An Integrative Glycomic Approach for Quantitative Meat Species Profiling
Abstract
:1. Introduction
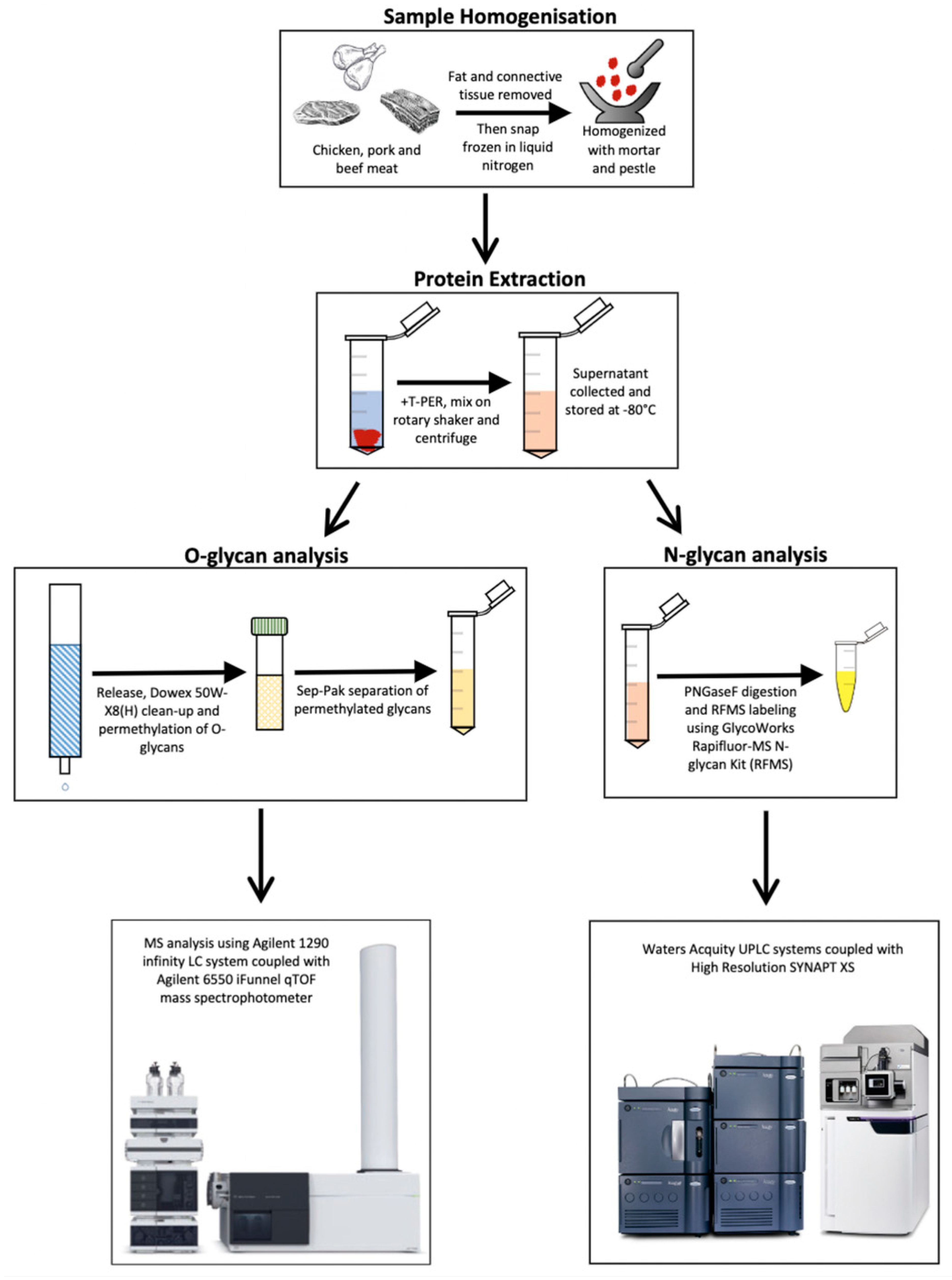
2. Materials and Methods
2.1. Meat Lysis and Protein Extraction
2.2. Release and Permethylation of O-Glycans
2.3. Sep-Pak Separation of Permethylated Glycans
2.4. Mass Spectrometry Analysis of O-Glycans
2.5. O-Glycan Assignment
2.6. N-Glycan Release and Labelling
2.7. Liquid Chromatography-Mass Spectrometry Analysis of RFMS-Labelled N-Glycan
2.8. N-Glycan Assignment
2.9. Statistical Analyses
3. Results
3.1. Framework for an Integrated Glycomic Study of Meat Samples from Different Species
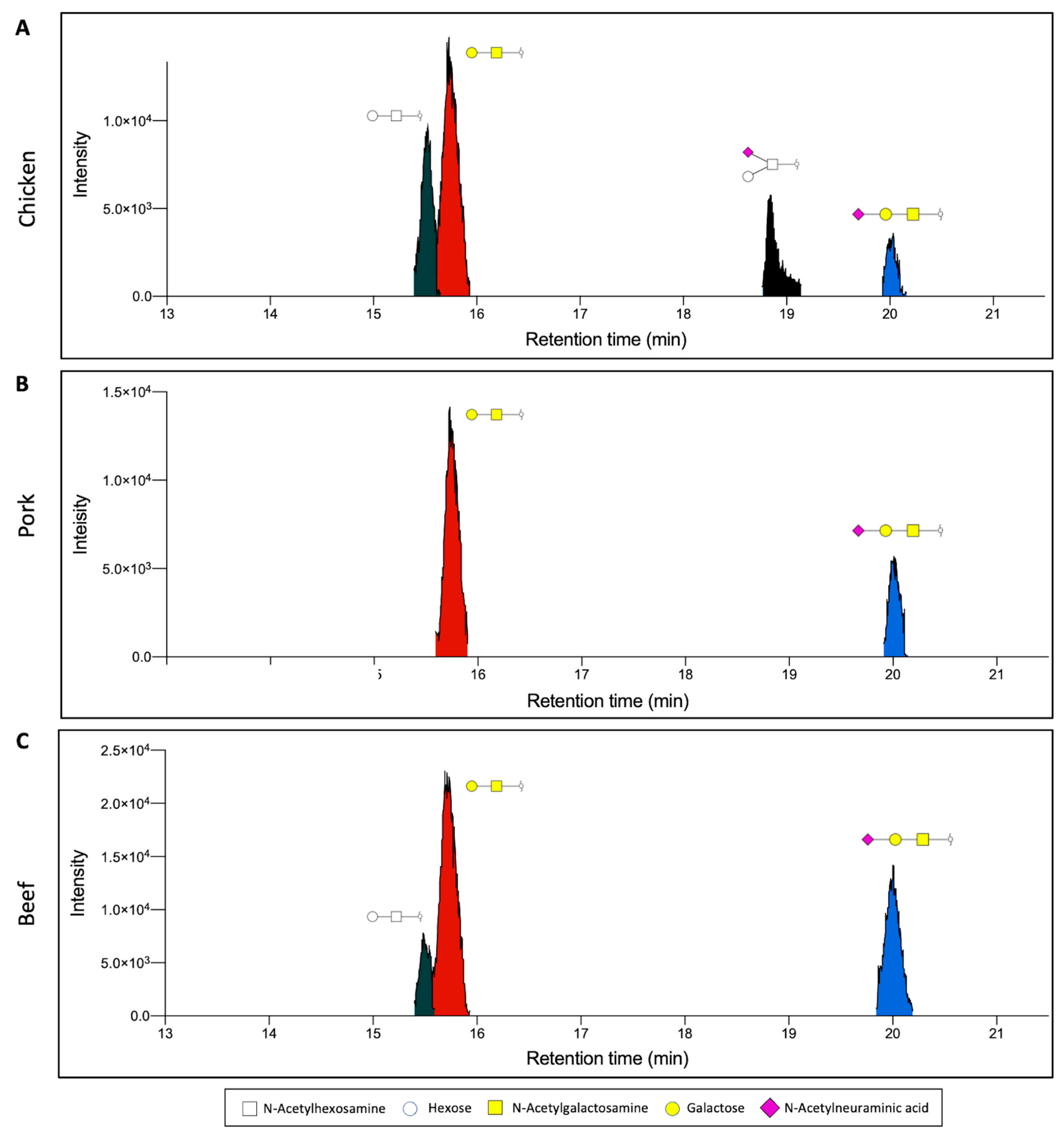
3.2. O-Glycan Characterisation of Meat
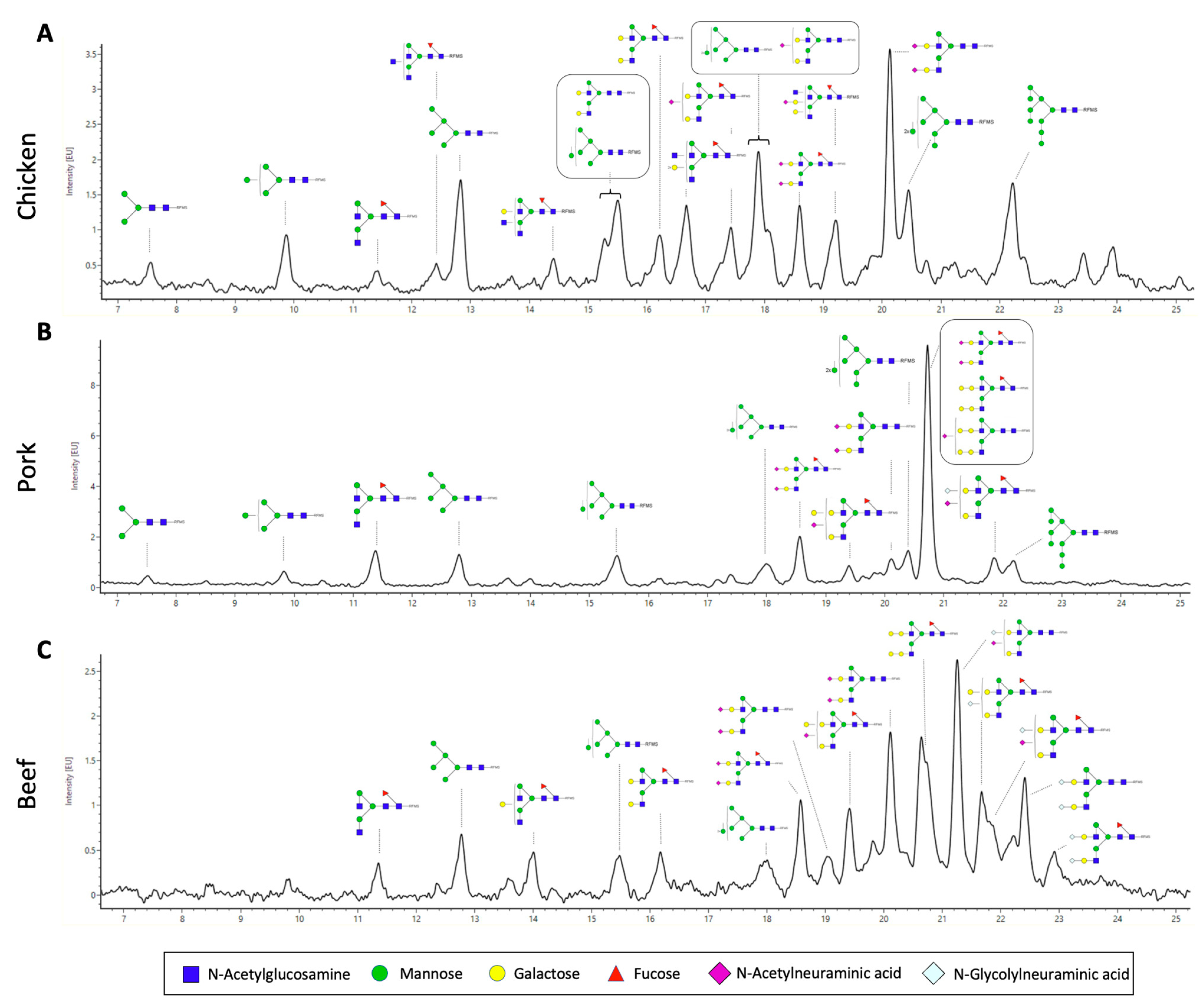
3.3. N-Glycan Characterisation of Meat
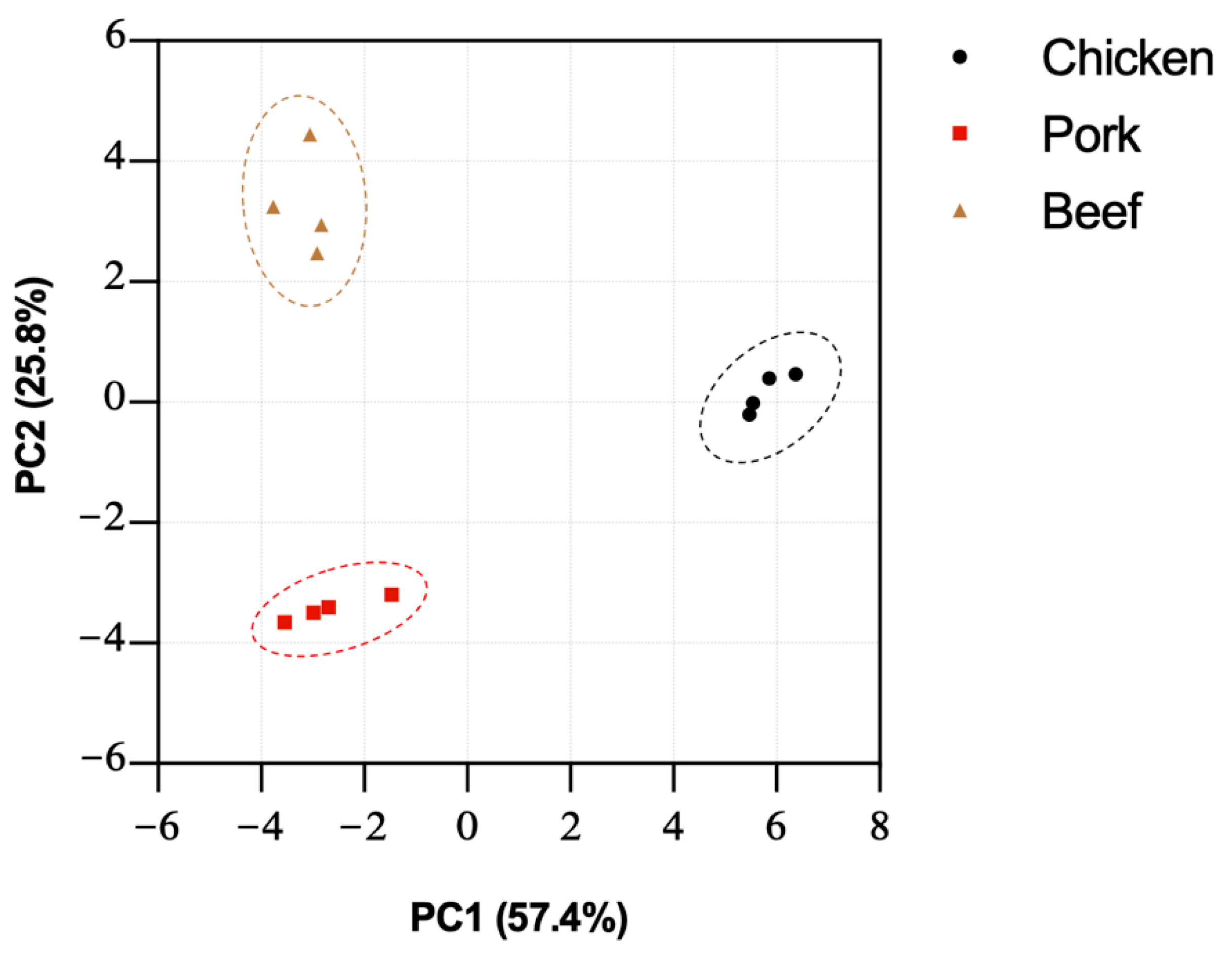
3.4. PCA Analysis
4. Discussion
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Black, C.; Chevallier, O.P.; Elliott, C.T. The current and potential applications of Ambient Mass Spectrometry in detecting food fraud. TrAC Trends Anal. Chem. 2016, 82, 268–278. [Google Scholar] [CrossRef] [Green Version]
- Sajali, N.; Wong, S.C.; Abu Bakar, S.; Khairil Mokhtar, N.F.; Manaf, Y.N.; Yuswan, M.H.; Mohd Desa, M.N. Analytical approaches of meat authentication in food. Int. J. Food Sci. Technol. 2021, 56, 1535–1543. [Google Scholar] [CrossRef]
- Ballin, N.Z.; Vogensen, F.K.; Karlsson, A.H. Species determination—Can we detect and quantify meat adulteration? Meat Sci. 2009, 83, 165–174. [Google Scholar] [CrossRef] [PubMed]
- Bansal, S.; Singh, A.; Mangal, M.; Mangal, A.K.; Kumar, S. Food adulteration: Sources, health risks, and detection methods. Crit. Rev. Food Sci. Nutr. 2017, 57, 1174–1189. [Google Scholar] [CrossRef]
- Hong, E.; Lee, S.Y.; Jeong, J.Y.; Park, J.M.; Kim, B.H.; Kwon, K.; Chun, H.S. Modern analytical methods for the detection of food fraud and adulteration by food category. J. Sci. Food Agric. 2017, 97, 3877–3896. [Google Scholar] [CrossRef]
- Huck, C.W.; Pezzei, C.K.; Huck-Pezzei, V.A. An industry perspective of food fraud. Curr. Opin. Food Sci. 2016, 10, 32–37. [Google Scholar] [CrossRef]
- Doosti, A.; Ghasemi Dehkordi, P.; Rahimi, E. Molecular assay to fraud identification of meat products. J. Food Sci. Technol. 2014, 51, 148–152. [Google Scholar] [CrossRef] [Green Version]
- Dirong, G.; Nematbakhsh, S.; Selamat, J.; Chong, P.P.; Idris, L.H.; Nordin, N.; Fatchiyah, F.; Razis, A.F.A. Omics-based analytical approaches for assessing chicken species and breeds in food authentication. Molecules 2021, 26, 6502. [Google Scholar] [CrossRef]
- Böhme, K.; Calo-Mata, P.; Barros-Velázquez, J.; Ortea, I. Recent applications of omics-based technologies to main topics in food authentication. TrAC Trends Anal. Chem. 2019, 110, 221–232. [Google Scholar] [CrossRef]
- Capozzi, F.; Bordoni, A. Foodomics: A new comprehensive approach to food and nutrition. Genes Nutr. 2013, 8, 1–4. [Google Scholar] [CrossRef] [Green Version]
- Barabási, A.L.; Menichetti, G.; Loscalzo, J. The unmapped chemical complexity of our diet. Nat. Food 2020, 1, 33–37. [Google Scholar] [CrossRef] [Green Version]
- Walsh, I.; Zhao, S.; Campbell, M.; Taron, C.H.; Rudd, P.M. Quantitative profiling of glycans and glycopeptides: An informatics’ perspective. Curr. Opin. Struct. Biol. 2016, 40, 70–80. [Google Scholar] [CrossRef] [PubMed]
- Shi, Z.; Yin, B.; Li, Y.; Zhou, G.; Li, C.; Xu, X.; Luo, X.; Zhang, X.; Qi, J.; Voglmeir, J.; et al. N-Glycan Profile as a Tool in Qualitative and Quantitative Analysis of Meat Adulteration. J. Agric. Food Chem. 2019, 67, 10543–10551. [Google Scholar] [CrossRef] [PubMed]
- Lauber, M.A.; Yu, Y.Q.; Brousmiche, D.W.; Hua, Z.; Koza, S.M.; Magnelli, P.; Guthrie, E.; Taron, C.H.; Fountain, K.J. Rapid preparation of released N -glycans for HILIC analysis using a labeling reagent that facilitates sensitive fluorescence and ESI-MS detection. Anal. Chem. 2015, 87, 5401–5409. [Google Scholar] [CrossRef]
- Pallister, E.G.; Choo, M.S.F.; Walsh, I.; Tai, J.N.; Tay, S.J.; Yang, Y.S.; Ng, S.K.; Rudd, P.M.; Flitsch, S.L.; Nguyen-Khuong, T. Utility of Ion-Mobility Spectrometry for Deducing Branching of Multiply Charged Glycans and Glycopeptides in a High-Throughput Positive ion LC-FLR-IMS-MS Workflow. Anal. Chem. 2020, 92, 15323–15335. [Google Scholar] [CrossRef]
- Goso, Y.; Sugaya, T.; Ishihara, K.; Kurihara, M. Comparison of Methods to Release Mucin-Type O-Glycans for Glycomic Analysis. Anal. Chem. 2017, 89, 8870–8876. [Google Scholar] [CrossRef]
- Reinhold, V.; Zhang, H.; Hanneman, A.; Ashline, D. Toward a platform for comprehensive glycan sequencing. Mol. Cell. Proteomics 2013, 12, 866–873. [Google Scholar] [CrossRef] [Green Version]
- Yusufi, F.N.K.; Lakshmanan, M.; Ho, Y.S.; Loo, B.L.W.; Ariyaratne, P.; Yang, Y.; Ng, S.K.; Tan, T.R.M.; Yeo, H.C.; Lim, H.L.; et al. Mammalian Systems Biotechnology Reveals Global Cellular Adaptations in a Recombinant CHO Cell Line. Cell Syst. 2017, 4, 530–542.e6. [Google Scholar] [CrossRef] [Green Version]
- Li, Y.C.; Liu, S.Y.; Meng, F.B.; Liu, D.Y.; Zhang, Y.; Wang, W.; Zhang, J.M. Comparative review and the recent progress in detection technologies of meat product adulteration. Compr. Rev. Food Sci. Food Saf. 2020, 19, 2256–2296. [Google Scholar] [CrossRef]
- Royle, L.; Mattu, T.S.; Hart, E.; Langridge, J.I.; Merry, A.H.; Murphy, N.; Harvey, D.J.; Dwek, R.A.; Rudd, P.M. An analytical and structural database provides a strategy for sequencing O-glycans from microgram quantities of glycoproteins. Anal. Biochem. 2002, 304, 70–90. [Google Scholar] [CrossRef]
- Zhao, S.; Walsh, I.; Abrahams, J.L.; Royle, L.; Nguyen-Khuong, T.; Spencer, D.; Fernandes, D.L.; Packer, N.H.; Rudd, P.M.; Campbell, M.P. GlycoStore: A database of retention properties for glycan analysis. Bioinformatics 2018, 34, 3231–3232. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pang, K.T.; Tay, S.J.; Wan, C.; Walsh, I.; Choo, M.S.F.; Yang, Y.S.; Choo, A.; Ho, Y.S.; Nguyen-Khuong, T. Semi-Automated Glycoproteomic Data Analysis of LC-MS Data Using GlycopeptideGraphMS in Process Development of Monoclonal Antibody Biologics. Front. Chem. 2021, 9, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Corfield, A.P.; Berry, M. Glycan variation and evolution in the eukaryotes. Trends Biochem. Sci. 2015, 40, 351–359. [Google Scholar] [CrossRef] [PubMed]
- Watson, M.E.; Diepeveen, L.A.; Stubbs, K.A.; Yeoh, G.C. Glycosylation-related diagnostic and therapeutic drug target markers in hepatocellular carcinoma. J. Gastrointest. Liver Dis. 2015, 24, 349–357. [Google Scholar] [CrossRef]
- Saldova, R.; Wilkinson, H. Current methods for the characterization of o-glycans. J. Proteome Res. 2020, 19, 3890–3905. [Google Scholar] [CrossRef]
- Li, Y.; Chen, X. Sialic acid metabolism and sialyltransferases: Natural functions and applications. Appl. Microbiol. Biotechnol. 2012, 94, 887–905. [Google Scholar] [CrossRef] [Green Version]
- Samraj, A.N.; Pearce, O.M.T.; Läubli, H.; Crittenden, A.N.; Bergfeld, A.K.; Band, K.; Gregg, C.J.; Bingman, A.E.; Secrest, P.; Diaz, S.L.; et al. A red meat-derived glycan promotes inflammation and cancer progression. Proc. Natl. Acad. Sci. USA 2015, 112, 542–547. [Google Scholar] [CrossRef] [Green Version]
- Commins, S.P.; Satinover, S.M.; Hosen, J.; Mozena, J.; Borish, L.; Lewis, B.D.; Woodfolk, J.A.; Platts-Mills, T.A.E. Delayed anaphylaxis, angioedema, or urticaria after consumption of red meat in patients with IgE antibodies specific for galactose-α-1,3-galactose. J. Allergy Clin. Immunol. 2009, 123, 426–433.e2. [Google Scholar] [CrossRef] [Green Version]
- Wilson, J.M.; Schuyler, A.J.; Workman, L.; Gupta, M.; James, H.R.; Posthumus, J.; McGowan, E.C.; Commins, S.P.; Platts-Mills, T.A.E. Investigation into the α-Gal Syndrome: Characteristics of 261 Children and Adults Reporting Red Meat Allergy. J. Allergy Clin. Immunol. Pract. 2019, 7, 2348–2358.e4. [Google Scholar] [CrossRef]
- Reis, C.A.; Osorio, H.; Silva, L.; Gomes, C.; David, L. Alterations in glycosylation as biomarkers for cancer detection. J. Clin. Pathol. 2010, 63, 322–329. [Google Scholar] [CrossRef]
- Rudman, N.; Gornik, O.; Lauc, G. Altered N-glycosylation profiles as potential biomarkers and drug targets in diabetes. FEBS Lett. 2019, 593, 1598–1615. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Seeling, M.; Brückner, C.; Nimmerjahn, F. Differential antibody glycosylation in autoimmunity: Sweet biomarker or modulator of disease activity? Nat. Rev. Rheumatol. 2017, 13, 621–630. [Google Scholar] [CrossRef] [PubMed]
- Van Leeuwen, S.S.; Schoemaker, R.J.W.; Timmer, C.J.A.M.; Kamerling, J.P.; Dijkhuizen, L. N -and O -Glycosylation of a Commercial Bovine Whey Protein Product. J. Agric. Food Chem. 2012, 60, 12553–12564. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Struwe, W.B.; Gough, R.; Gallagher, M.E.; Kenny, D.T.; Carrington, S.D.; Karlsson, N.G.; Rudd, P.M. Identification of O-glycan structures from chicken intestinal mucins provides insight into Campylobactor jejuni pathogenicity. Mol. Cell. Proteom. 2015, 14, 1464–1477. [Google Scholar] [CrossRef] [Green Version]
- Jin, C.; Padra, J.T.; Sundell, K.; Sundh, H.; Karlsson, N.G.; Lindén, S.K. Atlantic Salmon Carries a Range of Novel O-Glycan Structures Differentially Localized on Skin and Intestinal Mucins. J. Proteome Res. 2015, 14, 3239–3251. [Google Scholar] [CrossRef]
- Suzuki, N.; Abe, T.; Natsuka, S. Structural analysis of N-glycans in chicken trachea and lung reveals potential receptors of chicken influenza viruses. Sci. Rep. 2022, 12, 2081. [Google Scholar] [CrossRef]
- Jahan, M.; Thomson, P.C.; Wynn, P.C.; Wang, B. The non-human glycan, N-glycolylneuraminic acid (Neu5Gc), is not expressed in all organs and skeletal muscles of nine animal species. Food Chem. 2021, 343, 128439. [Google Scholar] [CrossRef]
- O’Mahony, P.J. Finding horse meat in beef products-a global problem. QJM 2013, 106, 595–597. [Google Scholar] [CrossRef] [Green Version]
- Post, M.J. Cultured beef: Medical technology to produce food. J. Sci. Food Agric. 2014, 94, 1039–1041. [Google Scholar] [CrossRef]
- Arshad, M.S.; Javed, M.; Sohaib, M.; Saeed, F.; Imran, A.; Amjad, Z. Tissue engineering approaches to develop cultured meat from cells: A mini review. Cogent Food Agric. 2017, 3, 1320814. [Google Scholar] [CrossRef]
- Datar, I.; Betti, M. Possibilities for an in vitro meat production system. Innov. Food Sci. Emerg. Technol. 2010, 11, 13–22. [Google Scholar] [CrossRef]
- Alston-Roberts, C.; Barallon, R.; Bauer, S.R.; Butler, J.; Capes-Davis, A.; Dirks, W.G.; Elmore, E.; Furtado, M.; Kerrigan, L.; Kline, M.C.; et al. Cell line misidentification: The beginning of the end. Nat. Rev. Cancer 2010, 10, 441–448. [Google Scholar] [CrossRef]
- Horbach, S.P.J.M.; Halffman, W. The ghosts of HeLa: How cell line misidentification contaminates the scientific literature. PLoS ONE 2017, 12, e0186281. [Google Scholar] [CrossRef] [PubMed]
- US Food and Drug Administration. FDA Approves First-of-Its-Kind Intentional Genomic Alteration in Line of Domestic Pigs for Both Human Food, Potential Therapeutic Uses. 2020. Available online: https://www.fda.gov/news-events/press-announcements/fda-approves-first-its-kind-intentional-genomic-alteration-line-domestic-pigs-both-human-food (accessed on 23 November 2021).
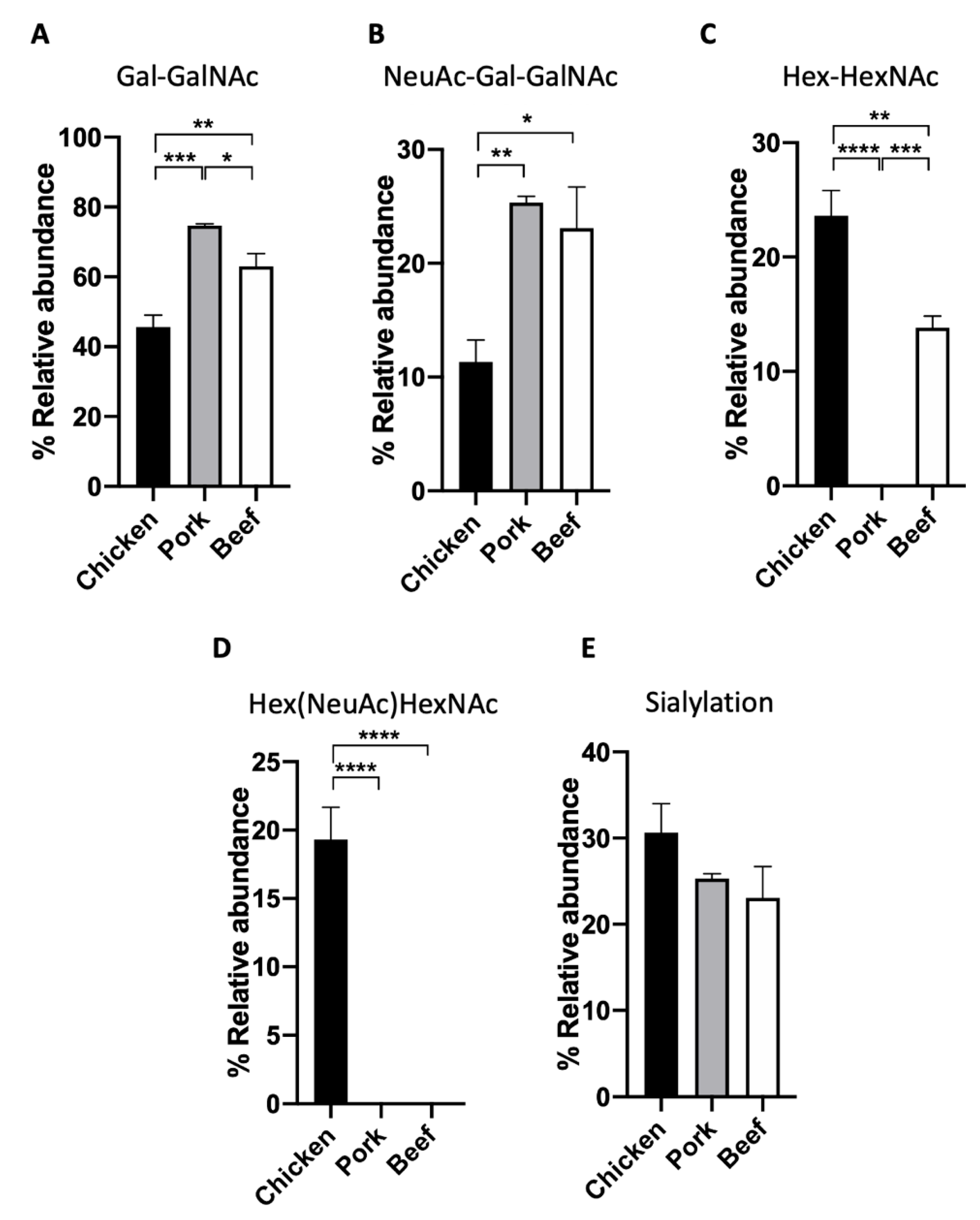

| Chicken (%) | Pork (%) | Beef (%) | |
|---|---|---|---|
| Hex-HexNAc | 23.6 ± 2.2 | ND | 13.8 ± 1.0 |
| Gal-GalNAc | 45.7 ± 3.3 | 74.7 ± 0.6 | 63.1 ± 3.6 |
| Hex(NeuAc)HexNAc | 19.3 ± 2.4 | ND | ND |
| NeuAc-Gal-GalNAc | 11.3 ± 1.9 | 25.3 ± 0.6 | 23.1 ± 3.6 |
| Chicken (%) | Pork (%) | Beef (%) | |
|---|---|---|---|
| HexNAc(2)Hex(3) | 1.0 ± 0.4 | 0.3 ± 0.3 | ND |
| HexNAc(2)Hex(4) | 2.9 ± 0.8 | 0.5 ± 0.5 | 1.0 ± 1.0 |
| HexNAc(4)Hex(3)Fuc(1) | 1.1 ± 0.2 | 6.3 ± 0.2 | 0.8 ± 0.5 |
| HexNAc(2)Hex(5) | 9.3 ± 0.2 | 4.3 ± 0.5 | 5.8 ± 0.3 |
| HexNAc(4)Hex(4)Fuc(1) | ND | 1.2 ± 0.4 | 1.8 ± 1.4 |
| HexNAc(5)Hex(4)Fuc(1) | 1.6 ± 0.1 | ND | ND |
| HexNAc(4)Hex(5) | 2.0 ± 0.2 | 0.3 ± 0.1 | ND |
| HexNAc(2)Hex(6) | 8.1 ± 0.8 | 4.5 ± 1.0 | 4.1 ± 1.4 |
| HexNAc(4)Hex(5)Fuc(1) | 2.2 ± 0.4 | ND | 2.5 ± 0.5 |
| HexNAc(5)Hex(5)Fuc(1) | 5.9 ± 0.5 | ND | ND |
| HexNAc(4)Hex(5)NeuAc(1) | 9.4 ± 0.5 | ND | ND |
| HexNAc(4)Hex(5)Fuc(1)NeuAc(1) | 6.2 ± 0.6 | 0.2 ± 0.2 | ND |
| HexNAc(2)Hex(7) | 6.6 ± 0.6 | 3.9 ± 0.6 | 4.1 ± 0.9 |
| HexNAc(4)Hex(5)Fuc(1)NeuAc(2) | 4.3 ± 0.5 | 60.7 ± 6.3 | 4.6 ± 0.1 |
| HexNAc(5)Hex(5)Fuc(1)NeuAc(1) | 6.0 ± 0.3 | ND | ND |
| HexNAc(4)Hex(6)Fuc(1)NeuAc(1) | ND | 1.2 ± 0.5 | 8.1 ± 0.7 |
| HexNAc(4)Hex(5)NeuAc(2) | 11.9 ± 1.0 | 2.3 ± 0.2 | 9.7 ± 1.0 |
| HexNAc(2)Hex(8) | 10.8 ± 1.1 | 2.2 ± 1.4 | 0.6 ± 0.1 |
| HexNAc(4)Hex(7)Fuc(1) | ND | 3.3 ± 0.3 | 17.8 ± 1.3 |
| HexNAc(4)Hex(6)Fuc(1)NeuGc(1) | ND | ND | 4.2 ± 0.4 |
| HexNAc(4)Hex(7)NeuAc(1) | ND | 1.1 ± 0.1 | 2.3 ± 0.2 |
| HexNAc(4)Hex(5)NeuAc(1)NeuGc(1) | ND | ND | 18.5 ± 1.4 |
| HexNAc(4)Hex(5)Fuc(1)NeuAc(1)NeuGc(1) | ND | 4.9 ± 0.3 | 2.2 ± 1.0 |
| HexNAc(2)Hex(9) | 9.2 ± 0.7 | 2.3 ± 1.4 | ND |
| HexNAc(4)Hex(5)NeuGc(2) | ND | ND | 5.8 ± 0.4 |
| HexNAc(4)Hex(5)Fuc(1)NeuGc(2) | ND | ND | 2.5 ± 1.0 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chia, S.; Teo, G.; Tay, S.J.; Loo, L.S.W.; Wan, C.; Sim, L.C.; Yu, H.; Walsh, I.; Pang, K.T. An Integrative Glycomic Approach for Quantitative Meat Species Profiling. Foods 2022, 11, 1952. https://doi.org/10.3390/foods11131952
Chia S, Teo G, Tay SJ, Loo LSW, Wan C, Sim LC, Yu H, Walsh I, Pang KT. An Integrative Glycomic Approach for Quantitative Meat Species Profiling. Foods. 2022; 11(13):1952. https://doi.org/10.3390/foods11131952
Chicago/Turabian StyleChia, Sean, Gavin Teo, Shi Jie Tay, Larry Sai Weng Loo, Corrine Wan, Lyn Chiin Sim, Hanry Yu, Ian Walsh, and Kuin Tian Pang. 2022. "An Integrative Glycomic Approach for Quantitative Meat Species Profiling" Foods 11, no. 13: 1952. https://doi.org/10.3390/foods11131952
APA StyleChia, S., Teo, G., Tay, S. J., Loo, L. S. W., Wan, C., Sim, L. C., Yu, H., Walsh, I., & Pang, K. T. (2022). An Integrative Glycomic Approach for Quantitative Meat Species Profiling. Foods, 11(13), 1952. https://doi.org/10.3390/foods11131952







