Isolation and Identification of Lactic Acid Bacteria from Natural Whey Cultures of Buffalo and Cow Milk
Abstract
:1. Introduction
2. Materials and Methods
2.1. Natural Whey Starter Cultures Sampling
2.2. Lactic Acid Bacteria Enumeration and Isolation
2.3. Genotypic Identification by Partial 16S rDNA Gene Sequence Analysis
2.4. Species- and Subspecies-Specific PCR
2.5. Random Amplified Polymorphic DNA-Polymerase Chain Reaction (RAPD-PCR) Analysis
2.6. High-Throughput Sequencing
3. Results
3.1. Enumeration and Isolation of Lactic Acid Bacteria from Natural Whey Starter Cultures
3.2. Taxonomic Identification of Species Isolated from Natural Whey Starters Cultures
3.2.1. Sequencing of V1–V3 rDNA Region
3.2.2. Species- and Subspecies-Specific PCR
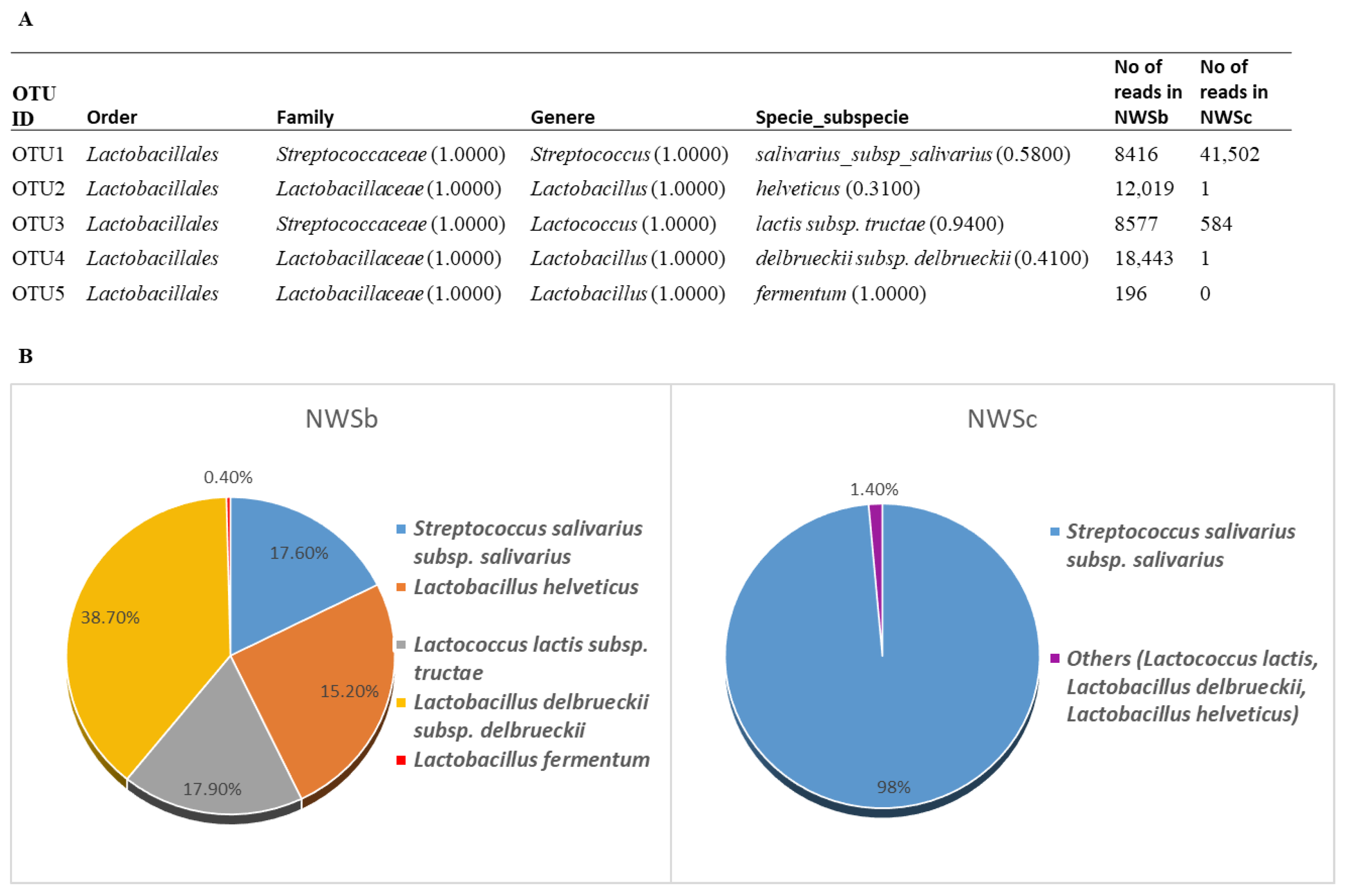
3.2.3. 16S rDNA Next-Generation Sequencing Analysis (NGS)
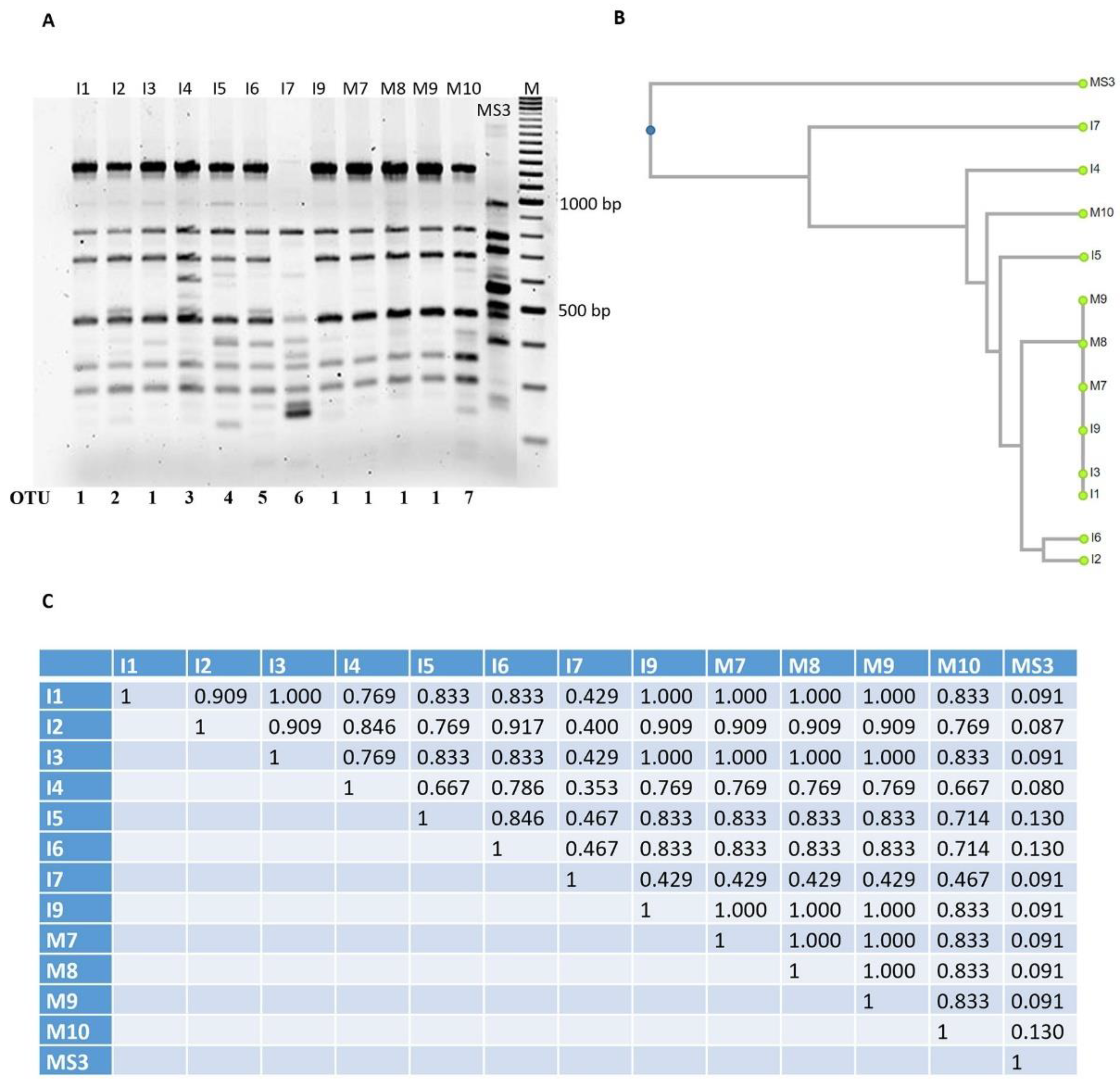
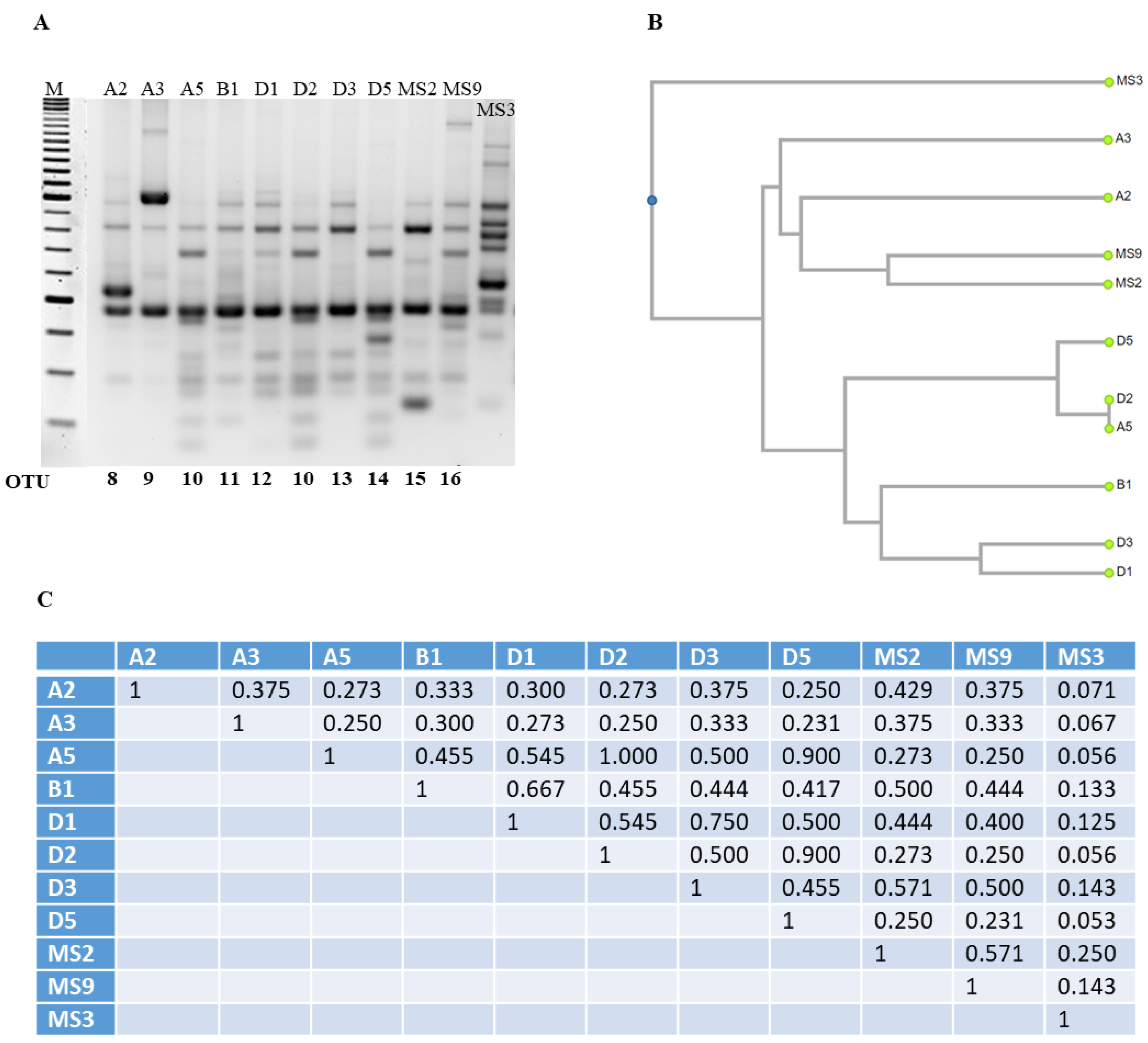
3.3. Optimization and Analysis of RAPD-PCR Profiles
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Fox, P.F.; Guinee, T.P.; Cogan, T.M.; McSweeney, P.L. Principal families of cheese. In Fundamentals of Cheese Science; Chapter 3; Springer: New York, NY, USA, 2016; pp. 27–69. [Google Scholar]
- Licitra, G.; Carpino, S. The microfloras and sensory profiles of selected protected designation of origin Italian cheeses. Microbiol. Spectr. 2012, 2, CM-0007-2012. [Google Scholar]
- Blaya, J.; Barzideh, Z.; Lapointe, G. Interaction of starter cultures and nonstarter lactic acid bacteria in the cheese environment. J. Dairy Sci. 2018, 101, 3611–3629. [Google Scholar] [CrossRef]
- Choi, J.; In Lee, S.; Rackerby, B.; Frojen, R.; Goddik, L.; Ha, S.D.; Park, S.H. Assessment of overall microbial community shift during cheddar cheese production from raw milk to aging. Appl. Microbiol. Biotechnol. 2020, 104, 6249–6260. [Google Scholar] [CrossRef] [PubMed]
- Jana, A.H.; Tagalpallewar, G.P. Functional properties of Mozzarella cheese for its end use application. J. Food Sci. Technol. 2017, 54, 3766–3778. [Google Scholar]
- Kindstedt, P.S. Symposium review: The mozzarella/pasta filata years: A tribute to David M. Barbano. J. Dairy Sci. 2019, 102, 10670–10676. [Google Scholar] [CrossRef] [PubMed]
- Mauriello, G.; Moio, L.; Genovese, A.; Ercolini, D. Relationships between flavouring capabilities, bacterial composition, and geographical origin of natural whey cultures used for traditional water-buffalo mozzarella cheese manufacture. J. Dairy Sci. 2003, 86, 486–497. [Google Scholar] [CrossRef]
- Gatti, M.; Bottari, B.; Lazzi, C.; Neviani, E.; Mucchetti, G. Microbial evolution in raw-milk, long ripened cheeses produced using undefined natural whey starters. J. Dairy Sci. 2014, 97, 573–591. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Losito, F.; Arienzo, A.; Bottini, G.; Priolisi, F.R.; Mari, A.; Antonini, G. Microbiological safety and quality of Mozzarella cheese assessed by the microbiological survey method. J. Dairy Sci. 2014, 97, 46–55. [Google Scholar] [CrossRef] [Green Version]
- Silva, L.F.; Casella, T.; Gomes-Pepe, E.S.; Nogueira, M.C.L.; Lindner, J.D.D.; Penna, A.L.B. Diversity of lactic acid bacteria isolated from Brazilian water buffalo mozzarella cheese. J. Food Sci. 2015, 80, M411–M417. [Google Scholar] [CrossRef] [PubMed]
- Ercolini, D.; Mauriello, G.; Blaiotta, G.; Moschetti, G.; Coppola, S. PCR-DGGE fingerprints of microbial succession during a manufacture of traditional water buffalo mozzarella cheese. J. Appl. Microbiol. 2004, 96, 263–270. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- de Candia, S.; De Angelis, M.; Dunlea, E.; Minervini, F.; McSweeney, P.L.; Faccia, M.; Gobbetti, M. Molecular identification and typing of natural whey starter cultures and microbiological and compositional proprieties of related traditional Mozzarella cheese. Int. J. Food Microbiol. 2007, 119, 182–191. [Google Scholar] [CrossRef]
- Ercolini, D.; Frisso, G.; Mauriello, G.; Salvatore, F.; Coppola, S. Microbial diversity in natural whey cultures used for the production of Caciocavallo Silano PDO cheese. Int. J. Food Microbiol. 2008, 124, 164–170. [Google Scholar] [CrossRef]
- Succi, M.; Aponte, M.; Tremonte, P.; Niro, S.; Sorrentino, E.; Iorizzo, M.; Tipaldi, L.; Pannella, G.; Panfili, G.; Fratianni, A.; et al. Variability in chemical and microbiological profiles of long-ripened Caciocavallo cheeses. J. Dairy Sci. 2016, 99, 9521–9533. [Google Scholar] [CrossRef] [Green Version]
- Pisano, M.B.; Scano, P.; Murgia, A.; Cosentino, S.; Caboni, P. Metabolomics and microbiological profile of Italian mozzarella cheese produced by buffalo and cow milk. Food Chem. 2016, 192, 618–624. [Google Scholar] [CrossRef] [PubMed]
- Marino, M.; Innocente, N.; Maifreni, M.; Mounier, J.; Cobo-Díaz, J.F.; Coton, E.; Carraro, L.; Cardazzo, B. Diversity within Italian cheesemaking brine-associated bacterial communities evidenced by massive parallel 16S rRNA gene tag sequencing. Front. Microbiol. 2017, 8, 2119. [Google Scholar] [CrossRef]
- Ercolini, D.; De Filippis, F.; La Storia, A.; Iacono, M. “Remake” by high-throughput sequencing of the microbiota involved in the production of water buffalo mozzarella cheese. Appl. Environ. Microbiol. 2012, 78, 8142–8145. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guidone, A.; Zotta, T.; Matera, A.; Ricciardi, A.; De Filippis, F.; Ercolini, D.; Parente, E. The microbiota of high-moisture mozzarella cheese produced with different acidification methods. Int. J. Food Microbiol. 2016, 216, 9–17. [Google Scholar] [CrossRef] [PubMed]
- Chen, G.; Chen, C.; Lei, Z. Meta-omics insight in the microbial community profiling and functional characterization of fermented foods. Trend Food Sci. Technol. 2017, 65, 23–31. [Google Scholar] [CrossRef]
- Marino, M.; Dubsky de Wittenau, G.; Saccà, E.; Cattonaro, F.; Spadotto, A.; Innocente, N.; Radovic, S.; Piasentier, E.; Marroni, F. Metagenomic profiles of different types of Italian high-moisture Mozzarella cheese. Food Microbiol. 2019, 79, 123–131. [Google Scholar] [CrossRef]
- Sanders, M.E.; Merenstein, D.J.; Reid, G.; Gibson, G.R.; Rastall, R.A. Probiotics and prebiotics in intestinal health and disease: From biology to the clinic. Nat. Rev. Gastroenterol. Hepatol. 2019, 16, 605–616. [Google Scholar] [CrossRef]
- Dehhaghi, M.; Panahi, H.K.S.; Heng, B.; Guillemin, G.J. The gut microbiota, kynurenine pathway, and immune system interaction in the development of brain cancer. Front. Cell Dev. Biol. 2020, 8, 562812. [Google Scholar] [CrossRef]
- Morais, L.H.; Schreiber, H.L.; Mazmanian, S.K. The gut microbiota-brain axis in behaviour and brain disorders. Nat. Rev. Microbiol. 2021, 19, 241–255. [Google Scholar] [CrossRef] [PubMed]
- Cruciata, M.; Sannino, C.; Ercolini, D.; Scatassa, M.L.; De Filippis, F.; Mancuso, I.; La Storia, A.; Moschetti, G.; Settanni, L. Animal rennets as sources of dairy lactic acid bacteria. Appl. Environ. Microbiol. 2014, 80, 2050–2061. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Altschul, S.F.; Madden, T.L.; Schäffer, A.A.; Zhang, J.; Zhang, Z.; Miller, W.; Lipman, D.J. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997, 25, 3389–3402. [Google Scholar] [CrossRef] [Green Version]
- Cremonesi, P.; Vanoni, L.; Morandi, S.; Silvetti, T.; Castiglioni, B.; Brasca, M. Development of a pentaplex PCR assay for the simultaneous detection of Streptococcus thermophilus, Lactobacillus delbrueckii subsp. bulgaricus, L. delbrueckii subsp. lactis, L. helveticus, L. fermentum in whey starter for Grana Padano cheese. Int. J. Food Microbiol. 2011, 146, 207–211. [Google Scholar] [CrossRef] [PubMed]
- Robert, H.; Gabriel, V.; Fontagné-Faucher, C. Biodiversity of lactic acid bacteria in French wheat sourdough as determined by molecular characterization using species-specific PCR. Int. J. Food Microbiol. 2009, 135, 53–59. [Google Scholar] [CrossRef] [PubMed]
- Klijn, N.; Weerkamp, A.H.; de Vos, W.M. Identification of mesophilic lactic acid bacteria by using polymerase chain reaction-amplified variable regions of 16S rRNA and specific DNA probes. Appl. Environ. Microbiol. 1991, 57, 3390–3393. [Google Scholar] [CrossRef] [Green Version]
- Tilsala-Timisjärvi, A.; Alatossava, T. Development of oligonucleotide primers from the 16S-23S rRNA intergenic sequences for identifying different dairy and probiotic lactic acid bacteria by PCR. Int. J. Food Microbiol. 1997, 35, 49–56. [Google Scholar] [CrossRef]
- Fontana, C.; Cocconcelli, P.S.; Vignolo, G. Monitoring the bacterial population dynamics during fermentation of artisanal Argentinean sausages. Int. J. Food Microbiol. 2005, 103, 131–142. [Google Scholar] [CrossRef]
- Rossetti, L.; Fornasaria, M.E.; Gatti, M.; Lazzi, C.; Neviani, E.; Giraffa, G. Grana Padano cheese whey starters: Microbial composition and strain distribution. Int. J. Food Microbiol. 2008, 127, 168–171. [Google Scholar] [CrossRef]
- Parente, E.; Guidone, A.; Matera, A.; De Filippis, F.; Mauriello, G.; Ricciardi, A. Microbial community dynamics in thermophilic undefined milk starter cultures. Int. J. Food Microbiol. 2016, 217, 59–67. [Google Scholar] [CrossRef] [PubMed]
- Syrokou, M.K.; Themeli, C.; Paramithiotis, S.; Mataragas, M.; Bosnea, L.; Argyri, A.A.; Chorianopoulos, N.G.; Skandamis, P.N.; Drosinos, E.H. Microbial ecology of Greek wheat sourdoughs, identified by a culture-dependent and a culture-independent approach. Foods 2020, 9, 1603. [Google Scholar] [CrossRef] [PubMed]
- Kim, F.; Yang, S.M.; Lim, B.; Park, S.H.; Rackerby, B.; Kim, H.Y. Design of PCR assays to specifically detect and identify 37 Lactobacillus species in a single 96 well plate. BMC Microbiol. 2020, 20, 96. [Google Scholar] [CrossRef] [Green Version]
- Shirali Pandya, K.R.; Srinivas, V.; Jadhav, S.; Khan, A.; Arun, A.; Riley, L.W.; Madhivanan, P. Comparison of culture-dependent and culture-independent molecular methods for characterization of vaginal microflora. J. Med. Microbiol. 2017, 66, 149–153. [Google Scholar] [CrossRef] [PubMed]
- Pombert, J.F.; Sistek, V.; Boissinot, M.; Frenette, M. Evolutionary relationships among salivarius streptococci as inferred from multilocus phylogenies based on 16S rRNA-encoding, recA, secA, and secY gene sequences. BMC Microbiol. 2009, 30, 232. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Margalho, L.P.; Jorge, G.P.; Noleto, D.A.; Silva, C.E.; Abreu, J.S.; Piran, M.V.; Brocchi, M.; Sant’Ana, A.S. Biopreservation and probiotic potential of a large set of lactic acid bacteria isolated from Brazilian artisanal cheeses: From screening to in product approach. Microbiol. Res. 2021, 242, 126622. [Google Scholar] [CrossRef] [PubMed]
- Amarela, T.V.; Veljović, K.; Tolinački, M.; Živković, M.; Lukić, J.; Lozo, J.; Fira, Đ.; Jovčić, B.; Strahinić, I.; Begović, J.; et al. Diversity of non-starter lactic acid bacteria in autochthonous dairy products from Western Balkan Countries—Technological and probiotic properties. Review. Food Res. Int. 2020, 136, 109494. [Google Scholar]
- Linares, D.M.; Gomez, C.; Renes, E.; Fresno, J.M.; Tornadijo, M.E.; Ross, R.P.; Stanton, C. Lactic acid bacteria and Bifidobacteria with potential to design natural biofunctional health-promoting dairy foods. Front. Microbiol. 2017, 8, 846. [Google Scholar] [CrossRef]
- Plaza-Diaz, J.; Ruiz-Ojeda, F.J.; Gil-Campos, M.; Gil, A. Mechanism of action of probiotics. Adv. Nutr. 2019, 10 (Suppl. 1), S49–S66. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Indira, M.; Venkateswarulu, T.C.; Peele, K.A.; Bobby, N.; Krupanidhi, S. Bioactive molecules of probiotic bacteria and their mechanism of action: A review. Biotech 2019, 9, 306. [Google Scholar] [CrossRef] [PubMed]
- Basavaprabhu, H.N.; Syed, A.A.; Pradip, V.B.; Hariom, Y. Postbiotics-parabiotics: The new horizons in microbial biotherapy and functional foods. Microb. Cell Fact. 2020, 19, 168. [Google Scholar]
- Bogdan, P.B.; Georgescu, L.A.; Vasile, M.A.; Rocha, J.M.; Bahrim, G.E. Selection of wild lactic acid bacteria strains as promoters of postbiotics in gluten-free sourdoughs. Microorganisms 2020, 8, 643. [Google Scholar]
- Giello, M.; La Storia, A.; Masucci, F.; Di Francia, A.; Ercolini, D.; Villani, F. Dynamics of bacterial communities during manufacture and ripening of traditional Caciocavallo of Castelfranco cheese in relation to cows’ feeding. Food Microbiol. 2017, 63, 170–177. [Google Scholar] [CrossRef] [PubMed]
- Zotta, T.; Ricciardi, A.; Condelli, N.; Parente, E. Metataxonomic and metagenomic approaches for the study of undefined strain starters for cheese manufacture. Crit. Rev. Food Sci. Nutr. 2021, 18, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Li, T.T.; Tian, W.L.; Gu, C.T. Elevation of Lactococcus lactis subsp. cremoris to the species level as Lactococcus cremoris sp. nov. and transfer of Lactococcus lactis subsp. tructae to Lactococcus cremoris as Lactococcus cremoris subsp. tructae comb. nov. Int. J. Syst. Evol. Microbiol. 2021, 71, 004727. [Google Scholar] [CrossRef]
- Russo, R.; Valletta, M.; Rega, C.; Marasco, R.; Muscariello, L.; Pedone, P.V.; Sacco, M.; Chambery, A. Reliable identification of lactic acid bacteria by targeted and untargeted high-resolution tandem mass spectrometry. Food Chem. 2019, 285, 111–118. [Google Scholar] [CrossRef] [PubMed]

| Genus/Species/ Subspecies | Primer | Sequence | Target Gene | Amplicon Size (bp) | Ref. |
|---|---|---|---|---|---|
| S. thermophilus | Str-THER-F2116 Str-THER-R2693 | GCTTGGTTCTGAGGGAAGC CTTTCTTCTGCACCGTATCCA | lacZ | 578 | [26] |
| L. lactis subsp. lactis | Lactis F 23SRevLac | GCTGAAGGTTGGTACTTGTA AGTGCCAAGGCATCCACC | 16S rRNA (V1) & 16S/23S spacer region | 1824 | [27] |
| L. lactis subsp. cremoris | PLc(A)For 23SRevLac | GGTGCTTGCACCAATTTGAA AGTGCCAAGGCATCCACC | 16S rRNA (V1) & 16S/23S spacer region | 1814 | [28] |
| L. fermentum | Lac-FER-F753 Lac-FER-R1062 | CCAGATCAGCCAACTTCACA GGCAAACTTCAAGAGGACCA | Arginine-ornitine antiporter | 310 | [26] |
| L. delbrueckii | Del I Del II | ACGGATGGATGGAGAGCAG GCAAGTTTGTTCTTTCGAACTC | 16S-23S rRNA spacer region | 200 | [29] |
| E. faecium | EfaeciumF EfaeciumR | GCAAGGCTTCTTAGAGA CATCGTGTAAGCTAACTTC | D-Ala Ala ligase | 575 | [27] |
| Genus/Species/Subspecies | Melting | Annealing | Elongation | Cycles Number |
|---|---|---|---|---|
| S. thermophilus | 94 °C, 1 min | 58 °C, 1 min | 72 °C, 1 min | 30 |
| Lc. lactis subsp. lactis | 94 °C, 1 min | 58 °C, 1 min | 72 °C, 2 min | 30 |
| L. lactis subsp. cremoris | 94 °C, 1 min | 60 °C, 1 min | 72 °C, 2 min | 30 |
| L. fermentum | 94 °C, 1 min | 58 °C, 1 min | 72 °C, 1 min | 30 |
| L. delbrueckii | 95 °C, 30 s | 55 °C, 30 s | 72 °C, 30 s | 30 |
| E. faecium | 94 °C, 1 min | 54 °C, 1 min | 72 °C, 1 min | 30 |
| NWS Sample | Mesophilic Lactobacilli | Thermophilic Lactobacilli | Mesophilic Streptococci and Lactococci | Thermophilic Streptococci and Lactococci | Enterococci |
|---|---|---|---|---|---|
| Cow | 5.6 ± 0.1 | 4.7 ± 0.1 | 6.6 ± 0.2 | 6.5 ± 0.1 | 6.5 ± 0.1 |
| Buffalo | 7.0 ± 0.2 | 8.5 ± 0.3 | 8.1 ± 0.1 | 7.5 ± 0.1 | 8.0 ± 0.2 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Marasco, R.; Gazzillo, M.; Campolattano, N.; Sacco, M.; Muscariello, L. Isolation and Identification of Lactic Acid Bacteria from Natural Whey Cultures of Buffalo and Cow Milk. Foods 2022, 11, 233. https://doi.org/10.3390/foods11020233
Marasco R, Gazzillo M, Campolattano N, Sacco M, Muscariello L. Isolation and Identification of Lactic Acid Bacteria from Natural Whey Cultures of Buffalo and Cow Milk. Foods. 2022; 11(2):233. https://doi.org/10.3390/foods11020233
Chicago/Turabian StyleMarasco, Rosangela, Mariagiovanna Gazzillo, Nicoletta Campolattano, Margherita Sacco, and Lidia Muscariello. 2022. "Isolation and Identification of Lactic Acid Bacteria from Natural Whey Cultures of Buffalo and Cow Milk" Foods 11, no. 2: 233. https://doi.org/10.3390/foods11020233
APA StyleMarasco, R., Gazzillo, M., Campolattano, N., Sacco, M., & Muscariello, L. (2022). Isolation and Identification of Lactic Acid Bacteria from Natural Whey Cultures of Buffalo and Cow Milk. Foods, 11(2), 233. https://doi.org/10.3390/foods11020233






