Effects of Salt Treatment Time on the Metabolites, Microbial Composition, and Quality Characteristics of the Soy Sauce Moromi Extract
Abstract
:1. Introduction
2. Materials and Methods
2.1. Preparation of SSME and Salt Treatment
2.2. Determination of General Characteristics
2.3. Determination of Protease and Amylase Activities
2.4. Microbial Analysis
2.5. Sensory Evaluation
2.6. GC/MS-Based Metabolomic Analysis
2.7. UPLC-Q-TOF MS-Based Metabolomic Analysis
2.8. Organic Acid Analysis
2.9. Data Processing
2.10. Statistical Analysis
3. Results and Discussion
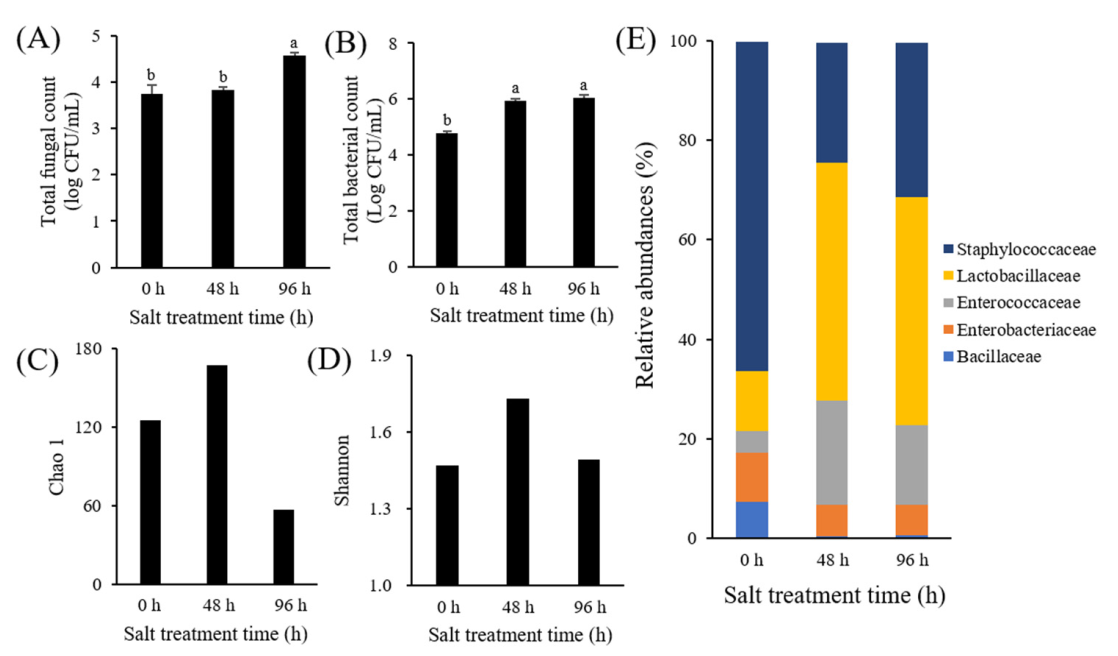
3.1. Microbial Analysis
3.2. General Characteristics
3.3. Enzymatic Activity
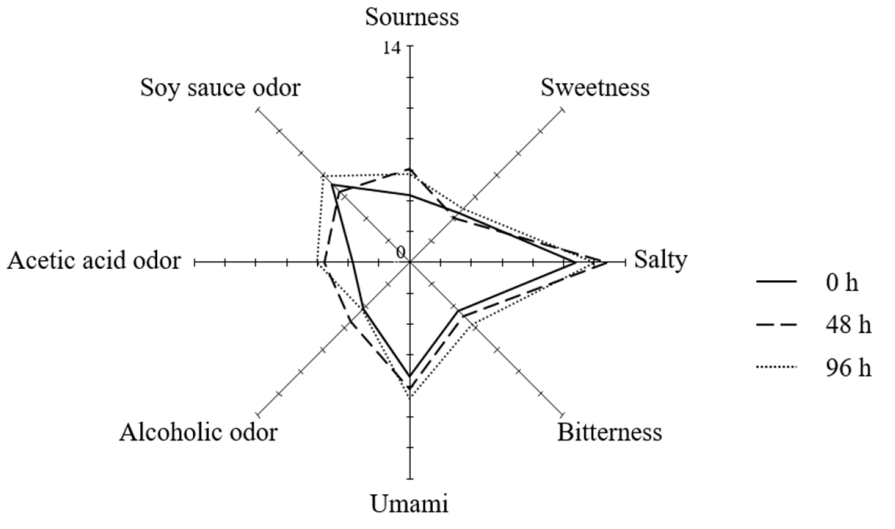
3.4. Sensory Evaluation
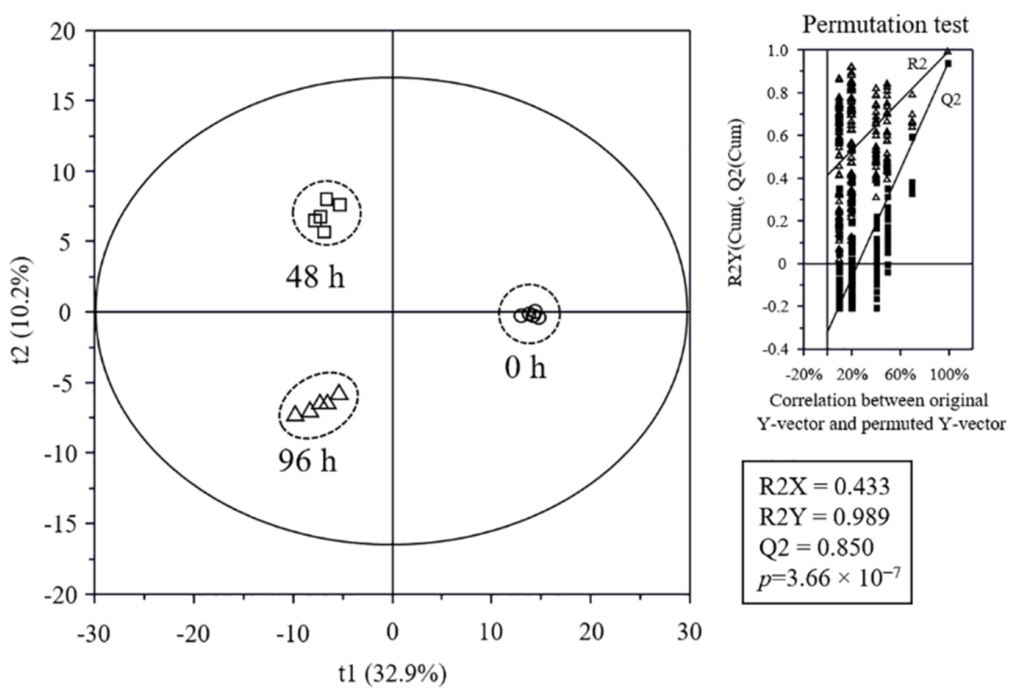
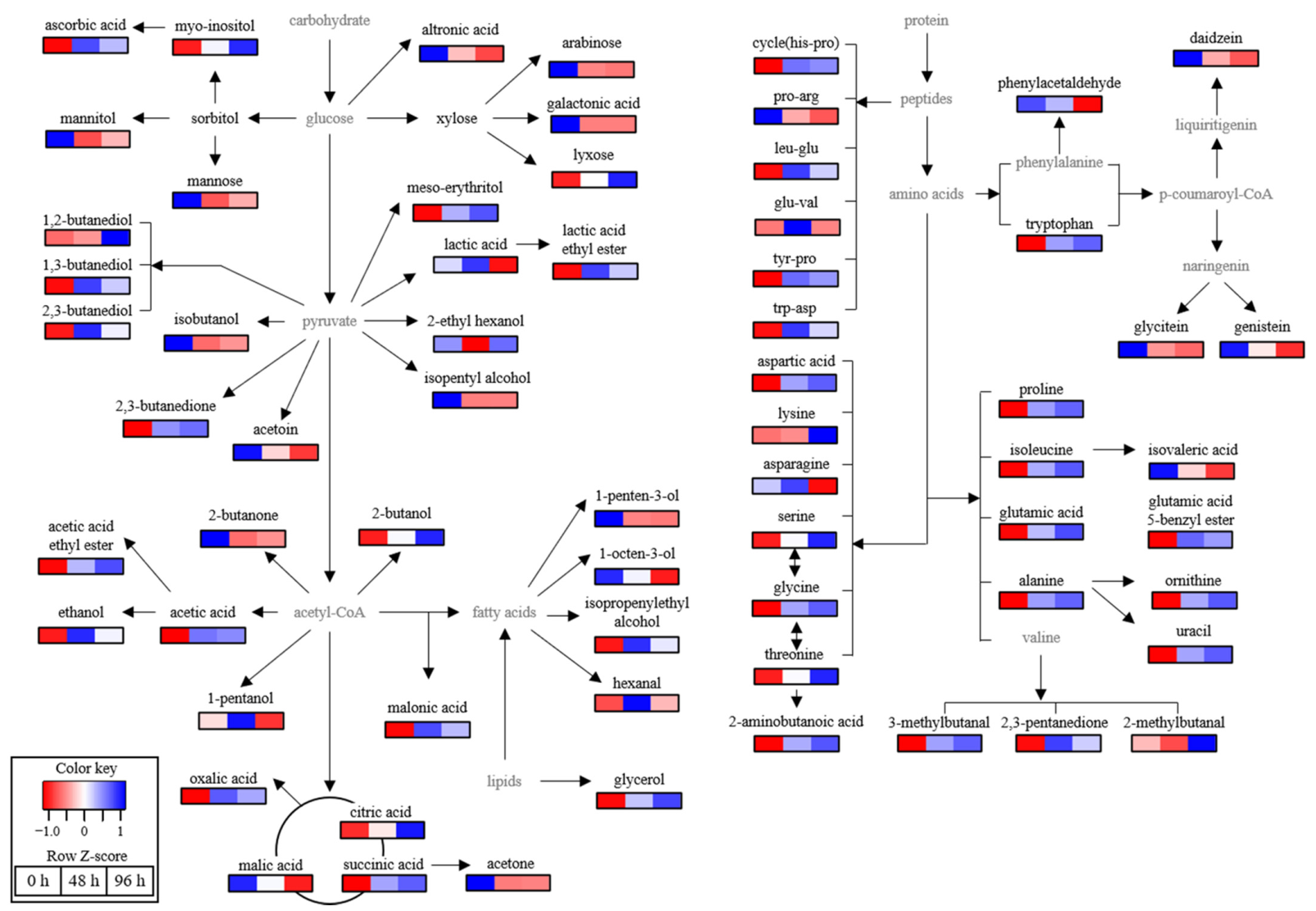
3.5. Metabolome Analysis
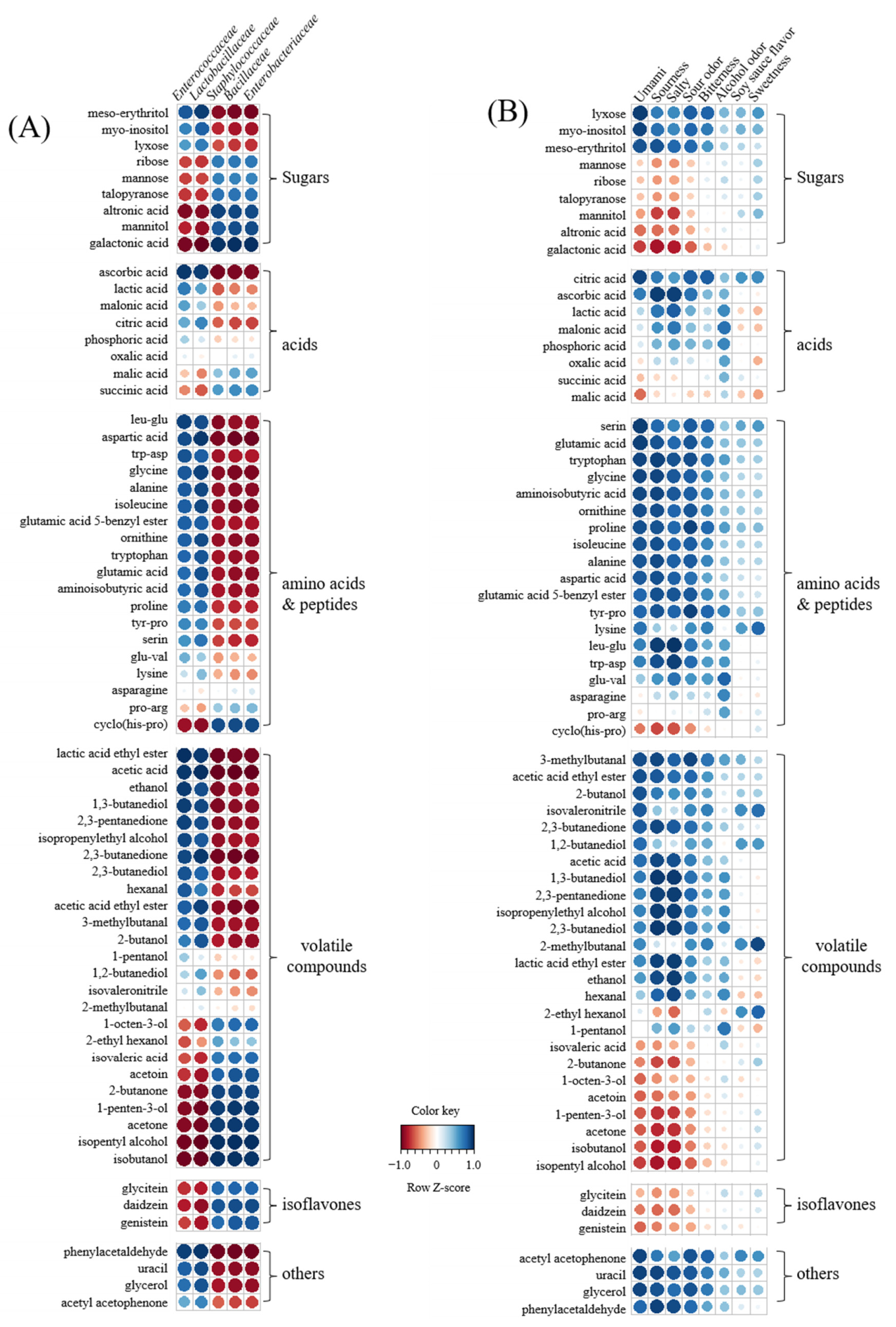
3.6. Correlation between Metabolites, Bacterial Populations, and Sensory Parameters
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Divya, J.B.; Varsha, K.K.; Nampoothiri, K.M.; Ismail, B.; Pandey, A. Probiotic fermented foods for health benefits. Eng. Life Sci. 2012, 12, 377–390. [Google Scholar] [CrossRef]
- Gille, D.; Schmid, A.; Walther, B.; Vergères, G. Fermented food and non-communicable chronic diseases: A review. Nutrients 2018, 10, 448. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, W.; Liu, Y.; Latta, M.; Ma, W.; Wu, Z.; Chen, P. Probiotics database: A potential source of fermented foods. Int. J. Food Prop. 2019, 22, 198–217. [Google Scholar] [CrossRef] [Green Version]
- Marco, M.L.; Heeney, D.; Binda, S.; Cifelli, C.J.; Cotter, P.D.; Folignè, B.; Ganzle, M.; Kort, R.; Pasin, G.; Pihlanto, A.; et al. Health benefits of fermented foods: Microbiota and beyond. Curr. Opin. Biotechnol. 2017, 44, 94–102. [Google Scholar] [CrossRef] [PubMed]
- Thapa, N.; Tamang, J.P. Functionality and Therapeutic Values of Fermented Foods. In Health Benefits of Fermented Foods; Tamang, J.P., Ed.; CRC Press: New York, NY, USA, 2015; pp. 128–130. [Google Scholar]
- Tamang, J.P.; Watanabe, K.; Holzapfel, W.H. Review: Diversity of microorganisms in global fermented foods and beverages. Front. Microbiol. 2016, 7, 377. [Google Scholar] [CrossRef] [Green Version]
- Couto, S.R.; Sanroman, M.A. Application of solid-state fermentation to food industry—A review. J. Food Eng. 2006, 76, 291–302. [Google Scholar] [CrossRef]
- Xiong, T.; Li, J.; Liang, F.; Wang, Y.; Guan, Q.; Xie, M. Effects of salt concentration on Chinese sauerkraut fermentation. LWT-Food Sci. Technol. 2016, 69, 169–174. [Google Scholar] [CrossRef]
- Kim, S.S.; Heo, J.A.; Kim, Y.; Kim, M.J.; Kwak, H.S. Salt contents and aging period effects on the physicochemical properties and sensory quality of Korean traditional fermented soybean paste (doenjang). Food Biosci. 2020, 36, 100645. [Google Scholar] [CrossRef]
- Hoang, N.X.; Ferng, S.; Ting, C.H.; Lu, Y.C.; Yeh, Y.F.; Lai, Y.R.; Chiou, R.Y.Y.; Hwang, J.Y.; Hsu, C.K. Effect of initial 5 days fermentation under low salt condition on the quality of soy sauce. LWT-Food Sci. Technol. 2018, 92, 234–241. [Google Scholar] [CrossRef]
- Feng, Y.; Cai, Y.; Su, G.; Zhao, H.; Wang, C.; Zhao, M. Evaluation of aroma differences between high-salt liquid-state fermentation and low-salt solid-state fermentation soy sauces from China. Food Chem. 2014, 145, 126–134. [Google Scholar] [CrossRef]
- Singh, D.; Lee, S.; Lee, C.H. Metabolomics for empirical delineation of the traditional Korean fermented foods and beverages. Trends Food Sci. Technol. 2017, 61, 103–115. [Google Scholar] [CrossRef]
- Seo, S.H.; Park, S.E.; Kim, E.J.; Lee, K.I.; Na, C.S.; Son, H.S. A GC-MS based metabolomics approach to determine the effect of salinity on Kimchi. Food Res. Int. 2018, 105, 492–498. [Google Scholar] [CrossRef]
- Han, D.M.; Chun, B.H.; Feng, T.; Kim, H.M.; Jeon, C.O. Dynamics of microbial communities and metabolites in ganjang, a traditional Korean fermented soy sauce, during fermentation. Food Microbiol. 2020, 92, 103591. [Google Scholar] [CrossRef]
- Hoang, N.X.; Ferng, S.; Ting, C.-H.; Huang, W.-H.; Chiou, R.Y.-Y.; Hsu, C.-K. Optimizing the initial moromi fermentation conditions to improve the quality of soy sauce. LWT-Food Sci. Technol. 2016, 74, 242–250. [Google Scholar] [CrossRef]
- AOAC. Official Methods of Analysis of AOAC; Association of Official Analytical Chemists: Arlington, TX, USA, 1990. [Google Scholar]
- Miller, G.L. Use of dinitrosalicylic acid reagent for determination of reducing sugar. Anal. Chem. 1959, 31, 426–428. [Google Scholar] [CrossRef]
- Bradford, M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Anson, M.L. The estimation of pepsin, trypsin, papain, and cathepsin with hemoglobin. J. Gen. Physiol. 1938, 22, 79–89. [Google Scholar] [CrossRef]
- Bernfeld, P. Amylase α and β. Methods Enzymol. 1955, 1, 149–158. [Google Scholar] [CrossRef]
- Jeong, S.-J.; Ryu, M.-S.; Yang, H.-J.; Wu, X.-H.; Jeong, D.-Y.; Park, S.-M. Bacterial Distribution, Biogenic Amine Contents, and Functionalities of Traditionally Made Doenjang, a Long-Term Fermented Soybean Food, from Different Areas of Korea. Microorganisms 2021, 9, 1348. [Google Scholar] [CrossRef]
- Wu, X.; Unno, T.; Kang, S.; Park, S. A Korean-style balanced diet has a potential connection with Ruminococcaceae enterotype and reduction of metabolic syndrome incidence in Korean adults. Nutrients 2021, 13, 495. [Google Scholar] [CrossRef]
- Kim, D.W.; Kim, B.-M.; Lee, H.-J.; Jang, G.-J.; Song, S.H.; Lee, J.-I.; Lee, S.B.; Shim, J.M.; Lee, K.W.; Kim, J.H.; et al. Effects of different salt treatments on the fermentation metabolites and bacterial profiles of kimchi. J. Food Sci. 2017, 82, 1124–1131. [Google Scholar] [CrossRef]
- He, B.; Ma, L.; Hu, Z.; Li, H.; Ai, M.; Long, C.; Zeng, B. Deep sequencing analysis of transcriptomes in Aspergillus oryzae in response to salinity stress. Appl. Microbiol. Biotechnol. 2018, 102, 897–906. [Google Scholar] [CrossRef]
- Savitri, M.; Kumar, V.; Kumari, A.; Angmo, K.; Bhalla, T.C. Isolation and characterization of lactic acid bacteria from traditional pickles of Himachal Pradesh, India. J. Food Sci. Technol. 2017, 54, 1945–1952. [Google Scholar] [CrossRef]
- Jeong, D.-W.; Kim, H.-R.; Jung, G.; Han, S.; Kim, C.-T.; Lee, J.-H. Bacterial community migration in the ripening of doenjang, a traditional Korean fermented soybean food. J. Microbiol. Biotechnol. 2014, 24, 648–660. [Google Scholar] [CrossRef] [Green Version]
- Wei, C.-L.; Chao, S.-H.; Tsai, W.-B.; Lee, P.-S.; Tsau, N.-H.; Chen, J.-S.; Lai, W.-L.; Tu, J.C.-Y.; Tsai, Y.-C. Analysis of bacterial diversity during the fermentation of inyu, a high-temperature fermented soy sauce, using nested PCR-denaturing gradient gel electrophoresis and the plate count method. Food Microbiol. 2013, 33, 252–261. [Google Scholar] [CrossRef]
- Shin, D.; Jeong, D. Korean traditional fermented soybean products: Jang. J. Ethn. Foods 2015, 2, 2–7. [Google Scholar] [CrossRef] [Green Version]
- Guan, L.; Cho, K.H.; Lee, J.-H. Analysis of the cultivable bacterial community in jeotgal, a Korean salted and fermented seafood, and identification of its dominant bacteria. Food Microbiol. 2011, 28, 101–113. [Google Scholar] [CrossRef]
- El-Ghaish, S.; Ahmadova, A.; Hadji-Sfaxi, I.; Mecherfi, K.E.E.; Bazukyan, I.; Choiset, Y.; Rabesona, H.; Sitohy, M.; Popov, Y.G.; Kuliev, A.A.; et al. Potential use of lactic acid bacteria for reduction of allergenicity and for longer conservation of fermented foods. Trends Food Sci. Technol. 2011, 22, 509–516. [Google Scholar] [CrossRef]
- M’hir, S.; Minervini, F.; Cagno, R.D.; Chammem, N.; Hamdi, M. Technological, functional and safety aspects of enterococci in fermented vegetable products: A mini-review. Ann. Microbiol. 2012, 62, 469–481. [Google Scholar] [CrossRef]
- Zeng, X.; Bai, W.; Zhu, X.; Dong, H. Browning intensity and taste change analysis of chicken protein–sugar maillard reaction system with antioxidants and different drying processes. J. Food Process. Preserv. 2016, 41, e13117. [Google Scholar] [CrossRef]
- Kim, M.J.; Kwak, H.S.; Kim, S.S. Effects of salinity on bacterial communities, Maillard reactions, isoflavone composition, antioxidation and antiproliferation in Korean fermented soybean paste (doenjang). Food Chem. 2018, 245, 402–409. [Google Scholar] [CrossRef] [PubMed]
- Kwak, E.-J.; Lim, S.-I. The effect of sugar, amino acid, metal ion, and NaCl on model Maillard reaction under pH control. Amino Acids 2004, 27, 85–90. [Google Scholar] [CrossRef] [PubMed]
- Yong, F.M.; Wood, B.J.B. Biochemical changes in experimental soy sauce Moromi. Int. J. Food Sci. Technol. 1977, 12, 263–273. [Google Scholar] [CrossRef]
- Jeon, H.H.; Jung, J.Y.; Chun, B.-H.; Kim, M.-D.; Baek, S.Y.; Moon, J.Y.; Yeo, S.-H.; Jeon, C.O. Screening and characterization of potential Bacillus starter cultures for fermenting low-salt soybean paste (Doenjang). J. Microbiol. Biotechnol. 2016, 26, 666–674. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fossi, B.T.; Tavea, F.; Fontem, A.; Ndjouenkeu, R.; Wanji, S. Microbial interactions for enhancement of α-amylase production by Bacillus amyloliquefaciens 04BBA15 and Lactobacillus fermentum 04BBA19. Biotechnol. Rep. 2014, 4, 99–106. [Google Scholar] [CrossRef] [Green Version]
- Kim, M.J.; Kwak, H.S.; Jung, H.Y.; Kim, S.S. Microbial communities related to sensory attributes in Korean fermented soy bean paste (doenjang). Food Res. Int. 2016, 89, 724–732. [Google Scholar] [CrossRef]
- Jiang, X.; Xu, Y.; Ye, J.; Yang, Z.; Huang, S.; Liu, Y.; Zhou, S. Isolation, identification and application on soy sauce fermentation flavor bacteria of CS1.03. J. Food Sci. Technol. 2019, 56, 2016–2026. [Google Scholar] [CrossRef]
- Feng, Y.; Cui, C.; Zhao, H.; Gao, X.; Zhao, M.; Sun, W. Effect of koji fermentation on generation of volatile compounds in soy sauce production. Int. J. Food Sci. Technol. 2013, 48, 609–619. [Google Scholar] [CrossRef]
- Zhao, C.J.; Schieber, A.; Gänzle, M.G. Formation of taste-active amino acids, amino acid derivatives and peptides in food fermentations—A review. Food Res. Int. 2016, 89, 39–47. [Google Scholar] [CrossRef]
- Liu, J.; Liu, M.; He, C.; Song, H.; Chen, F. Effect of thermal treatment on the flavor generation from Maillard reaction of xylose and chicken peptide. LWT-Food Sci. Technol. 2015, 64, 316–325. [Google Scholar] [CrossRef]
- Lee, K.E.; Lee, S.M.; Choi, Y.H.; Hurh, B.S.; Kim, Y.-S. Comparative volatile profiles in soy sauce according to inoculated microorganisms. Biosci. Biotechnol. Biochem. 2013, 77, 2192–2200. [Google Scholar] [CrossRef] [Green Version]
- Zhao, G.; Ding, L.-L.; Yao, Y.; Cao, Y.; Pan, Z.-H.; Kong, D.-H. Extracellular proteome analysis and flavor formation during soy sauce fermentation. Front. Microbiol. 2018, 9, 1872. [Google Scholar] [CrossRef]
- Zhang, Y.F.; Tao, W.Y. Flavour and taste compounds analysis in Chinese solid fermented soy sauce. Afr. J. Biotechnol. 2010, 8, 673–681. [Google Scholar]
- Wang, S.; Tamura, T.; Kyouno, N.; Liu, X.; Zhang, H.; Akiyama, Y.; Chen, J.Y. Effect of volatile compounds on the quality of Japanese fermented soy sauce. LWT-Food Sci. Technol. 2019, 111, 594–601. [Google Scholar] [CrossRef]
- Chun, B.H.; Kim, K.H.; Jeong, S.E.; Jeon, C.O. The effect of salt concentrations on the fermentation of doenjang, a traditional Korean fermented soybean paste. Food Microbiol. 2020, 86, 103329. [Google Scholar] [CrossRef]
| Salt Treatment Time (h) | ||||
|---|---|---|---|---|
| 0 | 48 | 96 | ||
| pH value | 5.12 ± 0.04 a | 4.71 ± 0.02 b | 4.72 ± 0.03 b | |
| Acid value (%) | 1.56 ± 0.010 c | 2.32 ± 0.05 b | 2.65 ± 0.20 a | |
| Amino-type nitrogen (mg %) | 389.84 ± 15.98 b | 781.98 ± 73.24 a | 735.85 ± 55.94 a | |
| Salt content (%) | 6.83 ± 0.42 b | 7.48 ± 0.17 a | 7.40 ± 0.14 a | |
| Reducing sugar (%) | 2.81 ± 0.19 a | 0.49 ± 0.08 b | 0.34 ± 0.04 b | |
| Color | L* | 24.70 ± 0.34 b | 26.73 ± 0.07 a | 26.74 ± 0.05 a |
| a* | 1.05 ± 0.15 b | 1.64 ± 0.03 a | 1.67 ± 0.03 a | |
| b* | 2.19 ± 0.18 c | 4.96 ± 0.06 b | 5.15 ± 0.04 a | |
| Brown index | 12.17 ± 1.37 c | 24.63 ± 0.30 b | 25.56 ± 0.18 a | |
| Enzymatic Activity (U/mg Protein) | Salt Treatment Time (h) | ||
|---|---|---|---|
| 0 | 48 | 96 | |
| Amylase activity | 1.99 ± 0.02 a | 0.52 ± 0.02 b | 0.45 ± 0.00 c |
| Protease activity | 1.27 ± 0.11 b | 1.70 ± 0.03 b | 2.51 ± 0.54 a |
| Compounds | VIP | p-Value | Fold Change (vs. 0 h) | ||
|---|---|---|---|---|---|
| 48 h | 96 h | ||||
| sugars | meso−erythritol | 1.25 | 2.27 × 10−8 | 2.32 | 2.69 |
| lyxose | 1.26 | 9.31 × 10−4 | 1.62 | 2.25 | |
| myo−inositol | 1.39 | 2.62 × 10−6 | 1.46 | 1.86 | |
| mannitol | 1.23 | 7.77 × 10−6 | −1.97 | −1.59 | |
| altronic acid | 1.22 | 9.74 × 10−6 | −2.06 | −3.46 | |
| mannose | 0.89 | 0.02 | − | −4.90 | |
| arabinose | 0.88 | 0.01 | −5.06 | −5.97 | |
| talopyranose | 0.92 | 9.15 × 10−3 | −7.96 | −6.09 | |
| galactonic acid | 1.23 | 1.58 × 10−11 | − | − | |
| acidic compounds | acetic acid | 1.24 | 3.93 × 10−12 | 7.09 | 6.74 |
| ascorbic acid | 1.29 | 1.66 × 10−7 | 6.86 | 5.34 | |
| malonic acid | 1.86 | 2.33 × 10−4 | 2.18 | 1.87 | |
| citric acid | 1.32 | 1.23 × 10−3 | 1.18 | 1.43 | |
| succinic acid | 1.14 | 9.15 × 10−3 | 1.54 | 1.04 | |
| oxalic acid | 1.66 | 6.62 × 10−4 | 1.38 | −1.08 | |
| phosphoric acid | 1.43 | 0.04 | 1.62 | −1.34 | |
| lactic acid | 1.83 | 1.92 × 10−5 | 1.17 | −1.41 | |
| isovaleric acid | 1.13 | 1.15 × 10−3 | −1.37 | −1.72 | |
| malic acid | 1.83 | 2.56 × 10−6 | −1.36 | −2.26 | |
| amino acids and peptides | glutamic acid 5-benzyl ester | 1.05 | 9.37 × 10−4 | 21.65 | 19.00 |
| trp−asp | 1.23 | 2.72 × 10−4 | 8.40 | 5.74 | |
| ornithine | 1.16 | 1.75 × 10−5 | 2.94 | 3.41 | |
| proline | 0.96 | 4.51 × 10−3 | 2.71 | 3.00 | |
| aspartic acid | 1.24 | 1.04 × 10−9 | 2.61 | 2.92 | |
| 2−aminobutyric acid | 1.11 | 1.98 × 10−4 | 2.29 | 2.61 | |
| lysine | 1.67 | 4.50 × 10−4 | 1.10 | 2.59 | |
| isoleucine | 1.18 | 5.68 × 10−6 | 2.26 | 2.57 | |
| glutamic acid | 1.22 | 9.45 × 10−6 | 2.12 | 2.53 | |
| alanine | 1.15 | 1.84 × 10−5 | 2.29 | 2.50 | |
| tyr-pro | 0.83 | 3.23 × 10−2 | 2.64 | 2.46 | |
| leu-glu | 1.30 | 8.52 × 10−6 | 3.00 | 2.33 | |
| glycine | 1.19 | 1.22 × 10−6 | 1.60 | 1.72 | |
| serin | 1.31 | 2.17 × 10−4 | 1.25 | 1.48 | |
| tryptophan | 1.10 | 1.68 × 10−4 | 1.37 | 1.44 | |
| glu-val | 1.32 | 3.69 × 10−2 | 2.57 | 1.05 | |
| pro-arg | 1.13 | 4.88 × 10−2 | −1.21 | −3.16 | |
| asparagine | 1.62 | 8.61 × 10−3 | 1.34 | −4.29 | |
| cyclo(his−pro) | 1.11 | 6.26 × 10−5 | − | − | |
| alcohols | 1,2-butanediol | 1.92 | 1.76 × 10−12 | 8.36 | 67.38 |
| 2-butanol | 1.49 | 4.76 × 10−3 | 3.69 | 6.16 | |
| 1,3-butanediol | 1.31 | 2.09 × 10−6 | 7.58 | 5.46 | |
| glycerol | 1.21 | 2.14 × 10−5 | 1.71 | 2.02 | |
| isopropenylethyl alcohol | 1.35 | 2.06 × 10−5 | 2.15 | 1.67 | |
| ethanol | 1.48 | 2.46 × 10−4 | 2.09 | 1.59 | |
| 2,3-butanediol | 1.34 | 1.21 × 10−4 | 1.61 | 1.33 | |
| 2-ethyl hexanol | 2.02 | 2.32 × 10−7 | −2.03 | 1.05 | |
| 1-pentanol | 1.93 | 2.23 × 10−4 | 1.19 | −1.14 | |
| 1-octen-3-ol | 1.46 | 5.54 × 10−6 | −1.20 | −1.57 | |
| isobutanol | 1.24 | 1.07 × 10−9 | −3.25 | −2.62 | |
| 1-penten-3-ol | 1.19 | 1.06 × 10−7 | −3.28 | −3.49 | |
| isopentyl alcohol | 1.23 | 3.22 × 10−13 | −5.01 | −4.93 | |
| ketones | 2,3-butanedione | 1.21 | 7.17 × 10−9 | 5.07 | 5.49 |
| 2,3-pentanedione | 1.29 | 1.08 × 10−5 | 4.58 | 3.40 | |
| acetyl acetophenone | 1.36 | 6.85 × 10−4 | 1.11 | 1.27 | |
| acetoin | 1.27 | 1.32 × 10−5 | −1.36 | −1.69 | |
| 2-butanone | 1.15 | 1.75 × 10−5 | −2.21 | −1.99 | |
| acetone | 1.18 | 3.79 × 10−7 | −3.20 | −3.12 | |
| aldehydes | 3-methylbutanal | 1.08 | 3.05 × 10−4 | 1.65 | 1.77 |
| hexanal | 1.77 | 2.61 × 10−7 | 2.96 | 1.49 | |
| 2-methylbutanal | 1.45 | 2.92 × 10−2 | −1.12 | 1.31 | |
| phenylacetaldehyde | 1.20 | 9.06 × 10−8 | ++ | + | |
| esters | acetic acid ethyl ester | 1.26 | 1.00 × 10−6 | 4.18 | 5.26 |
| lactic acid ethyl ester | 1.37 | 2.05 × 10−11 | 3.48 | 2.69 | |
| isoflavones | glycitein | 1.00 | 2.14 × 10−3 | −4.37 | −7.78 |
| genistein | 1.33 | 8.49 × 10−6 | −1.83 | −4.69 | |
| daidzein | 1.16 | 3.15 × 10−5 | −2.04 | −2.77 | |
| other | isovaleronitrile | 1.80 | 5.65 × 10−5 | 1.01 | 1.73 |
| uracil | 1.15 | 2.45 × 10−5 | 1.33 | 1.40 | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lee, S.; Kim, D.-S.; Son, Y.; Le, H.-G.; Jo, S.W.; Lee, J.; Song, Y.; Kim, H.-J. Effects of Salt Treatment Time on the Metabolites, Microbial Composition, and Quality Characteristics of the Soy Sauce Moromi Extract. Foods 2022, 11, 63. https://doi.org/10.3390/foods11010063
Lee S, Kim D-S, Son Y, Le H-G, Jo SW, Lee J, Song Y, Kim H-J. Effects of Salt Treatment Time on the Metabolites, Microbial Composition, and Quality Characteristics of the Soy Sauce Moromi Extract. Foods. 2022; 11(1):63. https://doi.org/10.3390/foods11010063
Chicago/Turabian StyleLee, Sun, Dong-Shin Kim, Yejin Son, Huong-Giang Le, Seung Wha Jo, Jungmi Lee, Yeji Song, and Hyun-Jin Kim. 2022. "Effects of Salt Treatment Time on the Metabolites, Microbial Composition, and Quality Characteristics of the Soy Sauce Moromi Extract" Foods 11, no. 1: 63. https://doi.org/10.3390/foods11010063
APA StyleLee, S., Kim, D.-S., Son, Y., Le, H.-G., Jo, S. W., Lee, J., Song, Y., & Kim, H.-J. (2022). Effects of Salt Treatment Time on the Metabolites, Microbial Composition, and Quality Characteristics of the Soy Sauce Moromi Extract. Foods, 11(1), 63. https://doi.org/10.3390/foods11010063






