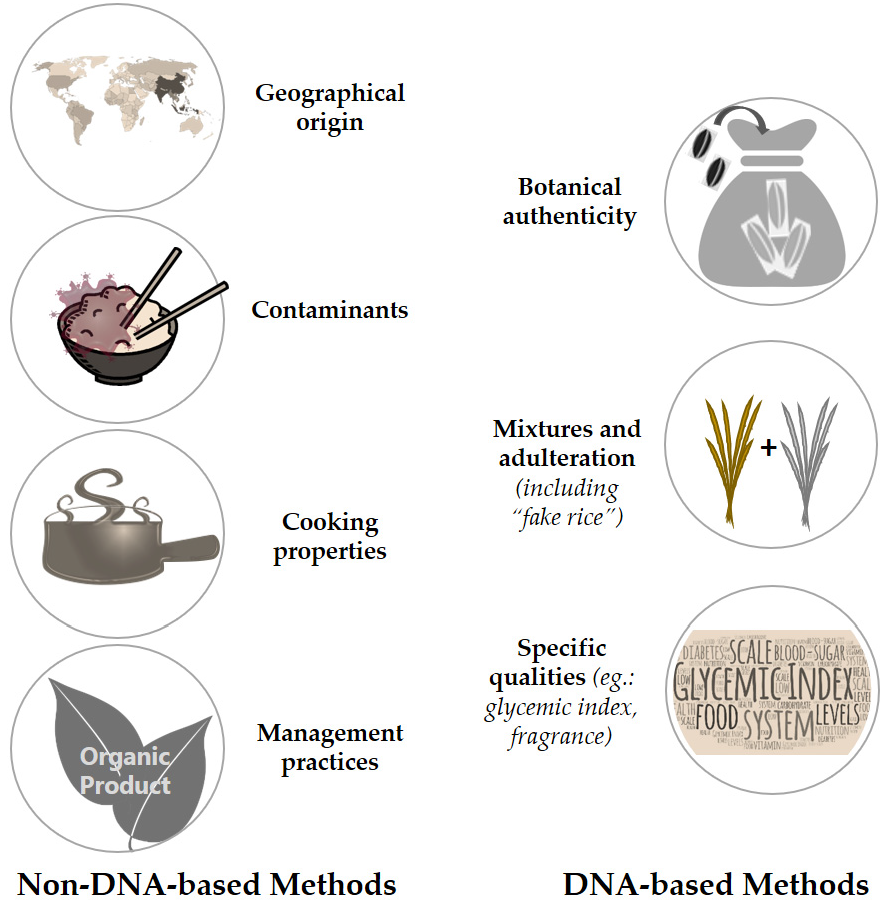
2. Brief Overview of Non-DNA Based Methods for Rice Certification
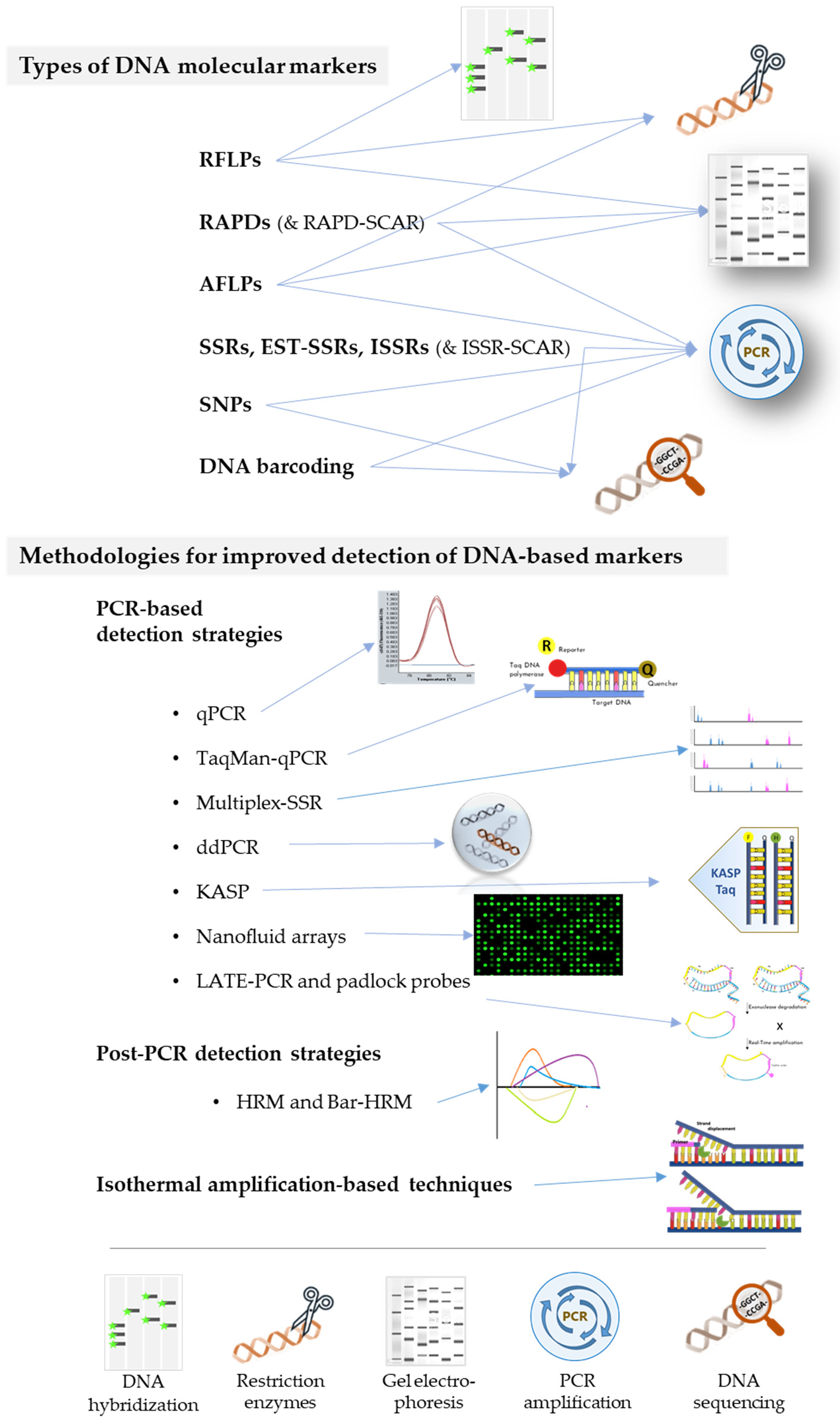
3. DNA-Based Tools for Rice Varietal Certification
3.1. Types of DNA Molecular Markers
3.1.1. Restriction Fragment Length Polymorphisms (RFLPs)
3.1.2. Random Amplification of Polymorphic DNAs (RAPDs)
3.1.3. Amplified Fragment Length Polymorphism (AFLPs)
3.1.4. Simple Sequence Repeats (SSRs)
3.1.5. Single Nucleotide Polymorphisms (SNPs)
3.1.6. DNA Barcoding
3.2. Methodologies for Improved Detection of DNA-Based Markers for Rice Authentication
3.2.1. PCR-Based Detection Strategies (qPCR, TaqMan-qPCR, Multiplex-SSR, ddPCR, KASP, Nanofluid Arrays, LATE-PCR and Padlock Probes)
3.2.2. Post-PCR Detection Strategies (HRM, Bar-HRM)
3.2.3. Isothermal Amplification-Based Techniques
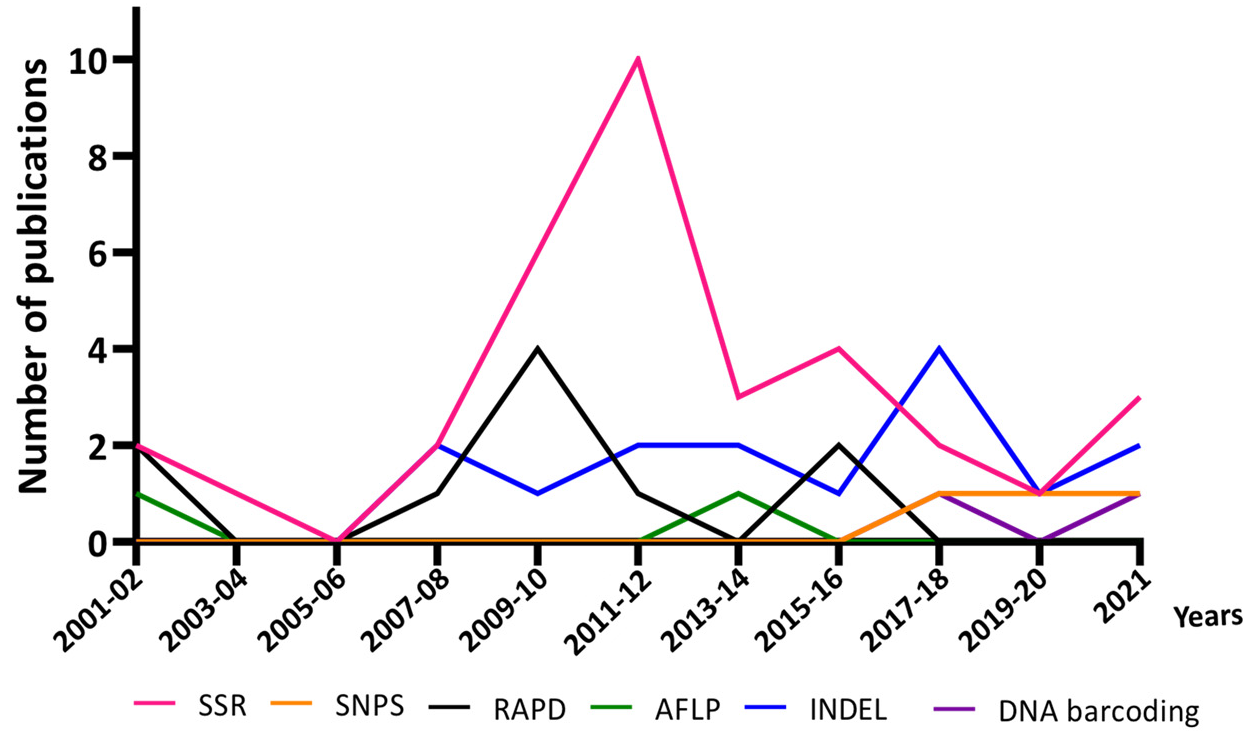
3.3. Progress in DNA-Based Methods for Rice Authentication
| Genotyping Methods | Type of Molecular Marker | Cultivars | References |
|---|---|---|---|
| Probe/enzyme | RFLP | indica vs. japonica | [63] |
| PCR | PCR-RFLP | Aromatic varieties | [64] |
| RAPD | Aromatic varieties | [67] | |
| 48 rice lines | [149] | ||
| RAPD/SCAR | Jasmine rice | [54] | |
| AFLP | 6 glutinous rice varieties | [89] | |
| SSR | Basmati vs. NB | [80] | |
| Indian rice hybrids | [137] | ||
| Basmati vs. advanced lines | [142] | ||
| 60 varieties | [155] | ||
| EST-SSR | Fine-grain varieties | [86] | |
| ISSR | japonica vs. indica | [88] | |
| ISSR, SSR | Basmati vs. EB vs. NB | [87] | |
| SSR, ISSR, RAPD | 30 indica varieties | [148] | |
| InDel | japonica vs. indica | [129] | |
| Multiplex PCR | SSR | Basmati vs. NB | [57] *,1 |
| Long and medium-grain rice varieties | [53] | ||
| 13 rice cultivars | [83] | ||
| Variety: Samba Mahsuri | [84] | ||
| STS | 130 varieties | [100] | |
| Duplex PCR | InDel | Basmati vs. NB | [58] * |
| SSR | Basmati vs. NB | [81] *,1 | |
| Duplex Digital Droplet PCR | InDel | Basmati vs. NB | [58] * |
| TaqMan PCR | SNP and InDel | Basmati vs. NB | [60] * |
| KASP | SNP | indica vs. japonica | [98] |
| SNP and InDel | Basmati varieties; Basmati vs. NB | [114] | |
| Fluidigm (PCR in Dynamic array IFCs) | SNP | indica vs. japonica | [101] |
| HRM | SSR and InDel | Basmati vs. NB | [59] * |
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- Wang, W.; Mauleon, R.; Hu, Z.; Chebotarov, D.; Tai, S.; Wu, Z.; Li, M.; Zheng, T.; Fuentes, R.R.; Zhang, F.; et al. Genomic Variation in 3,010 Diverse Accessions of Asian Cultivated Rice. Nature 2018, 557, 43–49. [Google Scholar] [CrossRef] [PubMed]
- United States Department of Agriculture. Foreign Agricultural Services, PSD Reports, World Rice Production, Consumption and Stocks. 12 January 2022. Available online: https://apps.fas.usda.gov/psdonline/app/index.html#/app/downloads (accessed on 15 November 2021).
- Genesys. Available online: https://www.genesys-pgr.org/c/rice (accessed on 15 November 2021).
- The 3000 Rice Genomes Project. GigaScience 2014, 3, 7. [CrossRef]
- Li, J.-Y.; Wang, J.; Zeigler, R.S. The 3000 Rice Genomes Project: New Opportunities and Challenges for Future Rice Research. GigaScience 2014, 3, 8. [Google Scholar] [CrossRef] [PubMed]
- IRRI International Rice Genebank. Available online: https://www.irri.org/international-rice-genebank (accessed on 15 January 2021).
- Garris, A.J.; Tai, T.H.; Coburn, J.; Kresovich, S.; McCouch, S. Genetic Structure and Diversity in Oryza sativa L. Genetics 2005, 169, 1631–1638. [Google Scholar] [CrossRef]
- Zhao, K.; Wright, M.; Kimball, J.; Eizenga, G.; McClung, A.; Kovach, M.; Tyagi, W.; Ali, M.L.; Tung, C.-W.; Reynolds, A.; et al. Genomic Diversity and Introgression in O. sativa Reveal the Impact of Domestication and Breeding on the Rice Genome. PLoS ONE 2010, 5, e10780. [Google Scholar] [CrossRef] [PubMed]
- Courtois, B.; Frouin, J.; Greco, R.; Bruschi, G.; Droc, G.; Hamelin, C.; Ruiz, M.; Clément, G.; Evrard, J.-C.; van Coppenole, S.; et al. Genetic Diversity and Population Structure in a European Collection of Rice. Crop. Sci. 2012, 52, 1663–1675. [Google Scholar] [CrossRef]
- Gutaker, R.M.; Groen, S.C.; Bellis, E.S.; Choi, J.Y.; Pires, I.S.; Bocinsky, R.K.; Slayton, E.R.; Wilkins, O.; Castillo, C.C.; Negrão, S.; et al. Genomic History and Ecology of the Geographic Spread of Rice. Nat. Plants 2020, 6, 492–502. [Google Scholar] [CrossRef] [PubMed]
- Khush, G.S. Origin, Dispersal, Cultivation and Variation of Rice. Plant Mol. Biol. 1997, 35, 25–34. [Google Scholar] [CrossRef]
- FAO. RMM FAO Rice Market Monitor (RMM), 1st ed.; Food and Agriculture Organization of the United Nations: Rome, Italy, 2018; Volume XXI. [Google Scholar]
- EC Agricultural Markets—Rice. Available online: https://agridata.ec.europa.eu/extensions/DataPortal/rice.html (accessed on 19 November 2021).
- Elbasiouny, H.; Elbehiry, F. Rice Production in Egypt: The Challenges of Climate Change and Water Deficiency. In Climate Change Impacts on Agriculture and Food Security in Egypt; Omran, E., Negm, A., Eds.; Springer Water: Cham, Switzerland, 2020; pp. 295–319. [Google Scholar]
- FAO. GIEWS Global Information and Early Warning System. Available online: https://www.fao.org/giews/country-analysis/country-briefs/country.jsp?lang=en&code=EGY (accessed on 15 November 2021).
- Courtois, B.; Greco, R.; Bruschi, G.; Frouin, J.; Ahmadi, N.; Droc, G.; Hamelin, C.; Ruiz, M.; Evrard, J.-C.; Katsantonis, D.; et al. Molecular Characterization of the European Rice Collection in View of Association Mapping. Plant Genet. Resour. 2011, 9, 233–235. [Google Scholar] [CrossRef][Green Version]
- Negrão, S.; Palaniappan, J.; Maroco, J.; Lourenço, T.; Mackill, D.; Oliveira, M.M. Bridging Sd1 Molecular Knowledge with Recent Breeding Strategies for the Improvement of Traditional Rice Varieties—A Japonica Case-Study. Afr. J. Biotechnol. 2010, 9, 2192–2200. [Google Scholar]
- EC. EU Agricultural Outlook for Markets and Income 2020–2030. 2020. Available online: https://data.europa.eu/doi/10.2762/252413 (accessed on 15 November 2021).
- FAO. RPU Rice Price Update. Available online: https://www.fao.org/markets-and-trade/commodities/rice/fao-rice-price-update/en/ (accessed on 15 November 2021).
- EC. Sustainable EU Rice. Available online: https://ec.europa.eu/chafea/agri/en/campaigns/sustainable-eu-rice (accessed on 15 November 2021).
- Steinberg, P.; Engert, S. A Daring Task: The Battle against Food Crime. J. Consum. Prot. Food Saf. 2019, 14, 4. [Google Scholar] [CrossRef]
- FAO. Food Fraud—Intention, Detection and Management: Food Safety Technical Toolkit for Asia and the Pacific; FAO: Bangkok, Thailand, 2021. [Google Scholar]
- Interpol Illicit Food and Drink Worth EUR 53 Million Seized in Global Operation. Available online: https://www.interpol.int/en/News-and-Events/News/2021/Illicit-food-and-drink-worth-EUR-53-million-seized-in-global-operation (accessed on 16 November 2021).
- Śliwińska-Bartel, M.; Burns, D.T.; Elliott, C. Rice Fraud a Global Problem: A Review of Analytical Tools to Detect Species, Country of Origin and Adulterations. Trends Food Sci. Technol. 2021, 116, 36–46. [Google Scholar] [CrossRef]
- Attaviroj, N.; Noomhorm, A. Discriminant Analysis of Multiple Physicochemical Properties for Thai Rough Rice Varietal Authentication. Int. J. Food Prop. 2014, 17, 1136–1149. [Google Scholar] [CrossRef]
- Tariq, A.; Bhawani, S.A.; Husaini, A.; Moheman, A. Authentication and Tracebility of Rice. In Fingerprinting Techniques in Food Authentication and Traceability; Siddiq, E.A., Nollet, L.M.L., Eds.; CRC Press: Boca Raton, FL, USA, 2018. [Google Scholar]
- Vemireddy, L.R.; Satyavathi, V.V.; Siddiq, E.A.; Nagaraju, J. Review of Methods for the Detection and Quantification of Adulteration of Rice: Basmati as a Case Study. J. Food Sci. Technol. 2015, 52, 3187–3202. [Google Scholar] [CrossRef]
- Carter, R.M.; Yan, Y.; Tomlins, K. Digital Imaging Based Classification and Authentication of Granular Food Products. Meas. Sci. Technol. 2005, 17, 235–240. [Google Scholar] [CrossRef]
- Kim, S.S.; Jo, J.S.; Kim, Y.J.; Sung, N.K. Authentication of Rice by Three-Sided Image Analysis of Kernels Using Two Mirrors. Cereal Chem. 1997, 74, 212–215. [Google Scholar] [CrossRef]
- Ortea, I.; O’Connor, G.; Maquet, A. Review on Proteomics for Food Authentication. J. Proteom. 2016, 147, 212–225. [Google Scholar] [CrossRef]
- Nguyen, Q.C.; Doan Duy, L.N.; Marini, F.; Biancolillo, A. Authentication of Rice (Oryza sativa L.) Using Near Infrared Spectroscopy Combined with Different Chemometric Classification Strategies. Appl. Sci. 2021, 11, 362. [Google Scholar]
- Maione, C.; Barbosa, R.M. Recent Applications of Multivariate Data Analysis Methods in the Authentication of Rice and the Most Analyzed Parameters: A Review. Crit. Rev. Food Sci. Nutr. 2019, 59, 1868–1879. [Google Scholar] [CrossRef]
- Lim, D.K.; Mo, C.; Lee, D.-K.; Long, N.P.; Lim, J.; Kwon, S.W. Non-Destructive Profiling of Volatile Organic Compounds Using HS-SPME/GC–MS and Its Application for the Geographical Discrimination of White Rice. J. Food Drug Anal. 2018, 26, 260–267. [Google Scholar] [CrossRef]
- Bagur-González, M.G.; Jiménez-Carvelo, A.M.; Ortega-Gavilán, F.; González-Casado, A. Chromatographic Methods. In Food Authentication and Traceability; Galanakis, C.M., Ed.; Academic Press: London, UK, 2021; pp. 65–99. [Google Scholar]
- Cheajesadagul, P.; Arnaudguilhem, C.; Shiowatana, J.; Siripinyanond, A.; Szpunar, J. Discrimination of Geographical Origin of Rice Based on Multi-Element Fingerprinting by High Resolution Inductively Coupled Plasma Mass Spectrometry. Food Chem. 2013, 141, 3504–3509. [Google Scholar] [CrossRef]
- Melo, M.G.; Carqueijo, A.; Freitas, A.; Barbosa, J.; Silva, A.S. Modified QuEChERS Extraction and HPLC-MS/MS for Simultaneous Determination of 155 Pesticide Residues in Rice (Oryza sativa L.). Foods 2019, 9, 18. [Google Scholar] [CrossRef]
- Shannon, M.; Ratnasekhar, C.H.; McGrath, T.F.; Kapil, A.P.; Elliott, C.T. A Two-Tiered System of Analysis to Tackle Rice Fraud: The Indian Basmati Study. Talanta 2021, 225, 122038. [Google Scholar] [CrossRef]
- Cubadda, F. Inductively Coupled Plasma-Mass Spectrometry for the Determination of Elements and Elemental Species in Food: A Review. J. AOAC Int. 2004, 87, 173–204. [Google Scholar] [CrossRef] [PubMed]
- Pérez Rodríguez, M.; Dirchwolf, P.; Varao, T.; Villafañe, R.N.; Neto, A.; Pellerano, R.; Ferreira, E. Brown Rice Authenticity Evaluation by Spark Discharge-Laser-Induced Breakdown Spectroscopy. Food Chem. 2019, 297, 124960. [Google Scholar] [CrossRef]
- Osborne, B.; Mertens, B.; Thompson, M.; Fearn, T. The Authentication of Basmati Rice Using NIR Spectroscopy. J. Near Infrared Spectrosc. 1993, 1, 77–83. [Google Scholar] [CrossRef]
- Monakhova, Y.B.; Rutledge, D.N.; Roßmann, A.; Waiblinger, H.-U.; Mahler, M.; Ilse, M.; Kuballa, T.; Lachenmeier, D.W. Determination of Rice Type by 1H NMR Spectroscopy in Combination with Different Chemometric Tools. J. Chemom. 2014, 28, 83–92. [Google Scholar] [CrossRef]
- Sha, M.; Gui, D.; Zhang, Z.; Ji, X.; Shi, X.; Liu, J.; Zhang, D. Evaluation of Sample Pretreatment Method for Geographic Authentication of Rice Using Raman Spectroscopy. J. Food Meas. Charact. 2019, 13, 1705–1712. [Google Scholar] [CrossRef]
- Wang, W.; Kong, W.; Shen, T.; Man, Z.; Zhu, W.; He, Y.; Liu, F.; Liu, Y. Application of Laser-Induced Breakdown Spectroscopy in Detection of Cadmium Content in Rice Stems. Front. Plant Sci. 2020, 11, 2073. [Google Scholar] [CrossRef] [PubMed]
- Dachoupakan Sirisomboon, C.; Wongthip, P.; Sirisomboon, P. Potential of near Infrared Spectroscopy as a Rapid Method to Detect Aflatoxins in Brown Rice. J. Near Infrared Spectrosc. 2019, 27, 232–240. [Google Scholar] [CrossRef]
- Ramesh, V. Nuclear Magnetic Resonance; Royal Society of Chemistry: London, UK, 2016; Volume 45. [Google Scholar]
- Yue, X.; Tan, Y.; Fan, W.; Song, S.; Ji, H.; Li, B. Raman Spectroscopic Analysis of Paddy Rice Infected by Three Pests and Diseases Common in Northeast Asia. J. Phys. Conf. Ser. 2019, 1324, 012050. [Google Scholar] [CrossRef]
- Fanelli, V.; Mascio, I.; Miazzi, M.M.; Savoia, M.A.; De Giovanni, C.; Montemurro, C. Molecular Approaches to Agri-Food Traceability and Authentication: An Updated Review. Foods 2021, 10, 1644. [Google Scholar] [CrossRef]
- Mafra, I.; Ferreira, I.M.P.L.V.O.; Oliveira, M.B.P.P. Food Authentication by PCR-Based Methods. Eur. Food Res. Technol. 2008, 227, 649–665. [Google Scholar] [CrossRef]
- Fridez, F. Basmati Rice Fraud under the Magnifying Glass of DNA Analysis. Int. J. Chem. 2016, 70, 354–356. [Google Scholar] [CrossRef]
- Lidder, P.; Sonnino, A. Biotechnologies for the Management of Genetic Resources for Food and Agriculture. In Advances in Genetics; Goodwin, S.F., Friedmann, T., Dunlap, J.C., Eds.; Academic Press: Cambridge, MA, USA, 2012; Volume 78, pp. 1–167. [Google Scholar]
- Sepahian, N.; Noourmohammadi, Z.; Sheidai, M.; Zamanizadeh, H.-R. Authentication, Genetic Fingerprinting and Assessing Relatedness of Rice (Oryza sativa) Genotypes by SSR Molecular Markers. Caryologia 2021, 74, 13–22. [Google Scholar] [CrossRef]
- Razak, S.A.; Azman, N.H.E.N.; Kamaruzaman, R.; Saidon, S.A.; Yusof, M.F.M.; Ismail, S.N.; Jaafar, M.A.; Abdullah, N. Genetic Diversity of Released Malaysian Rice Varieties Based on Single Nucleotide Polymorphism Markers. Czech J. Genet. Plant Breed. 2020, 56, 62–70. [Google Scholar] [CrossRef]
- Satturu, V.; Rani, D.; Gattu, S.; Md, J.; Mulinti, S.; Nagireddy, R.K.; Eruvuri, R.; Yanda, R. DNA Fingerprinting for Identification of Rice Varieties and Seed Genetic Purity Assessment. Agric Res. 2018, 7, 379–390. [Google Scholar] [CrossRef]
- Wu, Y.; Zhang, Z.; Chen, Y.; Wang, B.; Yang, G.; Yang, W. Authentication of Thailand Jasmine Rice Using RAPD and SCAR Methods. Eur. Food Res. Technol. 2009, 3, 515–521. [Google Scholar] [CrossRef]
- Kingsakul, S.; Aoki, S.; Dechakhamphu, A. Genetic Polymorphism of Glutinous Rice (Oryza sativa L.) Using an Amplified Fragment Length Polymorphism (AFLP) Technique. Acta Hortic. 2013, 973, 225–229. [Google Scholar] [CrossRef]
- Scarano, D.; Rao, R. DNA Markers for Food Products Authentication. Diversity 2014, 6, 579–596. [Google Scholar] [CrossRef]
- Archak, S.; Lakshminarayanareddy, V.; Nagaraju, J. High-Throughput Multiplex Microsatellite Marker Assay for Detection and Quantification of Adulteration in Basmati Rice (Oryza sativa). Electrophoresis 2007, 28, 2396–2405. [Google Scholar] [CrossRef]
- Bucher, T.B.; Fridez, F.; Köppel, R. Duplex Real-Time PCR for the Determination of Non-Basmati Rice in Basmati Rice (Oryza sativa). Eur. Food Res. Technol. 2014, 238, 417–423. [Google Scholar] [CrossRef]
- Ganopoulos, I.; Argiriou, A.; Tsaftaris, A. Adulterations in Basmati Rice Detected Quantitatively by Combined Use of Microsatellite and Fragrance Typing with High Resolution Melting (HRM) Analysis. Food Chem. 2011, 129, 652–659. [Google Scholar] [CrossRef]
- Lopez, S.J. TaqMan Based Real Time PCR Method for Quantitative Detection of Basmati Rice Adulteration with Non-Basmati Rice. Eur. Food Res. Technol. 2008, 227, 619–622. [Google Scholar] [CrossRef]
- Agarwal, M.; Shrivastava, N.; Padh, H. Advances in Molecular Marker Techniques and Their Applications in Plant Sciences. Plant Cell Rep. 2008, 27, 617–631. [Google Scholar] [CrossRef]
- Shabir, G.; Aslam, K.; Khan, A.R.; Shahid, M.; Manzoor, H.; Noreen, S.; Khan, M.A.; Baber, M.; Sabar, M.; Shah, S.; et al. Rice Molecular Markers and Genetic Mapping: Current Status and Prospects. J. Integr. Agric. 2017, 16, 1879–1891. [Google Scholar] [CrossRef]
- Zhang, Q.; Maroof, M.A.S.; Lu, T.Y.; Shen, B.Z. Genetic Diversity and Differentiation of Indica and Japonica Rice Detected by RFLP Analysis. Theoret. Appl. Genet. 1992, 83, 495–499. [Google Scholar] [CrossRef]
- Balamurugan, S.; Murugan, S.B.; Varghese, I.P.; Harish, M.C.; Sathishkumar, R. Novel Method for the Detection of Adulteration in Expensive Aromatic Varieties Using PCR-RFLP. IN Patent 201711012732 A, 2017. Available online: https://b-u.ac.in/136/patent-filed (accessed on 15 November 2021).
- Martins-Lopes, P.; Gomes, S.; Pereira, L.; Guedes-Pinto, H. Molecular Markers for Food Traceability. Food Technol. Biotechnol. 2013, 51, 198–207. [Google Scholar]
- Hasan, A.; Rafii, M.; Harun, A.; Ahmad, F. Current Applicable DNA Markers for Marker Assisted Breeding in Rice (Oryza sativa L.). In Recent Advances in Rice Research; Ansari, M.R., Ed.; IntechOpen: London, UK, 2020. [Google Scholar]
- Choudhury, P.; Kohli, S.; Srinivasan, K.; Mohapatra, T.; Sharma, R.P. Identification and Classification of Aromatic Rices Based on DNA Fingerprinting. Euphytica 2001, 118, 243–251. [Google Scholar] [CrossRef]
- Ying, X. Detection of Purity of Thai Hom Mali Rice by RAPD. Agric. Sci. Technol. 2011, 12, 1565–1568. [Google Scholar]
- Zhu, J.; Gale, M.D.; Quarrie, S.; Jackson, M.T.; Bryan, G.J. AFLP Markers for the Study of Rice Biodiversity. Theor. Appl. Genet. 1998, 96, 602–611. [Google Scholar] [CrossRef]
- Kumar, P.; Gupta, V.K.; Misra, A.K.; Modi, D.; Pandey, B.K. Potential of Molecular Markers in Plant Biotechnology. Plant Omics 2009, 2, 141–162. [Google Scholar]
- Mackill, D.J.; Zhang, Z.; Redoña, E.D.; Colowit, P.M. Level of Polymorphism and Genetic Mapping of AFLP Markers in Rice. Genome 1996, 39, 969–977. [Google Scholar] [CrossRef]
- Miah, G.; Rafii, M.Y.; Ismail, M.R.; Puteh, A.B.; Rahim, H.A.; Islam, K.N.; Latif, M.A. A Review of Microsatellite Markers and Their Applications in Rice Breeding Programs to Improve Blast Disease Resistance. Int. J. Mol. Sci. 2013, 14, 22499–22528. [Google Scholar] [CrossRef]
- Paun, O.; Schönswetter, P. Amplified Fragment Length Polymorphism: An Invaluable Fingerprinting Technique for Genomic, Transcriptomic, and Epigenetic Studies. In Plant DNA Fingerprinting and Barcoding: Methods and Protocols; Sucher, N.J., Hennell, J.R., Carles, M.C., Eds.; Humana Press: Totowa, NJ, USA, 2012; pp. 75–87. [Google Scholar]
- Vos, P.; Hogers, R.; Bleeker, M.; Reijans, M.; van de Lee, T.; Hornes, M.; Friters, A.; Pot, J.; Paleman, J.; Kuiper, M.; et al. AFLP: A New Technique for DNA Fingerprinting. Nucleic Acids Res. 1995, 23, 4407–4414. [Google Scholar] [CrossRef]
- La Rota, M.; Kantety, R.V.; Yu, J.-K.; Sorrells, M.E. Nonrandom Distribution and Frequencies of Genomic and EST-Derived Microsatellite Markers in Rice, Wheat, and Barley. BMC Genom. 2005, 6, 23. [Google Scholar] [CrossRef]
- Lawson, M.J.; Zhang, L. Distinct Patterns of SSR Distribution in the Arabidopsis thaliana and Rice Genomes. Genome Biol. 2006, 7, R14. [Google Scholar] [CrossRef]
- Vemireddy, L.; Archak, S.; Nagaraju, J. Capillary Electrophoresis Is Essential for Microsatellite Marker Based Detection and Quantification of Adulteration of Basmati Rice (Oryza sativa). J. Agric. Food Chem. 2007, 55, 8112–8117. [Google Scholar] [CrossRef]
- Singh, H.; Deshmukh, R.K.; Singh, A.; Singh, A.K.; Gaikwad, K.; Sharma, T.R.; Mohapatra, T.; Singh, N.K. Highly Variable SSR Markers Suitable for Rice Genotyping Using Agarose Gels. Mol. Breed. 2010, 25, 359–364. [Google Scholar] [CrossRef]
- Temnykh, S.; DeClerck, G.; Lukashova, A.; Lipovich, L.; Cartinhour, S.; Mccouch, S. Computational and Experimental Analysis of Microsatellites in Rice (Oryza sativa L.): Frequency, Length Variation, Transposon Associations, and Genetic Marker Potential. Genome Res. 2001, 11, 1441–1452. [Google Scholar] [CrossRef]
- Bligh, H. Detection of Adulteration of Basmati Rice with Non-premium Long-grain Rice. Int. J. Food Sci. Technol. 2000, 35, 257–265. [Google Scholar] [CrossRef]
- Cassier, R.; Kozulic, M. Authentication of Basmati Rice Using SSR-PCR and the QIAxcel® Advanced System. 2014. Available online: https://www.qiagen.com/us/resources/download.aspx?id=7303c7f0-9681-4105-bc79-4bba045871e9&lang=en (accessed on 15 November 2021).
- Rai, V.P.; Singh, A.K.; Jaiswal, H.; Singh, S.P.; Singh, R.; Waza, S.A. Evaluation of Molecular Markers Linked to Fragrance and Genetic Diversity in Indian Aromatic Rice. Turk. J. Bot. 2015, 39, 209–217. [Google Scholar] [CrossRef]
- Coburn, J.R.; Temnykh, S.V.; Paul, E.M.; McCouch, S.R. Design and Application of Microsatellite Marker Panels for Semiautomated Genotyping of Rice (Oryza sativa L.). Crop Sci. 2002, 42, 2092–2099. [Google Scholar] [CrossRef]
- Kumari, K.; Durgarani, C.; Vanisree, S.; Sivaramakrishnan, S.; Balachandran, S.M.; Sundaram, R.M. Molecular Fingerprinting of the Elite, Fine-Grain Type Rice Cultivar, Samba Mahsuri (BPT 5204) and Assessment of Genetic Purity in Seed-Lots of the Variety Using SSR Markers. Int. J. Sci. Res. Publ. 2011, 1, 1–13. [Google Scholar]
- Bonow, S.; Pinho, E.V.R.V.; Vieira, M.G.C.; Vosman, B. Microsatellite Markers in and around Rice Genes: Applications in Variety Identification and DUS Testing. Crop Sci. 2009, 49, 880–886. [Google Scholar] [CrossRef]
- Moorthy, K.K.; Babu, P.; Sreedhar, M.; Sama, V.S.A.K.; Kumar, P.N.; Balachandran, S.M.; Sundaram, R.M. Identification of Informative EST-SSR Markers Capable of Distinguishing Popular Indian Rice Varieties and Their Utilization in Seed Genetic Purity Assessments. Seed Sci. Technol. 2011, 39, 282–292. [Google Scholar] [CrossRef]
- Nagaraju, J.; Kathirvel, M.; Kumar, R.R.; Siddiq, E.A.; Hasnain, S.E. Genetic Analysis of Traditional and Evolved Basmati and Non-Basmati Rice Varieties by Using Fluorescence-Based ISSR-PCR and SSR Markers. Proc. Natl. Acad. Sci. USA 2002, 99, 5836–5841. [Google Scholar] [CrossRef]
- Blair, M.W.; Panaud, O.; McCouch, S.R. Inter-Simple Sequence Repeat (ISSR) Amplification for Analysis of Microsatellite Motif Frequency and Fingerprinting in Rice (Oryza sativa L.). Theor. Appl. Genet. 1999, 98, 780–792. [Google Scholar] [CrossRef]
- Lin, Y.B.; Zhang, Y.M.; Hang, Y.Y.; Li, M.M.; Zhou, G.C.; Shen, X.L.; Sun, X.Q. A Two-Step Method for Identification of the Chinese Glutinous Rice Suyunuo, Based on ISSR-SCAR and Allele-Specific Markers. Genet. Mol. Res. 2016, 15, 4. [Google Scholar] [CrossRef]
- Begum, H.; Spindel, J.E.; Lalusin, A.; Borromeo, T.; Gregorio, G.; Hernandez, J.; Virk, P.; Collard, B.; McCouch, S.R. Genome-Wide Association Mapping for Yield and Other Agronomic Traits in an Elite Breeding Population of Tropical Rice (Oryza sativa). PLoS ONE 2015, 10, e0119873. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Wang, K.; Wang, W.; Yin, T.; Dong, W.; Xu, C. A High-Throughput SNP Discovery Strategy for RNA-Seq Data. BMC Genom. 2019, 20, 160. [Google Scholar] [CrossRef] [PubMed]
- Varshney, R.K.; Nayak, S.N.; May, G.D.; Jackson, S.A. Next-Generation Sequencing Technologies and Their Implications for Crop Genetics and Breeding. Trends Biotechnol. 2009, 27, 522–530. [Google Scholar] [CrossRef] [PubMed]
- Yuan, H.; Yang, W.; Zou, J.; Cheng, M.; Fan, F.; Liang, T.; Yu, Y.; Qiu, R.; Li, S.; Hu, J. InDel Markers Based on 3K Whole-Genome Re-Sequencing Data Characterise the Subspecies of Rice (Oryza sativa L.). Agriculture 2021, 11, 655. [Google Scholar] [CrossRef]
- Fuentes, R.R.; Chebotarov, D.; Duitama, J.; Smith, S.; De la Hoz, J.; Mohiyuddin, M.; Wing, R.A.; McNally, K.L.; Tatarinova, T.; Grigoriev, A.; et al. Structural Variants in 3000 Rice Genomes. Genome Res. 2019, 29, 870–880. [Google Scholar] [CrossRef]
- Kamboj, R.; Singh, B.; Mondal, T.K.; Bisht, D.S. Current Status of Genomic Resources on Wild Relatives of Rice. Breed. Sci. 2020, 70, 135–144. [Google Scholar] [CrossRef]
- Mansueto, L.; Fuentes, R.R.; Borja, F.N.; Detras, J.; Abriol-Santos, J.M.; Chebotarov, D.; Sanciangco, M.; Palis, K.; Copetti, D.; Poliakov, A.; et al. Rice SNP-Seek Database Update: New SNPs, Indels, and Queries. Nucleic Acids Res. 2017, 45, D1075–D1081. [Google Scholar] [CrossRef]
- Karau, M. Fingerprinting and Assessing Relatedness of Selected Rice (Oryza sativa) Genotypes in Kenya. J. Biotechnol. Biochem. 2018, 4, 48–51. [Google Scholar]
- Gouda, A.C.; Warburton, M.L.; Djedatin, G.L.; Kpeki, S.B.; Wambugu, P.W.; Gnikoua, K.; Ndjiondjop, M.N. Development and Validation of Diagnostic SNP Markers for Quality Control Genotyping in a Collection of Four Rice (Oryza) Species. Sci. Rep. 2021, 11, 18617. [Google Scholar] [CrossRef]
- Thomson, M.J. High-Throughput SNP Genotyping to Accelerate Crop Improvement. Plant Breed. Biotechnol. 2014, 2, 195–212. [Google Scholar] [CrossRef]
- Shinmura, K.; Kanagawa, H.; Mikami, T.; Fukumori, T. Development of Multiplex PCR Primer Sets for the Identification of Rice Varieties. Breed. Res. 2005, 7, 87–94. [Google Scholar] [CrossRef]
- Seo, J.; Lee, G.; Jin, Z.; Kim, B.; Chin, J.H.; Koh, H.-J. Development and Application of Indica–Japonica SNP Assays Using the Fluidigm Platform for Rice Genetic Analysis and Molecular Breeding. Mol. Breed. 2020, 40, 39. [Google Scholar] [CrossRef]
- Hebert, P.D.N.; Cywinska, A.; Ball, S.L.; deWaard, J.R. Biological Identifications through DNA Barcodes. Proc. Biol. Sci. 2003, 270, 313–321. [Google Scholar] [CrossRef] [PubMed]
- Singh, J.; Kakade, D.; Wallalwar, M.; Raghuvanshi, R.; Kongbrailatpam, M.; Verulkar, S.; Banerjee, S. Evaluation of Potential DNA Barcoding Loci from Plastid Genome: Intraspecies Discrimination in Rice (Oryza species). Int. J. Curr. Microbiol. App. Sci. 2017, 6, 2746–2756. [Google Scholar] [CrossRef][Green Version]
- Zhang, W.; Sun, Y.; Liu, J.; Xu, C.; Zou, X.; Chen, X.; Liu, Y.; Wu, P.; Yang, X.; Zhou, S. DNA Barcoding of Oryza: Conventional, Specific, and Super Barcodes. Plant Mol. Biol. 2021, 105, 215–228. [Google Scholar] [CrossRef]
- Abbai, R.; Singh, V.K.; Nachimuthu, V.V.; Sinha, P.; Selvaraj, R.; Vipparla, A.K.; Singh, A.K.; Singh, U.M.; Varshney, R.K.; Kumar, A. Haplotype Analysis of Key Genes Governing Grain Yield and Quality Traits across 3K RG Panel Reveals Scope for the Development of Tailor-Made Rice with Enhanced Genetic Gains. Plant Biotechnol. J. 2019, 17, 1612–1622. [Google Scholar] [CrossRef]
- Sun, C.; Hu, Z.; Zheng, T.; Lu, K.; Zhao, Y.; Wang, W.; Shi, J.; Wang, C.; Lu, J.; Zhang, D.; et al. RPAN: Rice Pan-Genome Browser for ∼3000 Rice Genomes. Nucleic Acids Res. 2017, 45, 597–605. [Google Scholar] [CrossRef]
- Barcaccia, G.; Lucchin, M.; Cassandro, M. DNA Barcoding as a Molecular Tool to Track down Mislabeling and Food Piracy. Diversity 2016, 8, 2. [Google Scholar] [CrossRef]
- Campos, M.D.; Valadas, V.; Campos, C.; Morello, L.; Braglia, L.; Breviario, D.; Cardoso, H.G. A TaqMan Real-Time PCR Method Based on Alternative Oxidase Genes for Detection of Plant Species in Animal Feed Samples. PLoS ONE 2018, 13, e0190668. [Google Scholar] [CrossRef]
- Jain, M.; Nijhawan, A.; Tyagi, A.K.; Khurana, J.P. Validation of Housekeeping Genes as Internal Control for Studying Gene Expression in Rice by Quantitative Real-Time PCR. Biochem. Biophys. Res. Commun. 2006, 345, 646–651. [Google Scholar] [CrossRef]
- Di Domenico, M.; Di Giuseppe, M.; Wicochea Rodríguez, J.D.; Cammà, C. Validation of a Fast Real-Time PCR Method to Detect Fraud and Mislabeling in Milk and Dairy Products. J. Dairy Sci. 2017, 100, 106–112. [Google Scholar] [CrossRef]
- Bucher, T.B.; Köppel, R. Duplex Digital Droplet PCR for the Determination of Non-Basmati Rice in Basmati Rice (Oryza sativa) on the Base of a Deletion in the Fragrant Gene. Eur. Food Res. Technol. 2016, 242, 927–934. [Google Scholar] [CrossRef]
- Yu, N.; Xing, R.; Wang, P.; Deng, T.; Zhang, J.; Zhao, G.; Chen, Y. A Novel Duplex Droplet Digital PCR Assay for Simultaneous Authentication and Quantification of Panax Notoginseng and Its Adulterants. Food Control 2022, 132, 108493. [Google Scholar] [CrossRef]
- Quan, P.-L.; Sauzade, M.; Brouzes, E. DPCR: A Technology Review. Sensors 2018, 18, 1271. [Google Scholar] [CrossRef]
- Steele, K.; Tulloch, M.Q.; Burns, M.; Nader, W. Developing KASP Markers for Identification of Basmati Rice Varieties. Food Anal. Methods 2021, 14, 663–673. [Google Scholar] [CrossRef]
- Prins, T.W.; van Dijk, J.P.; Beenen, H.G.; Van Hoef, A.A.; Voorhuijzen, M.M.; Schoen, C.D.; Aarts, H.J.; Kok, E.J. Optimised Padlock Probe Ligation and Microarray Detection of Multiple (Non-Authorised) GMOs in a Single Reaction. BMC Genom. 2008, 9, 584. [Google Scholar] [CrossRef]
- Voorhuijzen, M.M.; Van Dijk, J.P.; Prins, T.W.; Van Hoef, A.M.A.; Seyfarth, R.; Kok, E.J. Development of a Multiplex DNA-Based Traceability Tool for Crop Plant Materials. Anal. Bioanal. Chem. 2012, 402, 693–701. [Google Scholar] [CrossRef]
- Zhu, C.; Wu, X.; Li, Z.; Zhao, J.; Liu, Y.; Wang, A.; Deng, G.; Zhu, L. A Microfluidic System Integrated One-Step PCR and High-Resolution Melting Analysis for Rapid Rice Mutant Detection. Biotechnol. Biotechnol. Equip. 2019, 33, 1164–1171. [Google Scholar] [CrossRef]
- Druml, B.; Cichna-Markl, M. High Resolution Melting (HRM) Analysis of DNA—Its Role and Potential in Food Analysis. Food Chem. 2014, 158, 245–254. [Google Scholar] [CrossRef]
- Jaakola, L.; Suokas, M.; Häggman, H. Novel Approaches Based on DNA Barcoding and High-Resolution Melting of Amplicons for Authenticity Analyses of Berry Species. Food Chem. 2010, 123, 494–500. [Google Scholar] [CrossRef]
- Sun, W.; Li, J.; Xiong, C.; Zhao, B.; Chen, S. The Potential Power of Bar-HRM Technology in Herbal Medicine Identification. Front. Plant Sci. 2016, 7, 367. [Google Scholar] [CrossRef]
- Ganopoulos, I.; Bazakos, C.; Madesis, P.; Kalaitzis, P.; Tsaftaris, A. Barcode DNA High-Resolution Melting (Bar-HRM) Analysis as a Novel Close-Tubed and Accurate Tool for Olive Oil Forensic Use. J. Sci. Food Agric. 2013, 93, 2281–2286. [Google Scholar] [CrossRef]
- Sorochynskyi, B.V. Detection of Genetically Modified Plants Using LAMP (Loop-Mediated Amplification) Technologies. Plant Var. Stud. Prot. 2021, 17, 51–59. [Google Scholar]
- Chen, X.; Wang, X.; Jin, N.; Zhou, Y.; Huang, S.; Miao, Q.; Zhu, Q.; Xu, J. Endpoint Visual Detection of Three Genetically Modified Rice Events by Loop-Mediated Isothermal Amplification. Int. J. Mol. Sci. 2012, 13, 14421–14433. [Google Scholar] [CrossRef]
- Ongom, P.O.; Fatokun, C.; Togola, A.; Salvo, S.; Oyebode, O.G.; Ahmad, M.S.; Jockson, I.D.; Bala, G.; Boukar, O. Molecular Fingerprinting and Hybridity Authentication in Cowpea Using Single Nucleotide Polymorphism Based Kompetitive Allele-Specific PCR Assay. Front. Plant Sci. 2021, 12, 2205. [Google Scholar] [CrossRef] [PubMed]
- Mammadov, J.; Aggarwal, R.; Buyyarapu, R.; Kumpatla, S. SNP Markers and Their Impact on Plant Breeding. Int. J. Plant Genom. 2012, 2012, e728398. [Google Scholar] [CrossRef] [PubMed]
- Morgil, H.; Gercek, Y.C.; Tulum, I. Single Nucleotide Polymorphisms (SNPs). In Plant Genetics and Breeding; IntechOpen: London, UK, 2020. [Google Scholar]
- Romero, G.O.; Falla, M.C.B.; Anglacer, A.C. Polymerase Chain Reaction-Based Techniques for DNA Fingerprinting of Philippine Modern Rice Varieties. Philipp. Agric. Sci. 2002, 85, 299–306. [Google Scholar]
- Pal, S.; Jain, S.; Saini, N.; Jain, R.K. Identification of Microsatellite Markers for Differentiating Some Basmati and Non-Basmati Rice Varieties. Indian J. Biotechnol. 2004, 3, 519–526. [Google Scholar]
- Cai, X.; Liu, J.; Qiu, Y.; Zhao, W.; Song, Z.; Lu, B. Differentiation of Indica-Japonica Rice Revealed by Insertion/Deletion (InDel) Fragments Obtained from the Comparative Genomic Study of DNA Sequences between 93-11 (Indica) and Nipponbare (Japonica). Front. Biol. China 2007, 2, 291–296. [Google Scholar] [CrossRef]
- Tamilkumar, P.; Jerlin, R.; Senthil, N.; Ganesan, K.N.; Jeevan, R.J.; Raveendran, M. Fingerprinting of Rice Hybrids and Their Parental Lines Using Microsatellite Markers and Their Utilization in Genetic Purity Assessment of Hybrid Rice. Res. J. Seed Sci. 2008, 2, 40–47. [Google Scholar] [CrossRef]
- Pani, D.R.; Arif, M.; Taj, G.; Kar, C.S.; Singh, U.S. Random Amplified Polymorphic DNA Analysis of Indigenous Small and Medium-Grained Scented Rices (Oryza sativa L.) of Orissa. Indian J. Genet. Plant Breed. 2008, 68, 360–365. [Google Scholar]
- Minh, H.K.Q.; Rakshit, S.K. Use of Specific PCR-Based Molecular Markers for Discrimination, Rapid Analysis of Purity and Identification of Six Fragrant Rice Varieties. Int. J. Food Sci. Technol. 2009, 44, 1959–1965. [Google Scholar] [CrossRef]
- Rahman, M.S.; Molla, M.R.; Alam, M.S.; Rahman, L. DNA Fingerprinting of Rice (Oryza sativa L.) Cultivars Using Microsatellite Markers. Aust. J. Crop Sci. 2009, 3, 122–128. [Google Scholar]
- Jena, R.C.; Samal, K.C.; Narela, S.K.; Priyadersini, A. DNA Fingerprinting of Promising Rice (Oryza sativa L) Accessions from India Using RAPD Markers. Int. J. Integr. Biol. 2010, 10, 142–146. [Google Scholar]
- Sarao, N.K.; Vikal, Y.; Singh, K.; Joshi, M.A.; Sharma, R.C. SSR Marker-Based DNA Fingerprinting and Cultivar Identification of Rice (Oryza sativa L.) in Punjab State of India. Plant Genet. Resour. Characterisation Util. 2010, 8, 42–44. [Google Scholar] [CrossRef]
- Wankhade, S.D.; Cornejo, M.J.; Mateu-Andres, I. Microsatellite Marker-Based Genetic Variability in Spanish Rice Cultivars and Landraces. Span. J. Agric. Res. 2010, 8, 995–1004. [Google Scholar] [CrossRef]
- Anand, D.; Prabhu, K.; Singh, A. Analysis of Molecular Diversity and Fingerprinting of Commercially Grown Indian Rice Hybrids. J. Plant Biochem. Biotechnol. 2011, 21, 173–179. [Google Scholar] [CrossRef]
- Bezugliy, M.D.; Sivolap, Y.M.; Galaev, A.V.; Dudchenko, V.V.; Vozhegova, R.A. DNA-Identification of Rice Varieties (Oryza sativa L.) of Ukrainian Breeding. Cytol. Genet. 2011, 45, 27–32. [Google Scholar] [CrossRef]
- Kaushik, A.; Jain, S.; McCouch, S.R.; Jain, R.K. Phylogenetic Relationships among Various Groups of Rice (Oryza sativa L.) as Revealed by Microsatellite and Transposable Element-Based Marker Analysis. Indian J. Genet. Plant Breed. 2011, 71, 139–150. [Google Scholar]
- Chuang, H.-Y.; Lur, H.-S.; Hwu, K.-K.; Chang, M.-C. Authentication of Domestic Taiwan Rice Varieties Based on Fingerprinting Analysis of Microsatellite DNA Markers. Bot. Stud. 2011, 52, 393–405. [Google Scholar]
- Rahman, M.M.; Rasaul, M.G.; Hossain, M.A.; Iftekharuddaula, K.M.; Hasegawa, H. Molecular Characterization and Genetic Diversity Analysis of Rice (Oryza sativa L.) Using SSR Markers. J. Crop Improv. 2012, 26, 244–257. [Google Scholar] [CrossRef]
- Ashfaq, M.; Khan, A.S. Genetic Diversity in Basmati Rice (Oryza sativa L.) Germplasm as Revealed by Microsatellite (SSR) Markers. Russ. J. Genet. 2012, 48, 53–62. [Google Scholar] [CrossRef]
- Yamaki, S.; Ohyanagi, H.; Yamasaki, M.; Eiguchi, M.; Miyabayashi, T.; Kubo, T.; Kurata, N.; Nonomura, K.-I. Development of INDEL Markers to Discriminate All Genome Types Rapidly in the Genus. Oryza. Breed. Sci. 2013, 63, 246–254. [Google Scholar] [CrossRef]
- Shah, S.M.; Naveed, S.A.; Arif, M. Genetic Diversity in Basmati and Non-Basmati Rice Varieties Based on Microsatellite Markers. Pak. J. Bot. 2013, 45, 423–431. [Google Scholar]
- Yesmin, N.; Elias, S.M.; Rahman, M.S.; Haque, T.; Mahbub Hasan, A.K.M.; Seraj, Z.I. Unique Genotypic Differences Discovered among Indigenous Bangladeshi Rice Landraces. Int. J. Genom. 2014, 2014, 210328. [Google Scholar] [CrossRef]
- Al-Turki, T.A.; Basahi, M.A. Assessment of ISSR Based Molecular Genetic Diversity of Hassawi Rice in Saudi Arabia. Saudi J. Biol. Sci. 2015, 22, 591–599. [Google Scholar] [CrossRef]
- Ashu, S.; Sengar, R.S. DNA Fingerprinting Based Identification and Classification of Indica Rice (Oryza sativa L) to Assess Genetic Diversity via ISSR and RAPD Markers. Res. J. Biotechnol. 2015, 10, 90–99. [Google Scholar]
- Singh, N.; Roy Choudhury, D.; Tiwari, G.; Singh, A.; Kumar, S.; Srinivasan, K.; Tyagi, R.; Sharma, A.D.; Singh, N.; Singh, R. Genetic Diversity Trend in Indian Rice Varieties: An Analysis Using SSR Markers. BMC Genet. 2016, 17, 127. [Google Scholar] [CrossRef]
- Bora, A.; Choudhury, P.R.; Pande, V.; Mandal, A.B. RAPD-Holds Promise to Identify Different Genotypes of Rice for Use in Breeding Programs of Diverse Genetic Stocks of Rice (Oryza sativa L.) Based on Genetic Diversity. Vegetos 2016, 29, 69–77. [Google Scholar] [CrossRef]
- Yadav, M.K.; Ngangkham, U.; Shubudhi, H.N.; Bag, M.K.; Adak, T.; Munda, S.; Samantaray, S.; Jena, M. Use of Molecular Markers in Identification and Characterization of Resistance to Rice Blast in India. PLoS ONE 2017, 12, e0176236. [Google Scholar]
- Inam, S.; Khan, M.R.; Rehman, N.; Abbas, Z.; Noor, S. Molecular Detection and Quantification of Non-Basmati Adulterants in Basmati Rice Using BADH2 Gene Marker. Int. J. Agric. Biol. 2017, 19, 1463–1468. [Google Scholar]
- Cheon, K.-S.; Baek, J.; Cho, Y.-I.; Jeong, Y.-M.; Lee, Y.-Y.; Oh, J.; Won, Y.J.; Kang, D.-Y.; Oh, H.; Kim, S.L.; et al. Single Nucleotide Polymorphism (SNP) Discovery and Kompetitive Allele-Specific PCR (KASP) Marker Development with Korean Japonica Rice Varieties. Plant Breed. Biotechnol. 2018, 6, 391–403. [Google Scholar] [CrossRef]
- Markkandan, K.; Yoo, S.-I.; Cho, Y.-C.; Lee, D.W. Genome-Wide Identification of Insertion and Deletion Markers in Chinese Commercial Rice Cultivars, Based on next-Generation Sequencing Data. Agronomy 2018, 8, 36. [Google Scholar] [CrossRef]
- Xiujie, Z.; Wujun, J.; Wentao, X.; Xiaying, L.; Ying, S.; Sha, L.; Hongsheng, O. Comparison of Five Endogenous Reference Genes for Specific PCR Detection and Quantification of Rice. Rice Sci. 2019, 26, 248–256. [Google Scholar] [CrossRef]
- Beşer, N.; Mutafçilar, Z.Ç. Identification of SSR Markers for Differentiating Rice (Oryza sativa L.) Varieties Marketed in Turkey. Tarım Bilim. Derg. 2020, 26, 357–362. [Google Scholar] [CrossRef]
- Kumar, S.P.J.; Susmita, C.; Agarwal, D.K.; Pal, G.; Rai, A.K.; Simal-Gandara, J. Assessment of Genetic Purity in Rice Using Polymorphic SSR Markers and Its Economic Analysis with Grow-Out-Test. Food Anal. Methods 2021, 14, 856–864. [Google Scholar] [CrossRef]
- Bhargavi, M.; Maneesha, K.; Withanawasam, D.M.; Aratikatla, K.R.1; Himabindu, S.; Prashanth, M.; Shanthi, P.; Kommana, M.L.; Reddy, D.M.; Reddy, B.R.; et al. A Novel Barcode System for Rapid Identification of Rice (Oryza sativa L.) Varieties Using Agro-Morphological Descriptors and Molecular Markers. Mol. Biol. Rep. 2021, 48, 2209–2221. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Vieira, M.B.; Faustino, M.V.; Lourenço, T.F.; Oliveira, M.M. DNA-Based Tools to Certify Authenticity of Rice Varieties—An Overview. Foods 2022, 11, 258. https://doi.org/10.3390/foods11030258
Vieira MB, Faustino MV, Lourenço TF, Oliveira MM. DNA-Based Tools to Certify Authenticity of Rice Varieties—An Overview. Foods. 2022; 11(3):258. https://doi.org/10.3390/foods11030258
Chicago/Turabian StyleVieira, Maria Beatriz, Maria V. Faustino, Tiago F. Lourenço, and M. Margarida Oliveira. 2022. "DNA-Based Tools to Certify Authenticity of Rice Varieties—An Overview" Foods 11, no. 3: 258. https://doi.org/10.3390/foods11030258
APA StyleVieira, M. B., Faustino, M. V., Lourenço, T. F., & Oliveira, M. M. (2022). DNA-Based Tools to Certify Authenticity of Rice Varieties—An Overview. Foods, 11(3), 258. https://doi.org/10.3390/foods11030258









