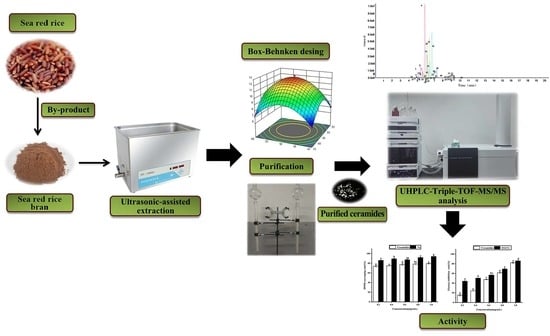
Extraction Optimization, UHPLC-Triple-TOF-MS/MS Analysis and Antioxidant Activity of Ceramides from Sea Red Rice Bran
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials and Reagents
2.2. Extraction of Cers
2.3. Procedures for Conventional Extraction and EAE
2.4. Single-Factor Experiment
2.5. Optimization of Ultrasonic-Assisted Extraction Via Box–Behnken Design
2.6. Thin-Layer Chromatography (TLC)
2.7. Purification of Cers
2.8. Structural Characterization of Cers by UHPLC-Triple-TOF-MS/MS
2.9. Software Data Analysis and Cers Identification
2.10. Antioxidant Activity Assays
2.10.1. DPPH Radical Scavenging Rate Experiment
2.10.2. Hydroxyl Radical Scavenging Rate Experiment
2.11. Anti-Aging Activity Assay
2.11.1. Tyrosinase Inhibition Test
2.11.2. Elastase Inhibition Test
2.12. Statistics and Analysis
3. Results
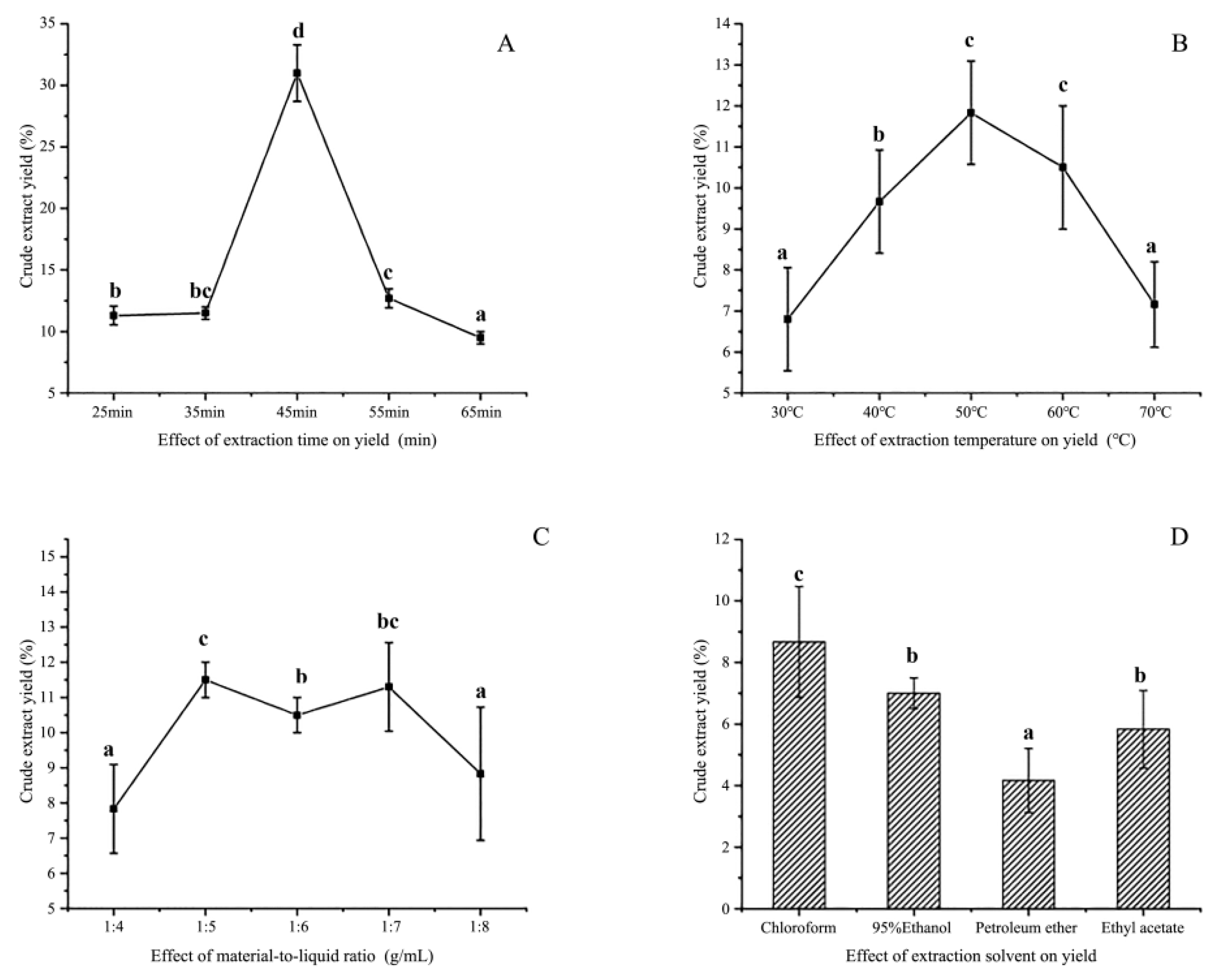
3.1. Single-Factor Experiment
3.1.1. Extraction Solvent
3.1.2. Extraction Time
3.1.3. Extraction Temperature
3.1.4. Material-to-Liquid Ratio
3.2. Optimised Design by RSM
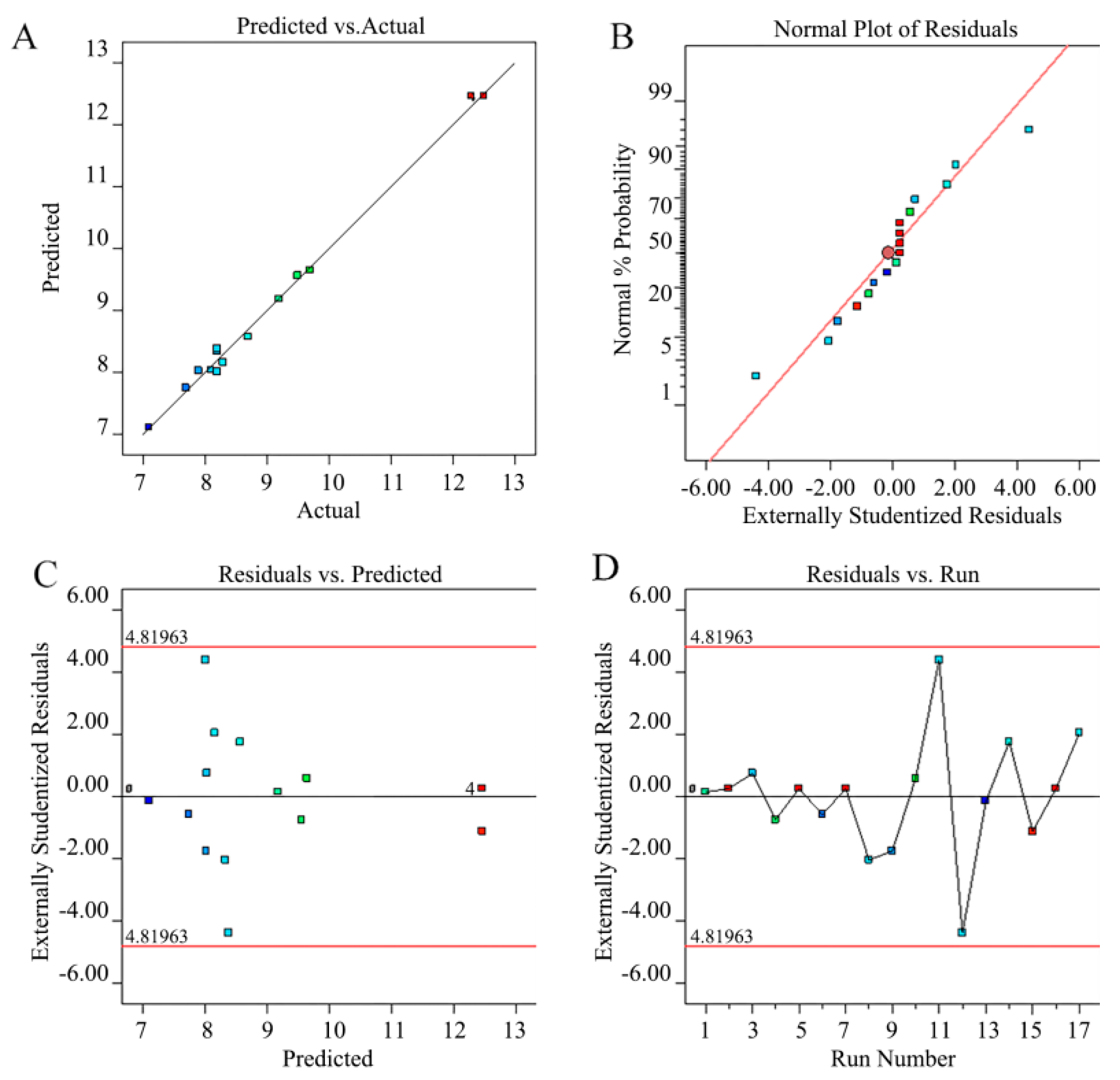
3.2.1. Model Fitting and Statistical Analysis
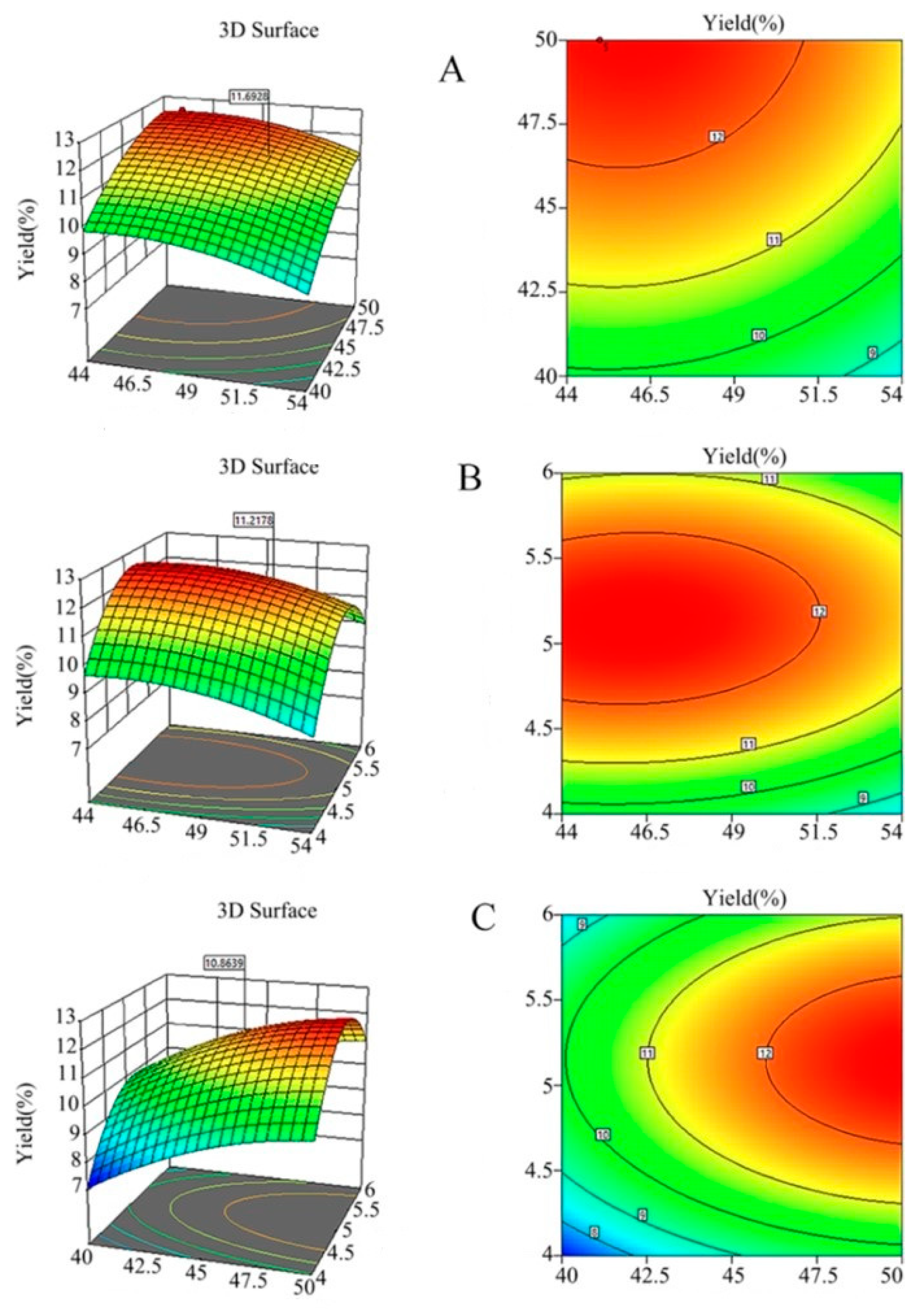
3.2.2. Response Surface Analysis
3.3. Parameter Optimization and Model Verification
3.4. Comparison of Different Extraction Methods
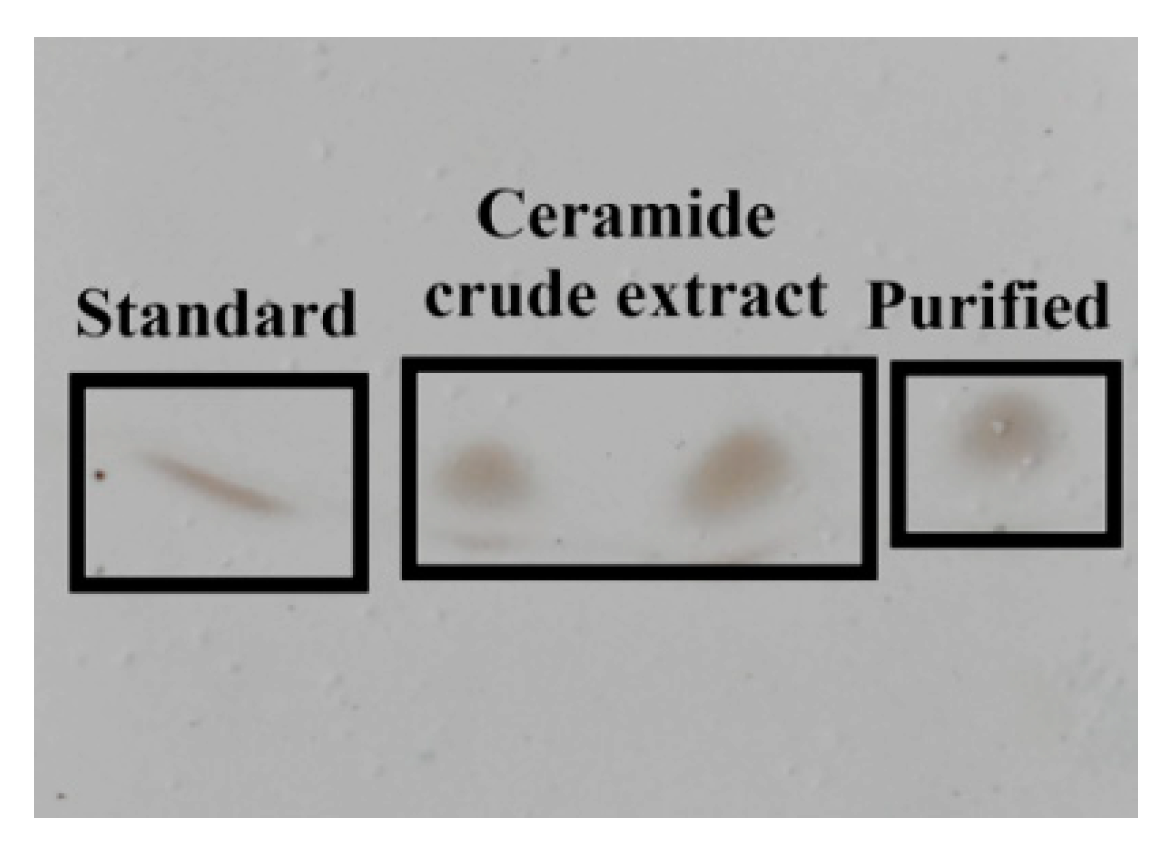
3.5. TLC and Cers Purification
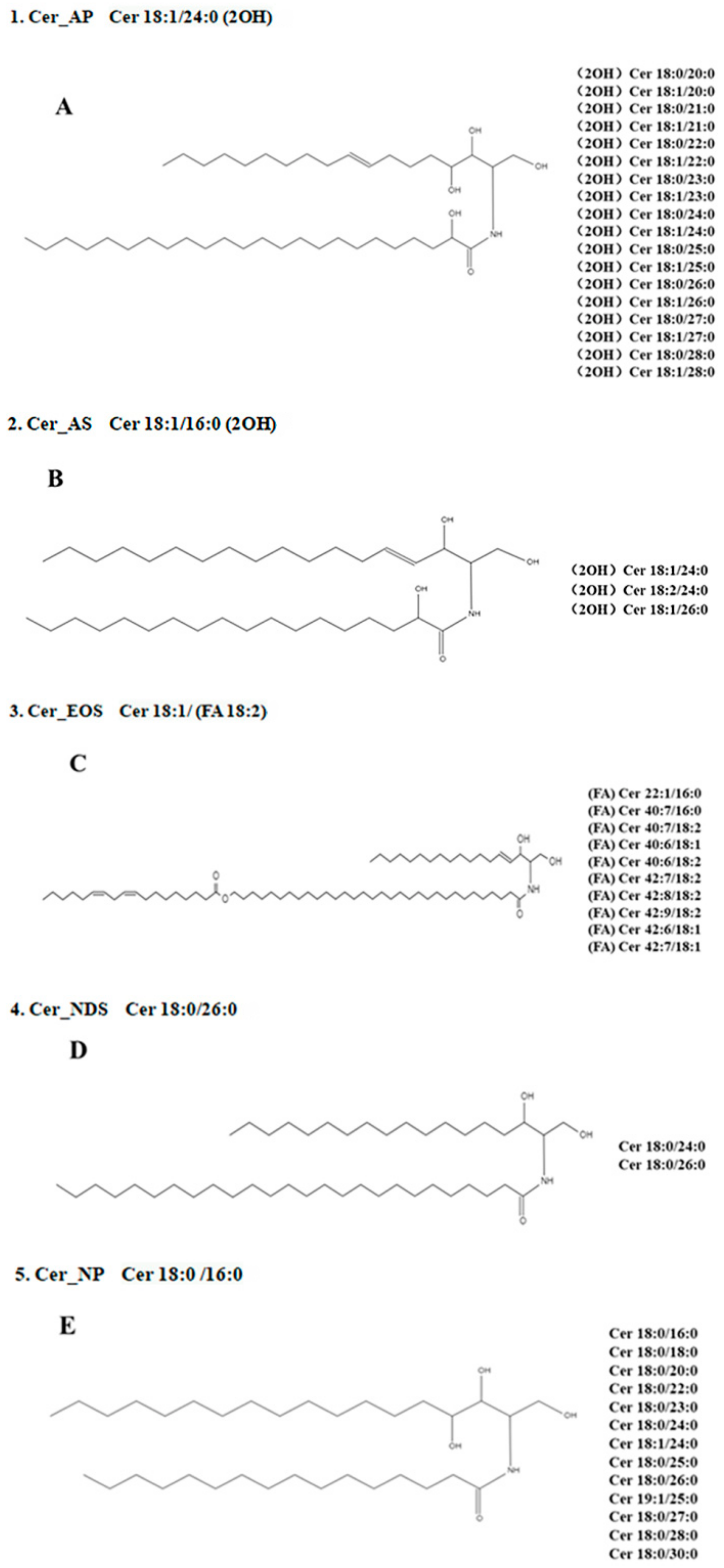
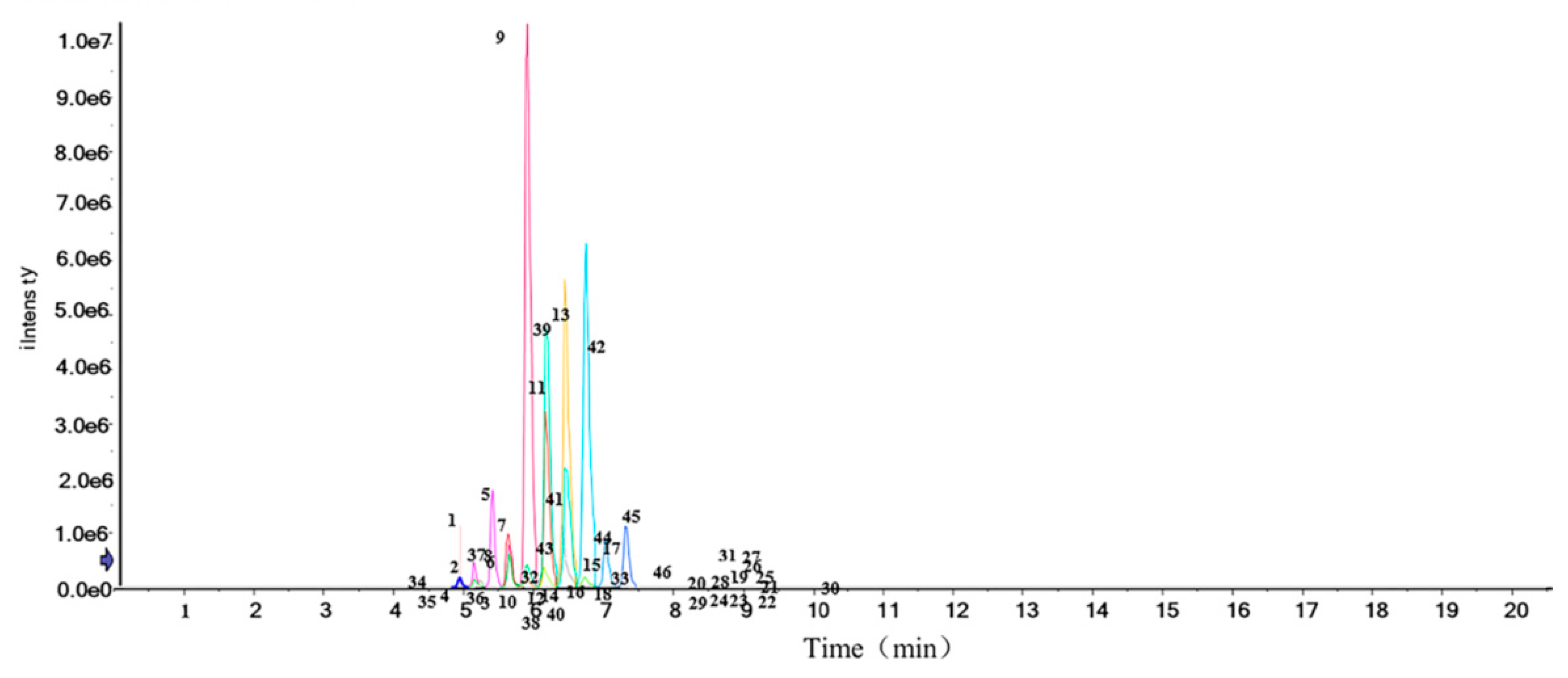
3.6. Structural Characterization of Cers by UHPLC-Triple-TOF-MS/MS
3.7. Cers Quantification Analysis
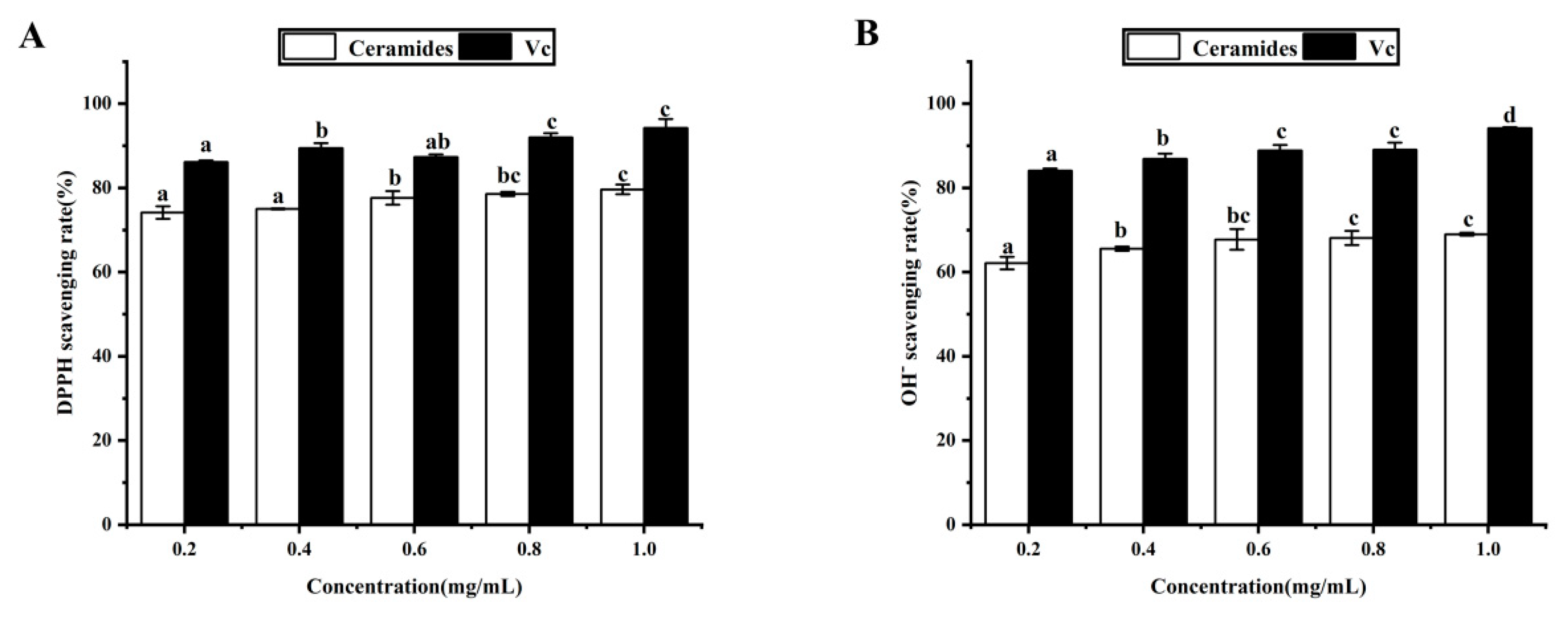
3.8. Antioxidant Activity Analysis In Vitro
3.9. Anti-Aging Activity of Purified Cers
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Chen, R.; Cheng, Y.; Han, S.; Van Handel, B.; Dong, L.; Li, X.; Xie, X. Whole genome sequencing and comparative transcriptome analysis of a novel seawater adapted, salt-resistant rice cultivar sea rice 86. BMC Genom. 2017, 18, 655. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, X.; Lin, R.; Zhang, W.; Xu, Y.; Wei, X.; Zhuo, C.; Li, H. Comparison of Cd subcellular distribution and Cd detoxification between low/high Cd-accumulative rice cultivars and sea rice. Ecotoxicol. Environ. Saf. 2019, 185, 109698. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Ma, Q.; Dong, L.; Jia, X.; Liu, L.; Huang, F.; Liu, G.; Sun, Z.; Zhang, R. Phenolic profiles and bioactivities of different milling fractions of rice bran from black rice. Food Chem. 2022, 378, 132035. [Google Scholar] [CrossRef] [PubMed]
- Shuvo, A.A.S.; Rahman, M.S.; Al-Mamum, M.; Islam, K.M.S. Cholesterol reduction and feed efficiency enhancement in broiler through the inclusion of nutritionally improved fermented rice bran. J. Appl. Poult. Res. 2022, 31, 100226. [Google Scholar] [CrossRef]
- Zhao, G.; Hu, M.; Lu, X.; Zhang, R. Soaking, heating and high hydrostatic pressure treatment degrade the flavonoids in rice bran. LWT 2022, 154, 112732. [Google Scholar] [CrossRef]
- Shimoda, H. Chapter 16—Natural Products Supporting the Extracellular Matrix: Rice Ceramide and Other Plant Extracts for Skin Health. In Nutritional Cosmetics; Tabor, A., Blair, R.M., Eds.; William Andrew Publishing: Boston, MA, USA, 2009; pp. 319–334. [Google Scholar] [CrossRef]
- Kumar, K.; Srivastav, S.; Sharanagat, V.S. Ultrasound assisted extraction (UAE) of bioactive compounds from fruit and vegetable processing by-products: A review. Ultrason. Sonochem. 2021, 70, 105325. [Google Scholar] [CrossRef]
- Ramírez, R.; Garay, I.; Álvarez, J.; Martí, M.; Parra, J.L.; Coderch, L. Supercritical fluid extraction to obtain ceramides from wool fibers. Sep. Purif. Technol. 2008, 63, 552–557. [Google Scholar] [CrossRef]
- Sepulveda, B.; Benites, D.; Albornoz, L.; Simirgiotis, M.; Castro, O.; Garcia-Beltran, O.; Areche, C. Green ultrasound-assisted extraction of lichen substances from Hypotrachyna cirrhata. Ethyl lactate, a better extracting agent than methanol toxic organic solvent? Nat. Prod. Res. 2021, 65, 1–5. [Google Scholar] [CrossRef]
- Singanusong, R.; Nipornram, S.; Tochampa, W.; Rattanatraiwong, P. Low Power Ultrasound-Assisted Extraction of Phenolic Compounds from Mandarin (Citrus reticulata Blanco cv. Sainampueng) and Lime (Citrus aurantifolia) Peels and the Antioxidant. Food Anal. Method. 2015, 8, 1112–1123. [Google Scholar] [CrossRef]
- Qian, S.; Fang, X.; Dan, D.; Diao, E.; Lu, Z. Ultrasonic-assisted enzymatic extraction of a water soluble polysaccharide from dragon fruit peel and its antioxidant activity. RSC Adv. 2018, 8, 42145–42152. [Google Scholar] [CrossRef] [Green Version]
- Charles Dorni, A.I.; Amalraj, A.; Gopi, S.; Varma, K.; Anjana, S.N. Novel cosmeceuticals from plants—An industry guided review. J. Appl. Res. Med. Aromat. Plants 2017, 7, 1–26. [Google Scholar] [CrossRef]
- Muñoz-Garcia, A.; Ro, J.; Brown, J.C.; Williams, J.B. Identification of complex mixtures of sphingolipids in the stratum corneum by reversed-phase high-performance liquid chromatography and atmospheric pressure photospray ionization mass spectrometry. J. Chromatogr. A 2006, 1133, 58–68. [Google Scholar] [CrossRef] [PubMed]
- Reisberg, M.; Arnold, N.; Porzel, A.; Neubert, R.H.H.; Dräger, B. Production of Rare Phyto-Ceramides from Abundant Food Plant Residues. J. Agric. Food Chem. 2017, 65, 1507–1517. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Wang, T.; Fehr, W.R. HPLC Quantification of Sphingolipids in Soybeans with Modified Palmitate Content. J. Agric. Food Chem. 2006, 54, 7422–7428. [Google Scholar] [CrossRef] [Green Version]
- Liang, J.; Lian, L.; Wang, X.; Li, L. Thymoquinone, extract from Nigella sativa seeds, protects human skin keratinocytes against UVA-irradiated oxidative stress, inflammation and mitochondrial dysfunction. Mol. Immunol. 2021, 135, 21–27. [Google Scholar] [CrossRef] [PubMed]
- Yamashita, S.; Soga, M.; Nguma, E.; Kinoshita, M.; Miyazawa, T. Protective Mechanism of Rice-Derived Lipids and Glucosylceramide in an In Vitro Intestinal Tract Model. J. Agric. Food Chem. 2021, 69, 10206–10214. [Google Scholar] [CrossRef]
- Jala, R.C.R.; Prasad, R.B.N. Rice Bran Lecithin: Compositional, Nutritional, and Functional Characteristics. In Polar Lipids; Ahmad, M.U., Xu, X., Eds.; Elsevier: London, UK, 2015; pp. 35–55. [Google Scholar] [CrossRef]
- Selmair, P.L.; Koehler, P. Molecular Structure and Baking Performance of Individual Glycolipid Classes from Lecithins. J. Agric. Food Chem. 2009, 57, 5597–5609. [Google Scholar] [CrossRef]
- López-Bascón, M.A.; Calderón-Santiago, M.; Sánchez-Ceinos, J.; Fernández-Vega, A.; Guzmán-Ruiz, R.; López-Miranda, J.; Malagon, M.M.; Priego-Capote, F. Influence of sample preparation on lipidomics analysis of polar lipids in adipose tissue. Talanta 2018, 177, 86–93. [Google Scholar] [CrossRef]
- Colsch, B.; Afonso, C.; Popa, I.; Portoukalian, J.; Fournier, F.; Tabet, J.; Baumann, N. Characterization of the ceramide moieties of sphingoglycolipids from mouse brain by ESI-MS/MS: Identification of ceramides containing sphingadienine. J. Lipid Res. 2004, 45, 281–286. [Google Scholar] [CrossRef] [Green Version]
- Tessema, E.N.; Gebre-Mariam, T.; Schmelzer, C.E.H.; Neubert, R.H.H. Isolation and structural characterization of glucosylceramides from Ethiopian plants by LC/APCI-MS/MS. J. Pharmaceut Biomed. 2017, 141, 241–249. [Google Scholar] [CrossRef]
- Chen, X.; Li, Z.; Wang, Z.; Liu, L.; Sun, T.; Ma, J.; Zhang, Y. Ultrasound-assisted extraction of total anthocyanins from Rubia sylvatica Nakai fruit and radical scavenging activity of the extract. Ind. Crops Prod. 2020, 150, 112420. [Google Scholar] [CrossRef]
- Qiu, K.; Li, Z.; Long, Y.; Lu, Z.; Zhu, W. Study on extraction methods of polysaccharides from a processed product of Aconitum carmichaeli Debx. RSC Adv. 2021, 11, 21259–21268. [Google Scholar] [CrossRef] [PubMed]
- Jang, M.; Park, H.; Nam, K. Whitening effects of 4-hydroxyphenethyl alcohol isolated from water boiled with Hizikia fusiformis. Food Sci. Biotechnol. 2014, 23, 555–560. [Google Scholar] [CrossRef]
- Angelis, A.; Mavros, P.; Nikolaou, P.E.; Mitakou, S.; Halabalaki, M.; Skaltsounis, L. Phytochemical analysis of olive flowers’ hydroalcoholic extract and in vitro evaluation of tyrosinase, elastase and collagenase inhibition activity. Fitoterapia 2020, 143, 104602. [Google Scholar] [CrossRef]
- Nutrizio, M.; Maltar-Strmečki, N.; Chemat, F.; Duić, B.; Jambrak, A.R. High-Voltage Electrical Discharges in Green Extractions of Bioactives from Oregano Leaves (Origanum vulgare L.) Using Water and Ethanol as Green Solvents Assessed by Theoretical and Experimental Procedures. Food Eng. Rev. 2021, 13, 161–174. [Google Scholar] [CrossRef]
- Kim, H.S.; Lee, A.Y.; Jo, J.E.; Moon, B.C.; Ji, Y.; Kim, H.K. Optimization of ultrasonic-assisted extraction of continentalic acid from the root of Aralia continentalis by using the response surface methodology. Arch. Pharm. Res. 2014, 37, 1437–1444. [Google Scholar] [CrossRef]
- Kralik, S.F.; Du, X.; Patel, C.; Walsh, J.P. A Method for Quantitative Extraction of Sphingosine 1-Phosphate into Organic Solvent. Anal. Bio-Chem. 2001, 294, 190–193. [Google Scholar] [CrossRef]
- Zhou, J.; Wang, M.; Saraiva, J.A.; Martins, A.P.; Pinto, C.A.; Prieto, M.A.; Simal-Gandara, J.; Cao, H.; Xiao, J.B.; Barba, F.J. Extraction of lipids from microalgae using classical and innovative approaches. Food Chem. 2022, 384, 132236. [Google Scholar] [CrossRef]
- Liu, Y.; Zhe, W.; Zhang, R.; Peng, Z.; Wang, Y.; Gao, H.; Xiao, J. Ultrasonic-assisted extraction of polyphenolic compounds from Paederia scandens (Lour.) Merr. Using deep eutectic solvent: Optimization, identification, and comparison with traditional methods. Ultrason. Sonochem. 2022, 86, 106005. [Google Scholar] [CrossRef]
- Zhong, M.; Huang, S.; Wang, H.; Huang, Y.; Xu, J.; Zhang, L. Optimization of ultrasonic-assisted extraction of pigment from Dioscorea cirrhosa by response surface methodology and evaluation of its stability. RSC Adv. 2019, 9, 1576–1585. [Google Scholar] [CrossRef] [Green Version]
- Das, S.; Nadar, S.S.; Rathod, V.K. Integrated strategies for enzyme assisted extraction of bioactive molecules: A review. Int. J. Biol. Macromol. 2021, 191, 899–917. [Google Scholar] [CrossRef]
- Esclapez, M.D.; García-Pérez, J.V.; Mulet, A.; Cárcel, J.A. Ultrasound-Assisted Extraction of Natural Products. Food Eng. Rev. 2011, 3, 108. [Google Scholar] [CrossRef]
- Gao, Y.; Shi, Y.; Miao, N.; Xing, W.; Yun, C.; Wang, S.; Wang, H. A Green Ultrasound-assisted Enzymatic Extraction Method for Efficient Extraction of Total Polyphenols from Empetrum nigrum and Determination of its Bioactivities. J. Ind. Eng. Chem. 2022, 3, 106. [Google Scholar] [CrossRef]
- Fuchs, B.; Süß, R.; Teuber, K.; Eibisch, M.; Schiller, J. Lipid analysis by thin-layer chromatography—A review of the current state. J. Chromatogr. A 2011, 1218, 2754–2774. [Google Scholar] [CrossRef] [PubMed]
- Pata, M.O.; Hannun, Y.A.; Ng, C.K. Plant sphingolipids: Decoding the enigma of the Sphinx. New Phytol. 2010, 185, 611–630. [Google Scholar] [CrossRef] [Green Version]
- Adem, A.A.; Belete, A.; Soboleva, A.; Frolov, A.; Tessema, E.N.; Gebre-Mariam, T.; Neubert, R.H.H. Structural characterization of plant glucosylceramides and the corresponding ceramides by UHPLC-LTQ-Orbitrap mass spectrometry. J. Pharm. Biomed. Anal. 2021, 192, 113677. [Google Scholar] [CrossRef]
- Blaas, N.; Schüürmann, C.; Bartke, N.; Stahl, B.; Humpf, H. Structural Profiling and Quantification of Sphingomyelin in Human Breast Milk by HPLC-MS/MS. J. Agric. Food Chem. 2011, 59, 6018–6024. [Google Scholar] [CrossRef]
- Motta, S.; Monti, M.; Sesana, S.; Caputo, R.; Carelli, S.; Ghidoni, R. Ceramide composition of the psoriatic scale. Biochim. Biophys. Acta (BBA)-Mol. Basis Dis. 1993, 1182, 147–151. [Google Scholar] [CrossRef]
- Markham, J.E.; Jaworski, J.G. Rapid measurement of sphingolipids from Arabidopsis thaliana by reversed-phase high-performance liquid chromatography coupled to electrospray ionization tandem mass spectrometry. Rapid Commun. Mass. Spectrom. 2007, 21, 1304–1314. [Google Scholar] [CrossRef]
- Contarini, G.; Pelizzola, V.; Scurati, S.; Povolo, M. Polar lipid of donkey milk fat: Phospholipid, ceramide and cholesterol composition. J. Food Compos. Anal. 2017, 57, 16–23. [Google Scholar] [CrossRef]
- Di Rienzi, S.C.; Johnson, E.L.; Waters, J.L.; Kennedy, E.A.; Jacobson, J.; Lawrence, P.; Ley, R.E. The microbiome affects liver sphingolipids and plasma fatty acids in a murine model of the Western diet based on soybean oil. J. Nutr. Biochem. 2021, 97, 108808. [Google Scholar] [CrossRef] [PubMed]
- Wieser, H.; Koehler, P.; Scherf, K.A. (Eds.) Chapter 2—Chemical composition of wheat grains. In Wheat—An Exceptional Crop; Woodhead Publishing: Sawston, UK, 2020; pp. 13–45. [Google Scholar] [CrossRef]
- Geng, P.; Harnlym, J.M.; Chen, P. Differentiation of Whole Grain from Refined Wheat (T. aestivum) Flour Using Lipid Profile of Wheat Bran, Germ, and Endosperm with UHPLC-HRAM Mass Spectrometry. Agric. Food Chem. 2015, 63, 6189–6211. [Google Scholar] [CrossRef] [PubMed]
- Mizutani, Y.; Mitsutake, S.; Tsuji, K.; Kihara, A.; Igarashi, Y. Ceramide biosynthesis in keratinocyte and its role in skin function. Biochimie 2009, 91, 784–790. [Google Scholar] [CrossRef] [PubMed]
- Valsecchi, M.; Mauri, L.; Casellato, R.; Ciampa, M.G.; Rizza, L.; Bonina, A.; Sonnino, S. Ceramides as Possible Nutraceutical Compounds: Characterization of the Ceramides of the Moro Blood Orange (Citrus sinensis). Agric. Food Chem. 2012, 60, 10103–10110. [Google Scholar] [CrossRef]
- Liang, X.; Wang, X.; Li, Z.; Hao, Q.; Wang, S. Improved in Vitro Assays of Superoxide Anion and 1,1-Diphenyl-2-picrylhydrazyl (DPPH) Radical-Scavenging Activity of Isoflavones and Isoflavone Metabolites. Agric. Food Chem. 2010, 58, 11548–11552. [Google Scholar] [CrossRef]
- Lee, J.; Kim, H.; Kim, J.; Jang, Y. Antioxidant Property of an Ethanol Extract of the Stem of Opuntia ficus-indica var. Saboten. Agric. Food Chem. 2002, 50, 6490–6496. [Google Scholar] [CrossRef]
- Leal, C.; Gouvinhas, I.; Santos, R.A.; Rosa, E.; Silva, A.M.; Saavedra, M.J.; Barros, A.I.R.N. Potential application of grape (Vitis vinifera L.) stem extracts in the cosmetic and pharmaceutical industries: Valorization of a by-product. Ind. Crops Prod. 2020, 154, 112675. [Google Scholar] [CrossRef]
- Ersoy, E.; Eroglu Ozkan, E.; Boga, M.; Yilmaz, M.A.; Mat, A. Anti-aging potential and anti-tyrosinase activity of three Hypericum species with focus on phytochemical composition by LC–MS/MS. Ind. Crops Prod. 2019, 141, 111735. [Google Scholar] [CrossRef]
- Blaas, N.; Humpf, H. Structural Profiling and Quantitation of Glycosyl Inositol Phosphoceramides in Plants with Fourier Transform Mass Spectrometry. J. Agric. Food Chem. 2013, 61, 4257–4269. [Google Scholar] [CrossRef]
| Level | |||
|---|---|---|---|
| Variable | −1 | 0 | 1 |
| Extraction time (min) | 35 | 45 | 55 |
| Extraction temperature (°C) | 40 | 50 | 60 |
| Material-to-liquid ratio (g/mL) | 1:4 | 1:5 | 1:6 |
| Test Number | Extraction Time A | Extraction Temperature B | Material-to-Liquid Ratio C | Yield |
|---|---|---|---|---|
| 1 | 0 | 1 | 1 | 9.2 |
| 2 | 0 | 0 | 0 | 12.5 |
| 3 | −1 | −1 | 0 | 8.1 |
| 4 | 1 | 1 | 0 | 9.5 |
| 5 | 0 | 0 | 0 | 12.5 |
| 6 | −1 | 0 | −1 | 7.7 |
| 7 | 0 | 0 | 0 | 12.5 |
| 8 | −1 | 1 | 0 | 8.2 |
| 9 | 1 | 0 | −1 | 7.9 |
| 10 | 1 | 0 | 1 | 9.7 |
| 11 | 0 | 1 | −1 | 8.2 |
| 12 | 0 | −1 | 1 | 8.2 |
| 13 | 0 | −1 | −1 | 7.1 |
| 14 | −1 | 0 | 1 | 8.7 |
| 15 | 0 | 0 | 0 | 12.3 |
| 16 | 0 | 0 | 0 | 12.5 |
| 17 | 1 | −1 | 0 | 8.3 |
| Source | Sum of Squares | Df | Mean Square | F-Value | p-Value | >F |
|---|---|---|---|---|---|---|
| Model | 64.30 | 9 | 7.14 | 271.08 | <0.0001 | significant |
| A-Extraction time | 0.9113 | 1 | 0.9113 | 34.57 | 0.0006 | |
| B-Temperature | 1.45 | 1 | 1.45 | 54.82 | 0.0001 | |
| C-Material-to-liquid ratio | 3.00 | 1 | 3.00 | 113.87 | <0.0001 | |
| AB | 0.3025 | 1 | 0.3025 | 11.48 | 0.0116 | |
| AC | 0.1600 | 1 | 0.1600 | 6.07 | 0.0432 | |
| BC | 0.0025 | 1 | 0.0025 | 0.0949 | 0.7671 | |
| A2 | 13.72 | 1 | 13.72 | 520.47 | <0.0001 | |
| B2 | 19.10 | 1 | 19.10 | 724.77 | <0.0001 | |
| C2 | 19.55 | 1 | 19.55 | 741.88 | <0.0001 | |
| Residual | 0.1845 | 7 | 0.0264 | |||
| Lack of Fit | 0.1525 | 3 | 0.0508 | 6.35 | 0.0530 | not significant |
| Pure Error | 0.0320 | 4 | 0.0080 | |||
| Cor Total | 64.49 | 16 | ||||
| R2 | 0.9971 | |||||
| Adj-R2 | 0.9935 | |||||
| Pred-R2 | 0.9614 | |||||
| Adeq precision | 42.9463 | |||||
| C.V.% | 1.69 | |||||
| p < 0.01 or p < 0.05 |
| NO | Compound | Formula | Rt (min) | Type | Ontology | Area | Percentage (%) | Classification Name | MS m/z | MS/MS |
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | Cer 18:0/20:0(2OH) | C38H77NO5 | 4.948 | [M-H]- | Cer_AP | 652,223 | 0.26 | Ceramide alpha-hydroxy fatty acid-phytospingosine | 626.57129 | 327.28967 |
| 2 | Cer 18:1/20:0(2OH) | C38H75NO5 | 4.725 | [M-H]- | Cer_AP | 254,409 | 0.10 | Ceramide alpha-hydroxy fatty acid-phytospingosine | 624.54907 | 370.33636 |
| 3 | Cer 18:0/21:0(2OH) | C39H79NO5 | 5.169 | [M-H]- | Cer_AP | 214,132 | 0.09 | Ceramide alpha-hydroxy fatty acid-phytospingosine | 640.58142 | 341.30014 |
| 4 | Cer 18:1/21:0(2OH) | C39H77NO5 | 4.934 | [M-H]- | Cer_AP | 123,692 | 0.05 | Ceramide alpha-hydroxy fatty acid-phytospingosine | 638.56104 | 384.33710 |
| 5 | Cer 18:0/22:0(2OH) | C40H81NO5 | 5.407 | [M-H]- | Cer_AP | 6,691,605 | 2.81 | Ceramide alpha-hydroxy fatty acid-phytospingosine | 654.60211 | 362.34216 |
| 6 | Cer 18:1/22:0(2OH) | C40H79NO5 | 5.164 | [M-H]- | Cer_AP | 1,680,557 | 0.70 | Ceramide alpha-hydroxy fatty acid-phytospingosine | 652.58353 | 309.30984 |
| 7 | Cer 18:0/23:0(2OH) | C41H83NO5 | 5.661 | [M-H]- | Cer_AP | 3,394,601 | 1.46 | Ceramide alpha-hydroxy fatty acid-phytospingosine | 668.62238 | 323.32379 |
| 8 | Cer 18:1/23:0(2OH) | C41H81NO5 | 5.401 | [M-H]- | Cer_AP | 509,753 | 0.22 | Ceramide alpha-hydroxy fatty acid-phytospingosine | 666.59741 | 369.33353 |
| 9 | Cer 18:0/24:0(2OH) | C42H85NO5 | 5.91 | [M-H]- | Cer_AP | 54,053,688 | 23.66 | Ceramide alpha-hydroxy fatty acid-phytospingosine | 682.63519 | 382.36456 |
| 10 | Cer 18:1/24:0(2OH) | C42H83NO5 | 5.656 | [M-H]- | Cer_AP | 4,853,410 | 2.12 | Ceramide alpha-hydroxy fatty acid-phytospingosine | 680.62280 | 382.37015 |
| 11 | Cer 18:0/25:0(2OH) | C43H87NO5 | 6.186 | [M-H]- | Cer_AP | 16,389,775 | 7.32 | Ceramide alpha-hydroxy fatty acid-phytospingosine | 696.65405 | 397.36826 |
| 12 | Cer 18:1/25:0(2OH) | C43H85NO5 | 5.901 | [M-H]- | Cer_AP | 747,527 | 0.33 | Ceramide alpha-hydroxy fatty acid-phytospingosine | 694.63794 | 397.36511 |
| 13 | Cer 18:0/26:0(2OH) | C44H89NO5 | 6.459 | [M-H]- | Cer_AP | 32,721,940 | 14.91 | Ceramide alpha-hydroxy fatty acid-phytospingosine | 710.66913 | 410.39723 |
| 14 | Cer 18:1/26:0(2OH) | C44H87NO5 | 6.177 | [M-H]- | Cer_AP | 1,769,607 | 0.80 | Ceramide alpha-hydroxy fatty acid-phytospingosine | 708.64484 | 411.38281 |
| 15 | Cer 18:0/27:0(2OH) | C45H91NO5 | 6.735 | [M-H]- | Cer_AP | 1,082,791 | 0.50 | Ceramide alpha-hydroxy fatty acid-phytospingosine | 724.67841 | 425.39743 |
| 16 | Cer 18:1/27:0(2OH) | C45H89NO5 | 6.436 | [M-H]- | Cer_AP | 131,391 | 0.06 | Ceramide alpha-hydroxy fatty acid-phytospingosine | 722.66553 | 425.39972 |
| 17 | Cer 18:0/28:0(2OH) | C46H93NO5 | 7.006 | [M-H]- | Cer_AP | 591,917 | 0.28 | Ceramide alpha-hydroxy fatty acid-phytospingosine | 738.69855 | 281.25009 |
| 18 | Cer 18:1/28:0(2OH) | C46H91NO5 | 6.725 | [M-H]- | Cer_AP | 164,664 | 0.08 | Ceramide alpha-hydroxy fatty acid-phytospingosine | 736.67798 | 438.43096 |
| 19 | Cer 18:1/24:0(2OH) | C42H83NO4 | 8.952 | [M-H]- | Cer_AS | 227,151 | 0.10 | Ceramide alpha-hydroxy fatty acid-sphingosine | 664.62946 | 438.39212 |
| 20 | Cer 18:2/24:0(2OH) | C42H81NO4 | 8.333 | [M-H]- | Cer_AS | 31,566 | 0.01 | Ceramide alpha-hydroxy fatty acid-sphingosine | 662.60376 | 438.3928 |
| 21 | Cer 18:1/26:0(2OH) | C44H87NO4 | 9.418 | [M-H]- | Cer_AS | 67,966 | 0.03 | Ceramide alpha-hydroxy fatty acid-sphingosine | 692.65619 | 466.42711 |
| 22 | Cer 22:1/(FA 16:0) | C56H99NO5 | 9.496 | [M-H]- | Cer_EOS | 34,843 | 0.02 | Ceramide Esterified omega-hydroxy fatty acid-sphingosine | 864.73761 | 255.2329 |
| 23 | Cer 40:7/(FA 16:0) | C56H97NO5 | 9.063 | [M-H]- | Cer_EOS | 59,792 | 0.03 | Ceramide Esterified omega-hydroxy fatty acid-sphingosine | 862.72430 | 255.23362 |
| 24 | Cer 40:7/(FA 18:2) | C58H97NO5 | 8.683 | [M-H]- | Cer_EOS | 107,728 | 0.06 | Ceramide Esterified omega-hydroxy fatty acid-sphingosine | 886.72754 | 279.23286 |
| 25 | Cer 40:6/(FA 18:1) | C58H101NO5 | 9.535 | [M-H]- | Cer_EOS | 160,669 | 0.09 | Ceramide Esterified omega-hydroxy fatty acid-sphingosine | 890.75922 | 281.24467 |
| 26 | Cer 40:6/(FA 18:2) | C58H99NO5 | 9.116 | [M-H]- | Cer_EOS | 202,413 | 0.12 | Ceramide Esterified omega-hydroxy fatty acid-sphingosine | 888.74298 | 251.23353 |
| 27 | Cer 42:7/(FA 18:2) | C60H101NO5 | 9.168 | [M-H]- | Cer_EOS | 261,037 | 0.15 | Ceramide Esterified omega-hydroxy fatty acid-sphingosine | 914.75830 | 281.24771 |
| 28 | Cer 42:8/(FA 18:2) | C60H99NO5 | 8.745 | [M-H]- | Cer_EOS | 191,375 | 0.11 | Ceramide Esterified omega-hydroxy fatty acid-sphingosine | 912.74392 | 279.23191 |
| 29 | Cer 42:9/(FA 18:2) | C60H97NO5 | 8.302 | [M-H]- | Cer_EOS | 91,233 | 0.05 | Ceramide Esterified omega-hydroxy fatty acid-sphingosine | 910.72479 | 279.23110 |
| 30 | Cer 42:6/(FA 18:1) | C60H105NO5 | 10.011 | [M-H]- | Cer_EOS | 26,952 | 0.02 | Ceramide Esterified omega-hydroxy fatty acid-sphingosine | 918.78662 | 281.24741 |
| 31 | Cer 42:7/(FA 18:1) | C60H103NO5 | 9.588 | [M-H]- | Cer_EOS | 202,406 | 0.12 | Ceramide Esterified omega-hydroxy fatty acid-sphingosine | 916.77295 | 281.24913 |
| 32 | Cer 18:0/24:0 | C42H85NO3 | 6.709 | [M-H]- | Cer_NDS | 92,443 | 0.04 | Ceramide non-hydroxyfatty acid-dihydrosphingosine | 650.64062 | 292.38466 |
| 33 | Cer 18:0/26:0 | C44H89NO3 | 7.295 | [M-H]- | Cer_NDS | 91,370 | 0.11 | Ceramide non-hydroxyfatty acid-dihydrosphingosine | 678.67194 | 377.37521 |
| 34 | Cer 18:0/16:0 | C34H69NO4 | 4.321 | [M-H]- | Cer_NP | 152,341 | 0.05 | Ceramide non-hydroxyfatty acid-phytospingosine | 554.51074 | 256.23243 |
| 35 | Cer 18:0/18:0 | C36H73NO4 | 4.746 | [M-H]- | Cer_NP | 251,333 | 0.09 | Ceramide non-hydroxyfatty acid-phytospingosine | 582.52173 | 338.30356 |
| 36 | Cer 18:0/20:0 | C38H77NO4 | 5.162 | [M-H]- | Cer_NP | 791,603 | 0.31 | Ceramide non-hydroxyfatty acid-phytospingosine | 610.57343 | 352.31653 |
| 37 | Cer 18:0/22:0 | C40H81NO4 | 5.655 | [M-H]- | Cer_NP | 2,629,922 | 1.08 | Ceramide non-hydroxyfatty acid-phytospingosine | 638.60986 | 382.37323 |
| 38 | Cer 18:0/23:0 | C41H83NO4 | 5.914 | [M-H]- | Cer_NP | 2,184,516 | 0.91 | Ceramide non-hydroxyfatty acid-phytospingosine | 652.62201 | 353.33591 |
| 39 | Cer 18:0/24:0 | C42H85NO4 | 6.192 | [M-H]- | Cer_NP | 27,966,104 | 11.96 | Ceramide non-hydroxyfatty acid-phytospingosine | 666.64271 | 408.38483 |
| 40 | Cer 18:1/24:0 | C42H83NO4 | 5.925 | [M-H]- | Cer_NP | 327,691 | 0.14 | Ceramide non-hydroxyfatty acid-phytospingosine | 664.62523 | 408.3831 |
| 41 | Cer 18:0/25:0 | C43H87NO4 | 6.469 | [M-H]- | Cer_NP | 13,976,958 | 6.10 | Ceramide non-hydroxyfatty acid-phytospingosine | 680.65839 | 424.40680 |
| 42 | Cer 18:0/26:0 | C44H89NO4 | 6.751 | [M-H]- | Cer_NP | 39,370,848 | 17.54 | Ceramide non-hydroxyfatty acid-phytospingosine | 694.67273 | 450.43082 |
| 43 | Cer 19:1/25:0 | C44H87NO4 | 6.479 | [M-H]- | Cer_NP | 679,806 | 0.30 | Ceramide non-hydroxyfatty acid-phytospingosine | 692.65485 | 424.41461 |
| 44 | Cer 18:0/27:0 | C45H91NO4 | 7.034 | [M-H]- | Cer_NP | 4,450,536 | 2.02 | Ceramide non-hydroxyfatty acid-phytospingosine | 708.68988 | 464.44794 |
| 45 | Cer 18:0/28:0 | C46H93NO4 | 7.325 | [M-H]- | Cer_NP | 5,419,656 | 2.51 | Ceramide non-hydroxyfatty acid-phytospingosine | 722.70349 | 423.42868 |
| 46 | Cer 18:0/30:0 | C48H97NO4 | 7.896 | [M-H]- | Cer_NP | 375,501 | 0.18 | Ceramide non-hydroxyfatty acid-phytospingosine | 750.73138 | 494.49304 |
| 47 | Cer d18:1/15:0 | C33 H67NO3 | 4.336 | [M-H]- | Cer_NDS | Internal standard | Ng | Ceramide non-hydroxyfatty acid-dihydrosphingosine | 589.55314 | 369.55314 |
| Clearance Rate (%) | Samples | 0.2 mg/mL | 0.4 mg/mL | 0.6 mg/mL | 0.8 mg/mL | 1.0 mg/mL |
|---|---|---|---|---|---|---|
| DPPH | Cers | 74.14 ± 1.47 a | 75.00 ± 0.12 a | 77.63 ± 1.60 b | 78.56 ± 0.49 b,c | 79.65 ± 1.13 c |
| VC | 86.12 ± 0.38 a | 89.45 ± 1.21 b | 87.36 ± 0.63 a,b | 92.01 ± 1.02 c | 94.23 ± 2.13 c | |
| OH– | Cers | 62.13 ± 1.49 a | 65.56 ± 0.18 b | 67.77 ± 2.47 b,c | 68.13 ± 1.67 c | 69.01 ± 0.32 c |
| VC | 84.10 ± 0.12 a | 86.93 ± 1.23 b | 88.89 ± 1.30 c | 89.06 ± 1.68 c | 92.18 ± 0.23 d |
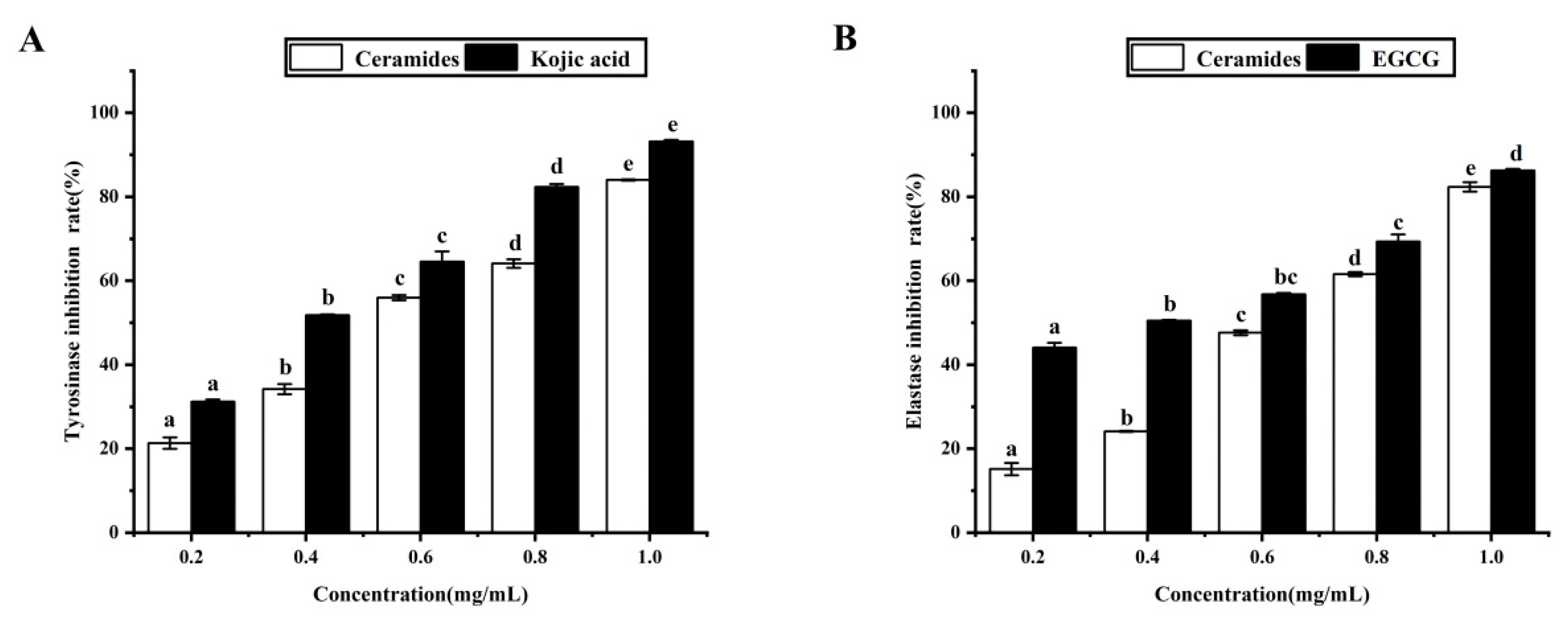
| Inhibition Rate (%) | Samples | 0.2 mg/mL | 0.4 mg/mL | 0.6 mg/mL | 0.8 mg/mL | 1.0 mg/mL |
|---|---|---|---|---|---|---|
| Tyrosinase inhibition | Cers | 21.31 ± 1.38 a | 34.20 ± 1.21 b | 56.0 ± 0.63 c | 64.12 ± 1.02 d | 84.01 ± 0.13 e |
| Kojic acid | 31.23 ± 0.49 a | 51.81 ± 0.18 b | 64.54 ± 2.47 c | 82.36 ± 0.67 d | 93.21 ± 0.32 e | |
| Elastase inhibition | Cers | 15.14 ± 1.47 a | 24.11 ± 0.12 b | 47.63 ± 0.60 c | 61.56 ± 0.49 d | 82.37 ± 1.13 e |
| EGCG | 44.12 ± 1.12 a | 51.48 ± 0.23 b | 56.10 ± 0.30 b,c | 69.36 ± 1.68 c | 86.30 ± 0.33 d |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, G.; Jia, X.-J.; Song, B.-B.; Li, R.; Liu, X.-F.; Chen, J.-P.; Zhong, S.-Y.; Zhou, H.-K. Extraction Optimization, UHPLC-Triple-TOF-MS/MS Analysis and Antioxidant Activity of Ceramides from Sea Red Rice Bran. Foods 2022, 11, 1399. https://doi.org/10.3390/foods11101399
Wang G, Jia X-J, Song B-B, Li R, Liu X-F, Chen J-P, Zhong S-Y, Zhou H-K. Extraction Optimization, UHPLC-Triple-TOF-MS/MS Analysis and Antioxidant Activity of Ceramides from Sea Red Rice Bran. Foods. 2022; 11(10):1399. https://doi.org/10.3390/foods11101399
Chicago/Turabian StyleWang, Gang, Xue-Jing Jia, Bing-Bing Song, Rui Li, Xiao-Fei Liu, Jian-Ping Chen, Sai-Yi Zhong, and Hong-Kai Zhou. 2022. "Extraction Optimization, UHPLC-Triple-TOF-MS/MS Analysis and Antioxidant Activity of Ceramides from Sea Red Rice Bran" Foods 11, no. 10: 1399. https://doi.org/10.3390/foods11101399
APA StyleWang, G., Jia, X.-J., Song, B.-B., Li, R., Liu, X.-F., Chen, J.-P., Zhong, S.-Y., & Zhou, H.-K. (2022). Extraction Optimization, UHPLC-Triple-TOF-MS/MS Analysis and Antioxidant Activity of Ceramides from Sea Red Rice Bran. Foods, 11(10), 1399. https://doi.org/10.3390/foods11101399