Frozen Vegetable Processing Plants Can Harbour Diverse Listeria monocytogenes Populations: Identification of Critical Operations by WGS
Abstract
:1. Introduction
2. Materials and Methods
2.1. Sampling
2.2. Detection and Isolation of Listeria monocytogenes Strains
2.3. Whole Genome Sequencing
2.3.1. Library Preparation
2.3.2. Quality Assurance and Genome Assembly
2.3.3. Serotyping, MLSTs, Core Genome MLST, Phylogenomic Analysis and MVLST
3. Results
3.1. Identification of Listeria monocytogenes Strains
3.2. Identification of Serotypes, Multi Locus Sequence Typing and Multi-Virulence-Locus Sequence Typing Analyses by Whole Genome Sequences
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Zhu, Q.; Gooneratne, R.; Hussain, M.A. Listeria monocytogenes in fresh produce: Outbreaks, prevalence and contamination levels. Foods 2017, 6, 21. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Walker, S.J.; Archer, P.; Banks, J.G. Growth of Listeria monocytogenes at refrigeration temperatures. J. Appl. Bacteriol. 1990, 68, 157–162. [Google Scholar] [CrossRef] [PubMed]
- EFSA Panel on Biological Hazards (BIOHAZ); Koutsoumanis, K.; Alvarez-Ordóñez, A.; Bolton, D.; Bover-Cid, S.; Chemaly, M.; Davies, R.; De Cesare, A.; Herman, L.; Hilbert, F.; et al. The public health risk posed by Listeria monocytogenes in frozen fruit and vegetables including herbs, blanched during processing. EFSA J. 2020, 18, e06092. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Allende, A.; Barre, L.; Jacxsens, L.; Liebana, E.; Messens, W.; Sarno, E.; da Silva Felicio, M.T. Urgent scientific and technical assistance to provide recommendations for sampling and testing in the processing plants of frozen vegetables aiming at detecting Listeria monocytogenes. EFSA Support. Publ. 2018, 15, 1445E. [Google Scholar] [CrossRef]
- European Food Safety Authority; European Centre for Disease Prevention and Control. Multi-country outbreak of Listeria monocytogenes serogroup IVb, multi-locus sequence type 6, infections linked to frozen corn and possibly to other frozen vegetables—First update. EFSA Support. Publ. 2018, 15, 1448E. [Google Scholar] [CrossRef]
- Chen, M.; Chen, Y.; Wu, Q.; Zhang, J.; Cheng, J.; Li, F.; Zeng, H.; Lei, T.; Pang, R.; Ye, Q.; et al. Genetic characteristics and virulence of Listeria monocytogenes isolated from fresh vegetables in China. BMC Microbiol. 2019, 19, 119. [Google Scholar] [CrossRef]
- Maćkiw, E.; Korsak, D.; Kowalska, J.; Felix, B.; Stasiak, M.; Kucharek, K.; Postupolski, J. Incidence and genetic variability of Listeria monocytogenes isolated from vegetables in Poland. Int. J. Food Microbiol. 2020, 339, 109023. [Google Scholar] [CrossRef]
- Muhterem-Uyar, M.; Ciolacu, L.; Wagner, K.H.; Wagner, M.; Schmitz-Esser, S.; Stessl, B. New Aspects on Listeria monocyto-genes ST5-ECVI predominance in a heavily contaminated cheese processing environment. Front. Microbiol. 2018, 9, 64. [Google Scholar] [CrossRef] [Green Version]
- Belias, A.; Sullivan, G.; Wiedmann, M.; Ivanek, R. Factors that contribute to persistent Listeria in food processing facilities and relevant interventions: A rapid review. Food Control 2021, 133, 108579. [Google Scholar] [CrossRef]
- Smith, A.; Moorhouse, E.; Monaghan, J.; Taylor, C.; Singleton, I. Sources and survival of Listeria monocytogenes on fresh, leafy produce. J. Appl. Microbiol. 2018, 125, 930–942. [Google Scholar] [CrossRef] [Green Version]
- Alvarez-Molina, A.; Cobo-Díaz, J.F.; López, M.; Prieto, M.; de Toro, M.; Alvarez-Ordóñez, A. Unraveling the emergence and population diversity of Listeria monocytogenes in a newly built meat facility through whole genome sequencing. Int. J. Food Microbiol. 2021, 340, 109043. [Google Scholar] [CrossRef] [PubMed]
- Dzieciol, M.; Schornsteiner, E.; Muhterem-Uyar, M.; Stessl, B.; Wagner, M.; Schmitz-Esser, S. Bacterial diversity of floor drain biofilms and drain waters in a Listeria monocytogenes contaminated food processing environment. Int. J. Food Microbiol. 2016, 223, 33–40. [Google Scholar] [CrossRef] [PubMed]
- Harrand, A.S.; Wiedmann, M. Listeria Develops Reduced Sanitizer Sensitivity but Not Resistance at Recommended Sanitizer Use Levels. CPS Research Symposium 2021. Available online: https://www.centerforproducesafety.org/funded-research-projects.php (accessed on 1 January 2020).
- Braga, V.; Vázquez, S.; Vico, V.; Pastorino, V.; Mota, M.I.; Legnani, M.; Schelotto, F.; Lancibidad, G.; Varela, G. Prevalence and serotype distribution of Listeria monocytogenes isolated from foods in Montevideo-Uruguay. Braz. J. Microbiol. 2017, 48, 689–694. [Google Scholar] [CrossRef] [PubMed]
- Jamshidi, A.; Zeinali, T. Significance and characteristics of Listeria monocytogenes in poultry products. Int. J. Food Sci. 2019, 7835253. [Google Scholar] [CrossRef] [Green Version]
- Ferreira, V.; Wiedmann, M.; Teixeira, P.; Stasiewicz, M.J. Listeria monocytogenes persistence in food-associated environ-ments: Epidemiology, strain characteristics, and implications for public health. J. Food Protec. 2014, 77, 150–170. [Google Scholar] [CrossRef] [PubMed]
- Buchanan, R.L.; Gorris, L.G.M.; Hayman, M.M.; Jackson, T.C.; Whiting, R.C. A review of Listeria monocytogenes: An update on outbreaks, virulence, dose-response, ecology, and risk assessments. Food Control 2017, 75, 236. [Google Scholar] [CrossRef]
- Schjørring, S.; Lassen, S.G.; Jensen, T.; Moura, A.; Kjeldgaard, J.S.; Müller, L.; Thielke, S.; Leclercq, A.; Maury, M.M.; Tourdjman, M.; et al. Cross-border outbreak of listeriosis caused by cold-smoked salmon, revealed by integrated surveillance and whole genome sequencing (WGS), Denmark and France, 2015 to 2017. Eurosurveillance 2017, 22, 17-00762. [Google Scholar] [CrossRef]
- Allard, M.W.; Bell, R.; Ferreira, C.M.; Gonzalez-Escalona, N.; Hoffmann, M.; Muruvanda, T.; Ottesen, A.; Ramachandran, P.; Reed, E.; Sharma, S.; et al. Genomics of foodborne pathogens for microbial food safety. Curr. Opin. Biotechnol. 2018, 49, 224–229. [Google Scholar] [CrossRef]
- Toledo, V.; Bakker, H.C.D.; Hormazábal, J.C.; González-Rocha, G.; Bello-Toledo, H.; Toro, M.; Moreno-Switt, A.I. Genomic Diversity of Listeria monocytogenes Isolated from Clinical and Non-Clinical Samples in Chile. Genes 2018, 9, 396. [Google Scholar] [CrossRef] [Green Version]
- Cabal, A.; Pietzka, A.; Huhulescu, S.; Allerberger, F.; Ruppitsch, W.; Schmid, D. Isolate-Based Surveillance of Listeria monocytogenes by Whole Genome Sequencing in Austria. Front. Microbiol. 2019, 10, 2282. [Google Scholar] [CrossRef]
- EFSA Panel on Biological Hazards (EFSA BIOHAZ Panel); Koutsoumanis, K.; Allende, A.; Alvarez-Ordóñez, A.; Bolton, D.; Bover-Cid, S.; Chemaly, M.; Davies, R.; De Cesare, A.; Hilbert, F.; et al. Whole genome sequencing and metagenomics for outbreak investigation, source attribution and risk assessment of food-borne microorganisms. EFSA J. 2019, 17, e05898. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moura, A.; Criscuolo, A.; Pouseele, H.; Maury, M.M.; Leclercq, A.; Tarr, C.; Björkman, J.T.; Dallman, T.; Reimer, A.; Enouf, V.; et al. Whole genome-based population biology and epidemiological surveillance of Listeria monocytogenes. Nat. Microbiol. 2016, 2, 16185. [Google Scholar] [CrossRef] [PubMed]
- Truchado, P.; Gil, M.I.; López, C.; Garre, A.; López-Aragón, R.F.; Böhme, K.; Allende, A. New standards at European Union level on water reuse for agricultural irrigation: Are the Spanish wastewater treatment plants ready to produce and distrib-ute reclaimed water within the minimum quality requirements? Int. J. Food Microbiol. 2021, 356, 109352. [Google Scholar] [CrossRef] [PubMed]
- Andrews, S. FastQC: A Quality Control Tool for High Throughput Sequence Data; Babraham Institute: Cambridge, UK, 2010; Available online: http://www.bioinformatics.babraham.ac.uk/projects/fastqc (accessed on 27 January 2017).
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [Green Version]
- Walker, B.J.; Abeel, T.; Shea, T.; Priest, M.; Abouelliel, A.; Sakthikumar, S.; Cuomo, C.A.; Zeng, Q.; Wortman, J.; Young, S.K.; et al. Pilon: An Integrated Tool for Comprehensive Microbial Variant Detection and Genome Assembly Improvement. PLoS ONE 2014, 9, e112963. [Google Scholar] [CrossRef]
- Gurevich, A.; Saveliev, V.; Vyahhi, N.; Tesler, G. QUAST: Quality assessment tool for genome assemblies. Bioinformatics 2013, 29, 1072–1075. [Google Scholar] [CrossRef]
- Jünemann, S.; Sedlazeck, F.J.; Prior, K.; Albersmeier, A.; John, U.; Kalinowski, J.; Mellmann, A.; Goesmann, A.; von Haeseler, A.; Stoye, J.; et al. Updating benchtop sequencing performance comparison. Nat. Biotechnol. 2013, 31, 294–296. [Google Scholar] [CrossRef] [Green Version]
- Doumith, M.; Buchrieser, C.; Glaser, P.; Jacquet, C.; Martin, P. Differentiation of the Major Listeria monocytogenes Serovars by Multiplex PCR. J. Clin. Microbiol. 2004, 42, 3819–3822. [Google Scholar] [CrossRef] [Green Version]
- Ruppitsch, W.; Pietzka, A.; Prior, K.; Bletz, S.; Fernandez, H.L.; Allerberger, F.; Harmsen, D.; Mellmann, A. Defining and eval-uating a core genome multilocus sequence typing scheme for whole-genome sequence-based typing of Listeria monocytogenes. J. Clin. Microbiol. 2015, 53, 2869–2876. [Google Scholar] [CrossRef] [Green Version]
- Kruskal, J.B. On the shortest spanning subtree of a graph and the traveling salesman problem. Proc. Am. Math. Soc. 1956, 7, 48–50. [Google Scholar] [CrossRef]
- Zhang, W.; Jayarao, B.M.; Knabel, S.J. Multi-Virulence-Locus Sequence Typing of Listeria monocytogenes. Appl. Environ. Microbiol. 2004, 70, 913–920. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- USFDA (US Food and Drug Administration). Control of Listeria Monocytogenes in Ready-To-Eat Foods: Guidance for Industry Draft Guidance. US Department of Health and Human Services, Food and Drug Administration. Center for Food Safety and Applied Nutrition. 2017. Available online: https://www.fda.gov/downloads/food/guidanceregulation/guidancedocumentsregulatoryinformation/ucm535981.pdf (accessed on 27 January 2017).
- Magdovitz, B.F.; Gummalla, S.; Thippareddi, H.; Harrison, M. Evaluating environmental monitoring protocols for Listeria spp. and Listeria monocytogenes in frozen food manufacturing facilities. J. Food Prot. 2020, 83, 172–187. [Google Scholar] [CrossRef] [PubMed]
- Estrada, E.; Hamilton, A.M.; Sullivan, G.B.; Wiedmann, M.; Critzer, F.J.; Strawn, L.K. Prevalence, Persistence, and Diversity of Listeria monocytogenes and Listeria Species in Produce Packinghouses in Three U.S. States. J. Food Prot. 2020, 83, 277–286. [Google Scholar] [CrossRef] [PubMed]
- Simmons, C.K.; Wiedmann, M. Identification and classification of sampling sites for pathogen environmental monitoring programs for Listeria monocytogenes: Results from an expert elicitation. Food Microbiol. 2018, 75, 2–17. [Google Scholar] [CrossRef]
- Magdovitz, B.F.; Gummalla, S.; Garren, D.M.; Thippareddi, H.; Berrang, M.E.; Harrison, M.A. Prevalence of Listeria Species and Listeria monocytogenes on Raw Produce Arriving at Frozen Food Manufacturing Facilities. J. Food Prot. 2021, 84, 1898–1903. [Google Scholar] [CrossRef]
- Swaminathan, B.; Gerner-Smidt, P. The epidemiology of human listeriosis. Microbes Infect. 2007, 9, 1236–1243. [Google Scholar] [CrossRef] [Green Version]
- Shen, J.; Rump, L.; Zhang, Y.; Chen, Y.; Wang, X.; Meng, J. Molecular subtyping and virulence gene analysis of Listeria mono-cytogenes isolates from food. Food Microbiol. 2013, 35, 58–64. [Google Scholar] [CrossRef]
- Ballesteros, L.; Moreno, Y.; Cuesta, G.; Rodrigo, A.; Tomas, D.; Hernández, M.; Ferrús, M.A.; García-Hernandez, J. Persistence of Listeria monocytogenes strains in a frozen vegetables processing plant determined by serotyping and REP-PCR. Int. J. Food Sci. Technol. 2011, 46, 1109–1112. [Google Scholar] [CrossRef]
- Ebner, R.; Stephan, R.; Althaus, D.; Brisse, S.; Maury, M.; Tasara, T. Phenotypic and genotypic characteristics of Listeria mono-cytogenes strains isolated during 2011–2014 from different food matrices in Switzerland. Food Control 2015, 57, 321–326. [Google Scholar] [CrossRef] [Green Version]
- Montero, D.; Bodero, M.; Riveros, G.; Lapierre, L.; Gaggero, A.; Vidal, R.M.; Vidal, M. Molecular epidemiology and genetic diversity of Listeria monocytogenes isolates from a wide variety of ready-to-eat foods and their relationship to clinical strains from listeriosis outbreaks in Chile. Front. Microbiol. 2015, 6, 384. [Google Scholar] [CrossRef] [Green Version]
- Skowron, K.; Kwiecińska-Piróg, J.; Grudlewska, K.; Świeca, A.; Paluszak, Z.; Bauza-Kaszewska, J.; Walecka-Zacharska, E.; Gospodarek-Komkowska, E. The occurrence, transmission, virulence and antibiotic resistance of Listeria monocytogenes in fish processing plant. Int. J. Food Microbiol. 2018, 282, 71–83. [Google Scholar] [CrossRef] [PubMed]
- Knudsen, G.M.; Nielsen, J.B.; Marvig, R.L.; Ng, Y.; Worning, P.; Westh, H.; Gram, L. Genome-wide-analyses of Listeria monocytogenes from food-processing plants reveal clonal diversity and date the emergence of persisting sequence types. Environ. Microbiol. Rep. 2017, 9, 428–440. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kurpas, M.; Osek, J.; Moura, A.; Leclercq, A.; Lecuit, M.; Wieczorek, K. Genomic Characterization of Listeria monocytogenes Isolated From Ready-to-Eat Meat and Meat Processing Environments in Poland. Front. Microbiol. 2020, 11, 1412. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Que, F.; Xu, B.; Sun, L.; Zhu, Y.; Chen, W.; Ye, Y.; Dong, Q.; Liu, H.; Zhang, X. Identification of Listeria mono-cytogenes contamination in a ready-to-eat meat processing plant in China. Front. Microbiol. 2021, 12, 628204. [Google Scholar] [CrossRef] [PubMed]
- Manso, B.; Melero, B.; Stessl, B.; Jaime, I.; Wagner, M.; Rovira, J.; Rodríguez-Lázaro, D. The Response to Oxidative Stress in Listeria monocytogenes Is Temperature Dependent. Microorganisms 2020, 8, 521. [Google Scholar] [CrossRef] [Green Version]
- Schmitz-Esser, S.; Gram, L.; Wagner, M. Complete Genome Sequence of the Persistent Listeria monocytogenes Strain R479a. Genome Announc. 2015, 3, e00150-15. [Google Scholar] [CrossRef] [Green Version]
- Fagerlund, A.; Langsrud, S.; Schirmer, B.C.; Møretrø, T.; Heir, E. Genome Analysis of Listeria monocytogenes Sequence Type 8 Strains persisting in salmon and poultry processing environments and comparison with related strains. PLoS ONE 2016, 11, e0151117. [Google Scholar] [CrossRef] [Green Version]
- Chau, M.L.; Aung, K.T.; Hapuarachchi, H.C.; Lee, P.S.V.; Lim, P.Y.; Kang, J.S.L.; Ng, Y.; Yap, H.M.; Yuk, H.-G.; Gutiérrez, R.A.; et al. Microbial survey of ready-to-eat salad ingredients sold at retail reveals the occurrence and the persistence of Listeria monocytogenes Sequence Types 2 and 87 in pre-packed smoked salmon. BMC Microbiol. 2017, 17, 1–13. [Google Scholar] [CrossRef] [Green Version]
- Sun, Q.; Cai, S.; Cheng, J.; Zhang, Y.; Lin, R.; Ye, Q.; Xue, L.; Zeng, H.; Lei, T.; Zhang, S.; et al. Distribution, contamination routes, and seasonal influence of persistent Listeria monocytogenes in a commercial fresh Hypsizigus marmoreus production facility. Food Control 2021, 127, 108118. [Google Scholar] [CrossRef]
- Zhang, Y.; Dong, S.; Chen, H.; Chen, J.; Zhang, J.; Zhang, Z.; Yang, Y.; Xu, Z.; Zhan, L.; Mei, L. Prevalence, Genotypic Characteristics and Antibiotic Resistance of Listeria monocytogenes From Retail Foods in Bulk in Zhejiang Province, China. Front. Microbiol. 2019, 10, 1710. [Google Scholar] [CrossRef]
- Lu, B.; Yang, J.; Gao, C.; Li, D.; Cui, Y.; Huang, L.; Chen, X.; Wang, D.; Wang, A.; Liu, Y.; et al. Listeriosis Cases and Genetic Diversity of Their L. monocytogenes Isolates in China, 2008–2019. Front. Cell. Infect. Microbiol. 2021, 11. [Google Scholar] [CrossRef] [PubMed]
- Chiaverini, A.; Guidi, F.; Torresi, M.; Acciari, V.A.; Centorotola, G.; Cornacchia, A.; Centorame, P.; Marfoglia, C.; Blasi, G.; Di Domenico, M.; et al. Phylogenetic Analysis and Genome-Wide As-sociation Study Applied to an Italian Listeria monocytogenes Outbreak. Front. Microbiol. 2021, 12, 2954. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Luo, L.; Zhang, Z.; Deng, J.; Wang, Y.; Miao, Y.; Zhang, L.; Chen, X.; Liu, X.; Sun, S.; et al. Preva-lence and molecular characteristics of Listeria monocytogenes in cooked products and its comparison with isolates from lis-teriosis cases. Front. Med. 2018, 12, 104–112. [Google Scholar] [CrossRef]
- Perez-Trallero, E.; Zigorraga, C.; Artieda, J.; Alkorta, M.; Marimon, J.M. Two outbreaks of Listeria monocytogenes infection, Northern Spain. Emerg. Infect. Dis. 2014, 20, 2155–2157. [Google Scholar] [CrossRef] [PubMed]
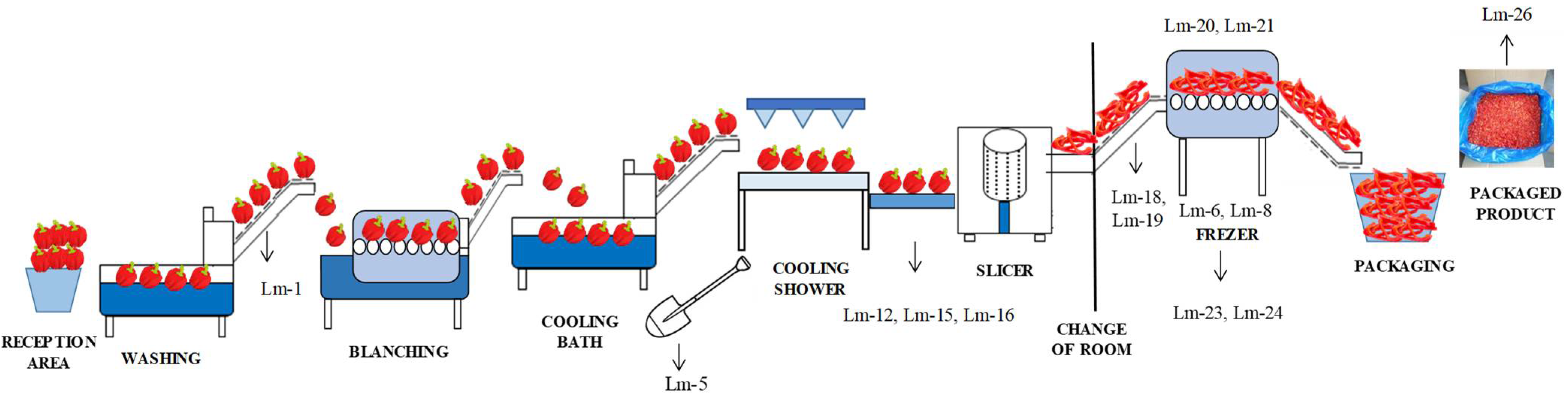

| Zone | Sample Type | No. of Samples | Present/Absent and Number of Positive Samples | Isolate Identification |
|---|---|---|---|---|
| FCS | ||||
| Conveyor-belt before blanching | 1 | + (1) | Lm-1 | |
| Blanching (surface of blanching tank) | 3 | - | ||
| Cooling bath (water) | 1 | - | ||
| A shovel used in the cooling bath | 1 | + (1) | Lm-5 | |
| Cooling shower (surface of the sprinkles and conveyor belt) | 4 | - | ||
| Conveyor belt for manual inspection | 2 | - | ||
| Slicer | 6 | - | ||
| Conveyor-belt from the slicer to the freezing tunnel | 2 | - | ||
| Freezing tunnel (surface) | 5 | + (2) | Lm-6/Lm-8 | |
| A shovel used in the freezing tunnel | 1 | - | ||
| Conveyor-belt after the freezing tunnel | 2 | - | ||
| Packaging (surface of the packaging machine) | 4 | - | ||
| SubTotal | 32 | 4 | ||
| n-FCS | ||||
| Conveyor-belt before blanching | 1 | - | ||
| Blanching | 1 | - | ||
| Cooling bath | 2 | - | ||
| Cooling shower | 4 | - | ||
| Floor and drains around the cooling shower | 8 | - | ||
| Floors and drains around the conveyor belt for manual inspection | 7 | + (3) | Lm-12/Lm-15/Lm-16 | |
| Slicer | 2 | - | ||
| The floor around the conveyor belt from the slicer to the freezing tunnel | 2 | + (2) | Lm-18/Lm-19 | |
| Surfaces around the freezing tunnel | 3 | - | ||
| Freezing tunnel | 2 | + (2) | Lm-20/Lm-21 | |
| Floor and drains around the freezing tunnel | 5 | + (2) | Lm-23/Lm-24 | |
| SubTotal | 37 | 9 | ||
| Product | ||||
| Raw material | 3 | |||
| Unpacked final product | 3 | + (1) | Lm-26 | |
| Packed final product | 3 | |||
| SubTotal | 9 | 1 | ||
| Total | 78 | 14 |
| Listeria monocytogenes | ||||
|---|---|---|---|---|
| Sample Type | Positive after Enrichment | Positive by Genedisc | Positive after OCLA/MALDI-TOF | Prevalence * (%) |
| FCS | 11/32 | 9/11 | 4/9 | 12.5% (4/32) |
| non-FCS | 17/37 | 15/17 | 9/15 | 24.3% (9/37) |
| Product | 6/9 | 3/6 | 1/3 | 11.1% (1/9) |
| TOTAL | 34/78 | 27/34 | 14/27 | 17.9% (14/78) |
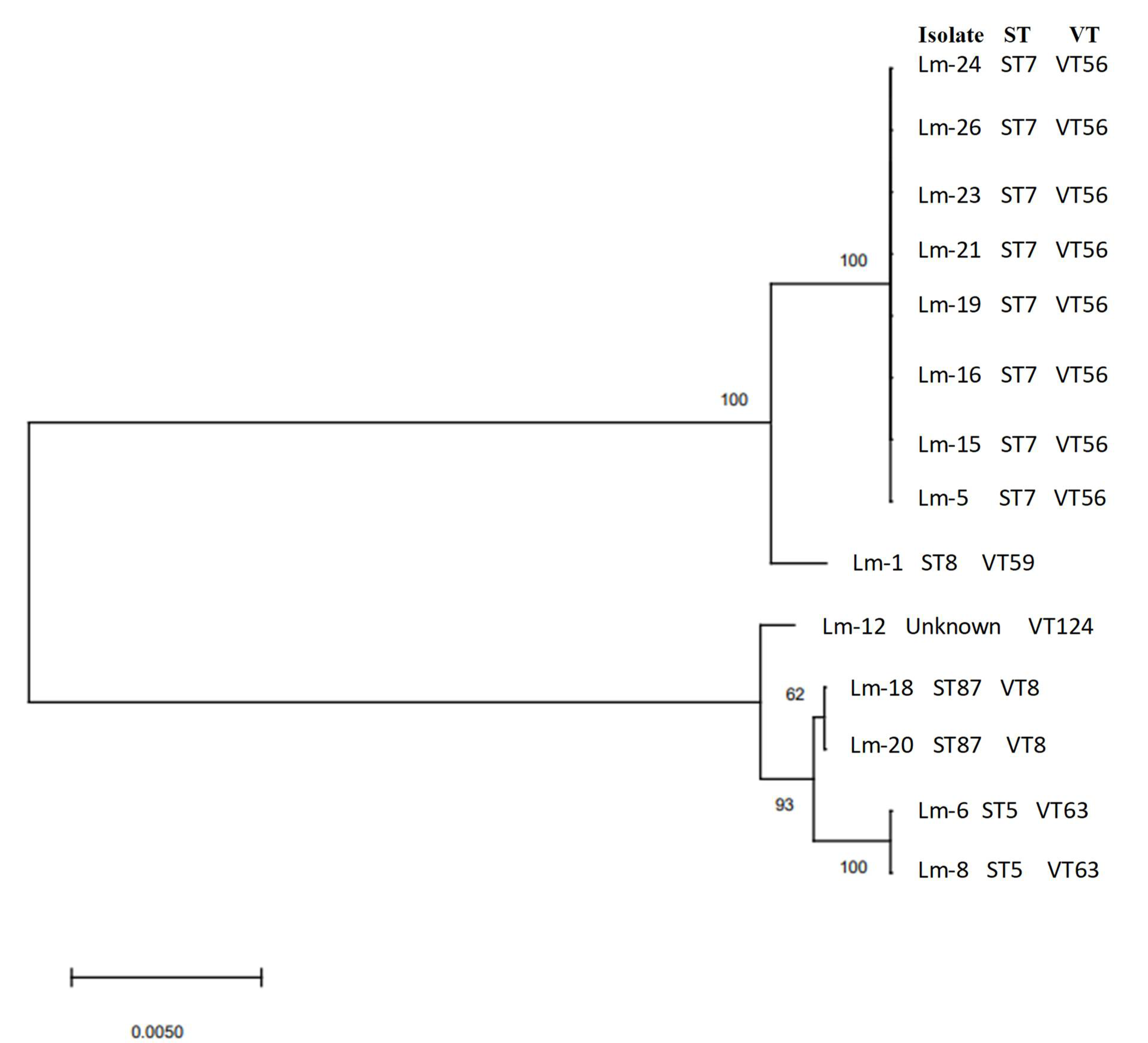
| Serogroup (Serotype) | Isolate Identification | Sequence Type (MLST) | Clonal Complex (MLST) | Complex Type (cgMLST) |
|---|---|---|---|---|
| IIa (1/2a and 3a) | Lm-1 | 8 | CC8 | - |
| IIa (1/2a and 3a) | Lm-5 | 7 | CC7 | - |
| IIa (1/2a and 3a) | Lm-15 | 7 | CC7 | - |
| IIa (1/2a and 3a) | Lm-16 | 7 | CC7 | - |
| IIa (1/2a and 3a) | Lm-20 | 7 | CC7 | - |
| IIa (1/2a and 3a) | Lm-21 | 7 | CC7 | - |
| IIa (1/2a and 3a) | Lm-23 | 7 | CC7 | |
| IIa (1/2a and 3a) | Lm-24 | 7 | CC7 | |
| IIa (1/2a and 3a) | Lm-26 | 7 | CC7 | |
| IIb (1/2b, 3b, and 7) | Lm-6 | 5 | CC5 | 7746 |
| IIb (1/2b, 3b, and 7) | Lm-8 | 5 | CC5 | 7746 |
| IIb (1/2b, 3b, and 7) | Lm-18 | 87 | CC87 | 6480 |
| IIb (1/2b, 3b, and 7) | Lm-19 | 87 | CC87 | 6480 |
| IIb (1/2b, 3b, and 7) | Lm-12 | Unknown | - | 3714 |
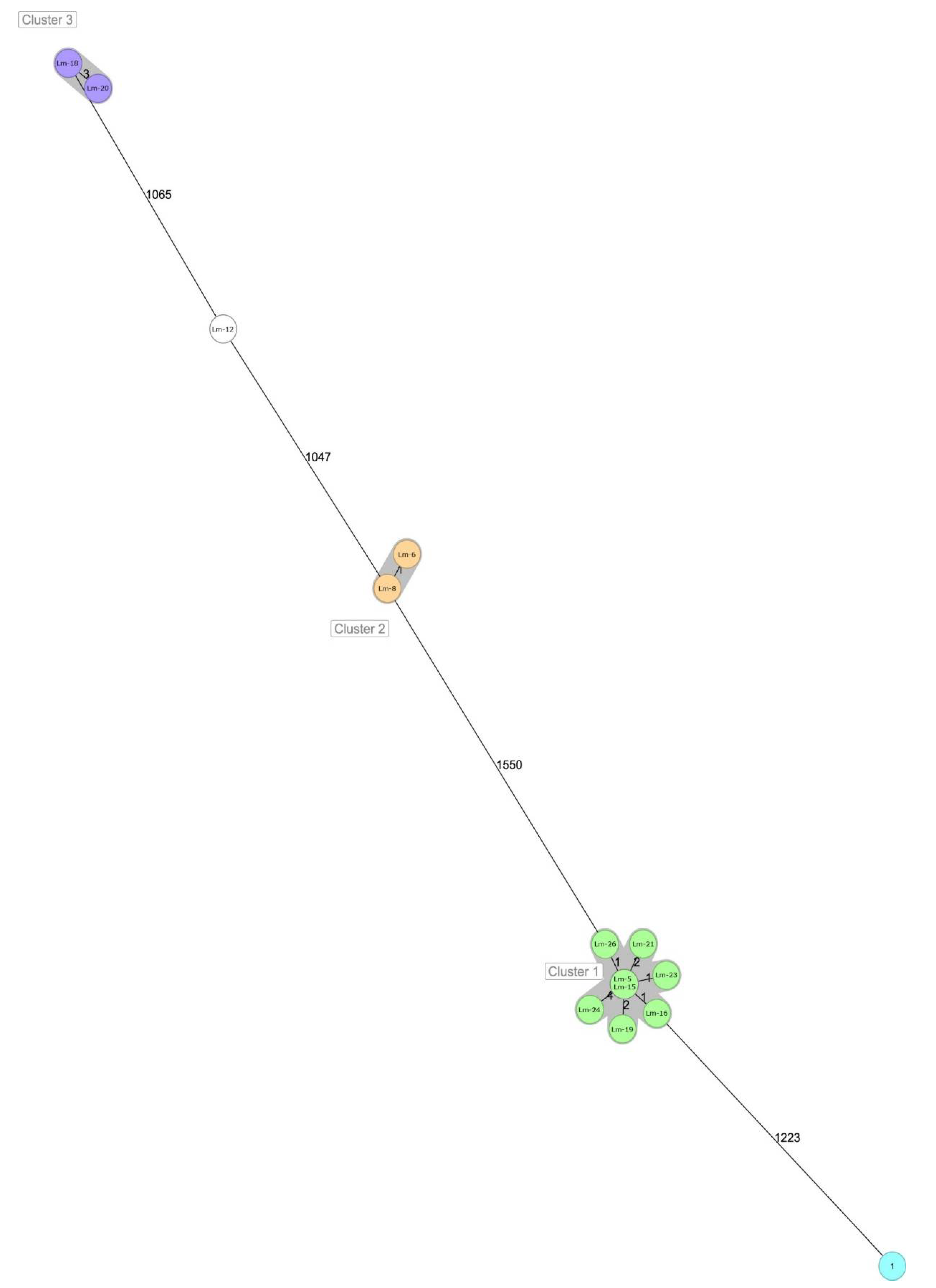
| Lm-12 | Lm-6 | Lm-8 | Lm-5 | Lm-15 | Lm-16 | Lm-19 | Lm-21 | Lm-23 | Lm-24 | Lm-26 | Lm-1 | Lm-18 | Lm-20 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lm-12 | 0 | 1065 | 1077 | 1047 | 1082 | 1627 | 1616 | 1618 | 1595 | 1627 | 1619 | 1614 | 1604 | 1596 |
| Lm-6 | 1065 | 0 | 1103 | 1074 | 3 | 1587 | 1580 | 1581 | 1560 | 1589 | 1583 | 1580 | 1566 | 1560 |
| Lm-8 | 1077 | 1103 | 0 | 1 | 1120 | 1622 | 1612 | 1617 | 1591 | 1623 | 1616 | 1612 | 1599 | 1596 |
| Lm-5 | 1047 | 1074 | 1 | 0 | 1095 | 1578 | 1566 | 1572 | 1550 | 1576 | 1574 | 1570 | 1557 | 1550 |
| Lm-15 | 1082 | 3 | 1120 | 1095 | 0 | 1606 | 1596 | 1600 | 1575 | 1608 | 1600 | 1598 | 1581 | 1576 |
| Lm-16 | 1627 | 1587 | 1622 | 1578 | 1606 | 0 | 1240 | 1235 | 1223 | 1247 | 1236 | 1235 | 1229 | 1225 |
| Lm-19 | 1616 | 1580 | 1612 | 1566 | 1596 | 1240 | 0 | 0 | 1 | 2 | 2 | 1 | 4 | 1 |
| Lm-21 | 1618 | 1581 | 1617 | 1572 | 1600 | 1235 | 0 | 0 | 1 | 2 | 2 | 1 | 4 | 1 |
| Lm-23 | 1595 | 1560 | 1591 | 1550 | 1575 | 1223 | 1 | 1 | 0 | 4 | 3 | 2 | 6 | 2 |
| Lm-24 | 1627 | 1589 | 1623 | 1576 | 1608 | 1247 | 2 | 2 | 4 | 0 | 4 | 3 | 6 | 3 |
| Lm-26 | 1619 | 1583 | 1616 | 1574 | 1600 | 1236 | 2 | 2 | 3 | 4 | 0 | 3 | 6 | 3 |
| Lm-1 | 1614 | 1580 | 1612 | 1570 | 1598 | 1235 | 1 | 1 | 2 | 3 | 3 | 0 | 5 | 2 |
| Lm-18 | 1604 | 1566 | 1599 | 1557 | 1581 | 1229 | 4 | 4 | 6 | 6 | 6 | 5 | 0 | 4 |
| Lm-20 | 1596 | 1560 | 1596 | 1550 | 1576 | 1225 | 1 | 1 | 2 | 3 | 3 | 2 | 4 | 0 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Truchado, P.; Gil, M.I.; Querido-Ferreira, A.P.; Capón, C.L.; Álvarez-Ordoñez, A.; Allende, A. Frozen Vegetable Processing Plants Can Harbour Diverse Listeria monocytogenes Populations: Identification of Critical Operations by WGS. Foods 2022, 11, 1546. https://doi.org/10.3390/foods11111546
Truchado P, Gil MI, Querido-Ferreira AP, Capón CL, Álvarez-Ordoñez A, Allende A. Frozen Vegetable Processing Plants Can Harbour Diverse Listeria monocytogenes Populations: Identification of Critical Operations by WGS. Foods. 2022; 11(11):1546. https://doi.org/10.3390/foods11111546
Chicago/Turabian StyleTruchado, Pilar, María I. Gil, Ania Pino Querido-Ferreira, Cecilia López Capón, Avelino Álvarez-Ordoñez, and Ana Allende. 2022. "Frozen Vegetable Processing Plants Can Harbour Diverse Listeria monocytogenes Populations: Identification of Critical Operations by WGS" Foods 11, no. 11: 1546. https://doi.org/10.3390/foods11111546
APA StyleTruchado, P., Gil, M. I., Querido-Ferreira, A. P., Capón, C. L., Álvarez-Ordoñez, A., & Allende, A. (2022). Frozen Vegetable Processing Plants Can Harbour Diverse Listeria monocytogenes Populations: Identification of Critical Operations by WGS. Foods, 11(11), 1546. https://doi.org/10.3390/foods11111546








