Intestinal Organoids: New Tools to Comprehend the Virulence of Bacterial Foodborne Pathogens
Abstract
:1. Introduction
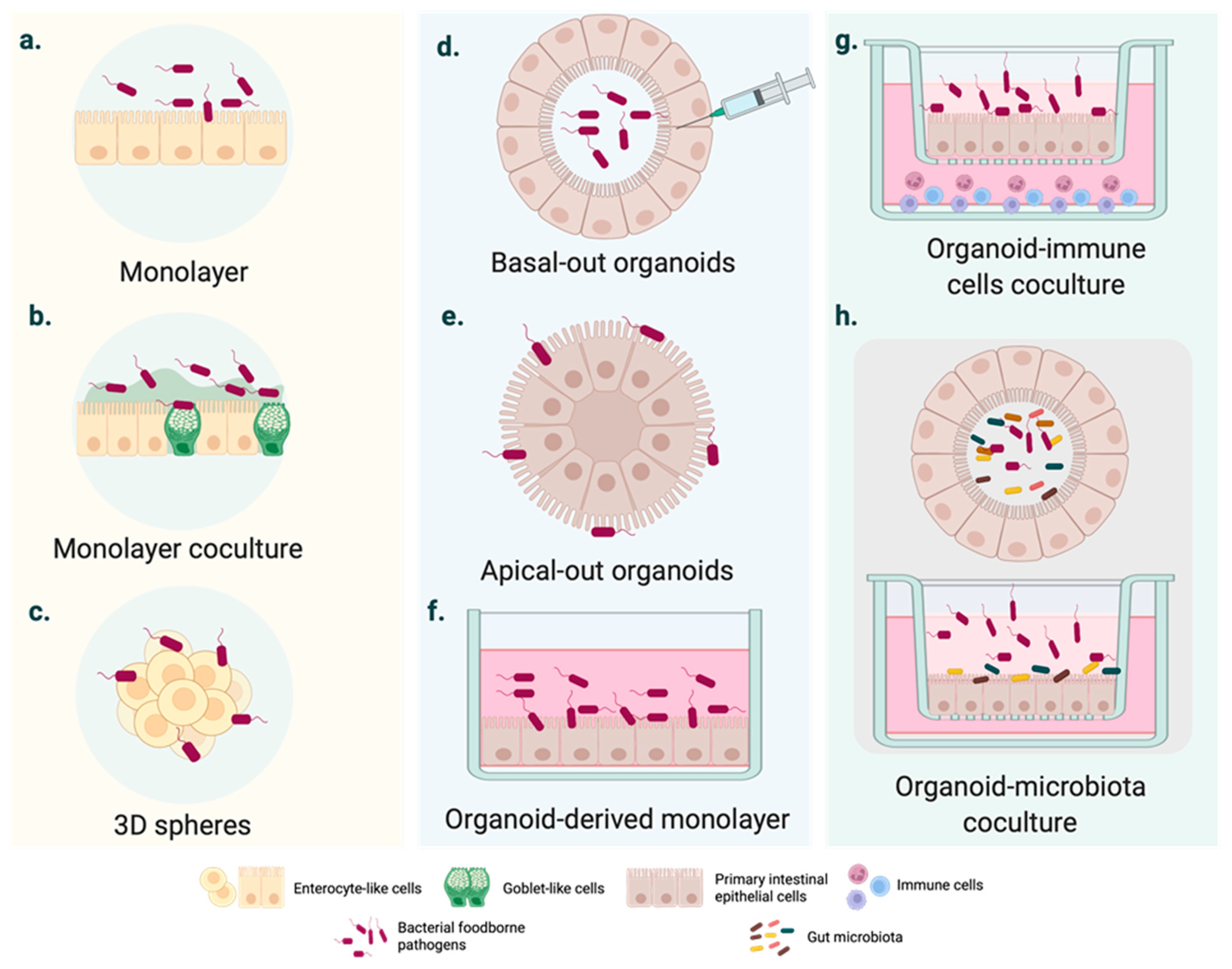
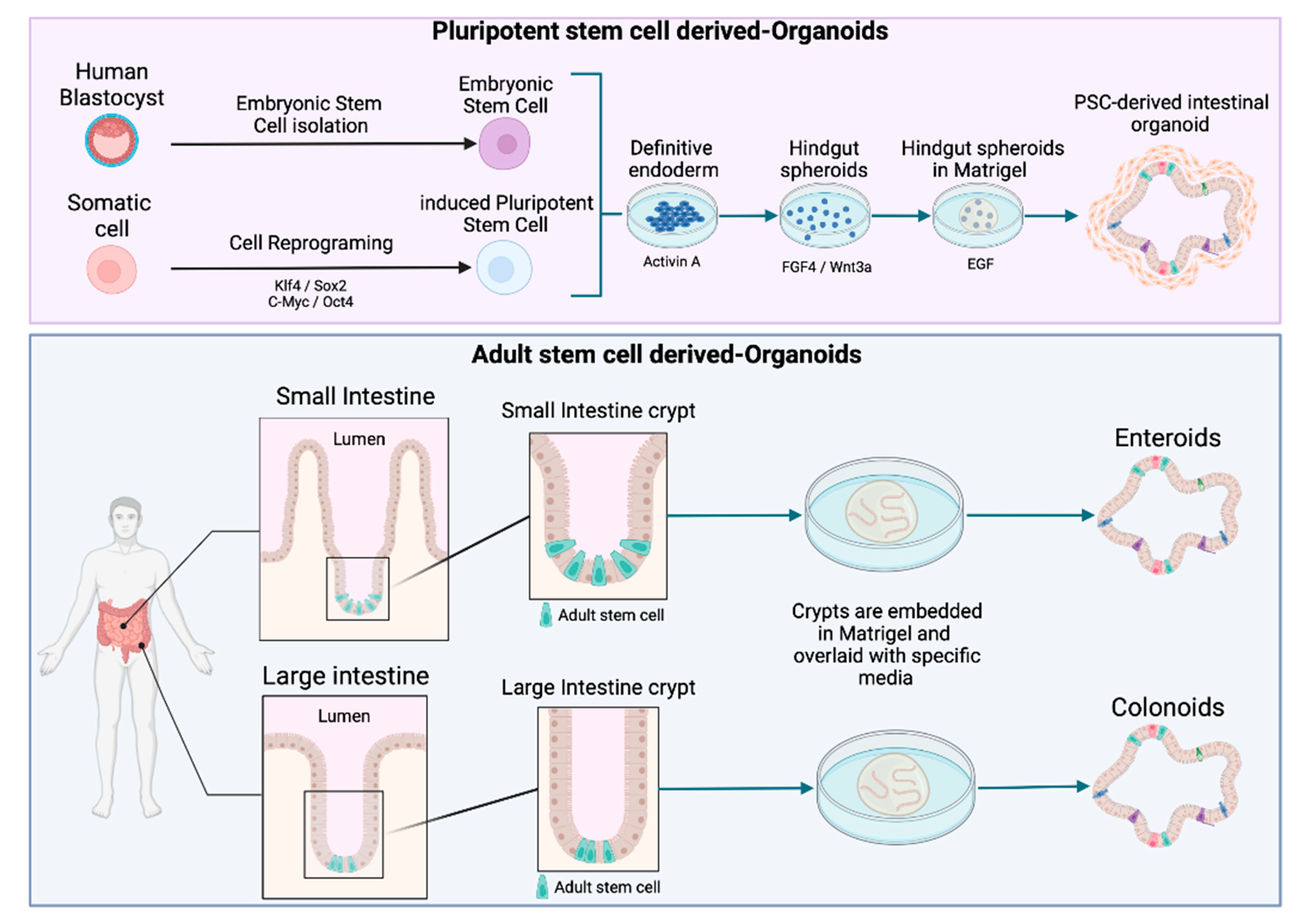
2. Moving from Cell Lines to Intestinal Organoids
3. Using Organoids to Explore the Cell and Tissue Tropism of FBPs
4. Organoids for Investigating the Host Immune Response Following Foodborne Infection
5. Organoids for Studying the Virulence Mechanisms of FBPs
6. Using Organoids to Investigate the Anti-FBP Activities of Probiotic (-like) Bacterial Strains
7. Current Challenges and Future Prospects
8. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- WHO. WHO Estimates of the Global Burden of Foodborne Diseases; World Health Organization: Geneva, Switzerland, 2015; p. 254. [Google Scholar]
- WHO. The Burden of Foodborne Diseases in the WHO European Region; World Health Organization: Copenhagen, Danemark, 2017. [Google Scholar]
- EFSA. EFSA Explains Zoonotic Diseases: Salmonella. Available online: https://data.europa.eu/doi/10.2805/61217 (accessed on 10 November 2021).
- Newell, D.G.; Koopmans, M.; Verhoef, L.; Duizer, E.; Aidara-Kane, A.; Sprong, H.; Opsteegh, M.; Langelaar, M.; Jacobson, A.; Scheutz, F.; et al. Food-borne diseases—The challenges of 20years ago still persist while new ones continue to emerge. Int. J. Food Microbiol. 2010, 139, S3–S15. [Google Scholar] [CrossRef]
- Drolia, R.; Bhunia, A.K. Crossing the intestinal barrier via Listeria adhesion protein and Internalin A. Trends Microbiol. 2019, 27, 408–425. [Google Scholar] [CrossRef]
- Abebe, E.; Gugsa, G.; Ahmed, M. Review on major food-borne zoonotic bacterial pathogens. J. Trop. Med. 2020, 2020, 4674235. [Google Scholar] [CrossRef] [PubMed]
- Sepe, L.P.; Hartl, K.; Iftekhar, A.; Berger, H.; Kumar, N.; Goosmann, C.; Chopra, S.; Schmidt, S.C.; Gurumurthy, R.K.; Meyer, T.F.; et al. Genotoxic effect of Salmonella Paratyphi A infection on human primary gallbladder cells. mBio 2020, 11, e01911-20. [Google Scholar] [CrossRef] [PubMed]
- Mayer, C.L.; Leibowitz, C.S.; Kurosawa, S.; Stearns-Kurosawa, D.J. Shiga toxins and the pathophysiology of hemolytic uremic syndrome in humans and animals. Toxins 2012, 4, 1261–1287. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McLauchlin, J.; Mitchell, R.T.; Smerdon, W.J.; Jewell, K. Listeria monocytogenes and listeriosis: A review of hazard characterisation for use in microbiological risk assessment of foods. Int. J. Food Microbiol. 2004, 92, 15–33. [Google Scholar] [CrossRef]
- Rees, J.H.; Soudain, S.E.; Gregson, N.A.; Hughes, R.A. Campylobacter jejuni infection and Guillain-Barre syndrome. N. Engl. J. Med. 1995, 333, 1374–1379. [Google Scholar] [CrossRef]
- Haddad, N.; Marce, C.; Magras, C.; Cappelier, J.M. An overview of methods used to clarify pathogenesis mechanisms of Campylobacter jejuni. J. Food Prot. 2010, 73, 786–802. [Google Scholar] [CrossRef]
- EFSA; ECDC. The European Union one health 2018 zoonoses report. EFSA J. 2019, 17, e05926. [Google Scholar] [CrossRef] [Green Version]
- Eskes, C. The usefulness of integrated strategy approaches in replacing animal experimentation. Ann. Ist. Super Sanita. 2019, 55, 400–404. [Google Scholar] [CrossRef]
- Simian, M.; Bissell, M.J. Organoids: A historical perspective of thinking in three dimensions. J. Cell Biol. 2017, 216, 31–40. [Google Scholar] [CrossRef]
- Ross, D.T.; Scherf, U.; Eisen, M.B.; Perou, C.M.; Rees, C.; Spellman, P.; Iyer, V.; Jeffrey, S.S.; Van de Rijn, M.; Waltham, M.; et al. Systematic variation in gene expression patterns in human cancer cell lines. Nat. Genet. 2000, 24, 227–235. [Google Scholar] [CrossRef] [PubMed]
- Langerholc, T.; Maragkoudakis, P.A.; Wollgast, J.; Gradisnik, L.; Cencic, A. Novel and established intestinal cell line models–An indispensable tool in food science and nutrition. Trends Food Sci. Technol. 2011, 22, S11–S20. [Google Scholar] [CrossRef] [PubMed]
- Ahmad, I.; Lamprokostopoulou, A.; Le Guyon, S.; Streck, E.; Barthel, M.; Peters, V.; Hardt, W.-D.; Römling, U. Complex c-di-GMP signaling networks mediate transition between virulence properties and biofilm formation in Salmonella enterica serovar Typhimurium. PLoS ONE 2011, 6, e28351. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bahrami, B.; Macfarlane, S.; Macfarlane, G.T. Induction of cytokine formation by human intestinal bacteria in gut epithelial cell lines. J. Appl. Microbiol. 2011, 110, 353–363. [Google Scholar] [CrossRef]
- Martinez-Argudo, I.; Jepson, M.A. Salmonella translocates across an in vitro M cell model independently of SPI-1 and SPI-2. Microbiology 2008, 154, 3887–3894. [Google Scholar] [CrossRef] [Green Version]
- Rodrigues, R.C.; Pocheron, A.-L.; Cappelier, J.-M.; Tresse, O.; Haddad, N. An adapted in vitro assay to assess Campylobacter jejuni interaction with intestinal epithelial cells: Taking into stimulation with TNFα. J. Microbiol. Met. 2018, 149, 67–72. [Google Scholar] [CrossRef]
- Zamora, C.Y.; Ward, E.M.; Kester, J.C.; Kelly Chen, W.L.; Velazquez, J.G.; Griffith, L.G.; Imperiali, B. Application of a gut-immune co-culture system for the study of N-glycan-dependent host-pathogen interactions of Campylobacter jejuni. Glycobiology 2020, 30, 374–381. [Google Scholar] [CrossRef]
- Nickerson, C.A.; Goodwin, T.J.; Terlonge, J.; Ott, C.M.; Buchanan, K.L.; Uicker, W.C.; Emami, K.; LeBlanc, C.L.; Ramamurthy, R.; Clarke, M.S.; et al. Three-dimensional tissue assemblies: Novel models for the study of Salmonella enterica serovar Typhimurium pathogenesis. Infect. Immun. 2001, 69, 7106–7120. [Google Scholar] [CrossRef] [Green Version]
- Höner zu Bentrup, K.; Ramamurthy, R.; Ott, C.M.; Emami, K.; Nelman-Gonzalez, M.; Wilson, J.W.; Richter, E.G.; Goodwin, T.J.; Alexander, J.S.; Pierson, D.L.; et al. Three-dimensional organotypic models of human colonic epithelium to study the early stages of enteric salmonellosis. Microb. Infect. 2006, 8, 1813–1825. [Google Scholar] [CrossRef]
- Fujii, M.; Sato, T. Somatic cell-derived organoids as prototypes of human epithelial tissues and diseases. Nat. Mater. 2020, 20, 156–169. [Google Scholar] [CrossRef]
- Clevers, H. Modeling development and disease with organoids. Cell 2016, 165, 1586–1597. [Google Scholar] [CrossRef] [Green Version]
- Flatres, C.; Loffet, E.; Neunlist, M.; Mahe, M.M. From human pluripotent stem cells to custom-made intestinal organoids. Med. Sci. 2019, 35, 549–555. [Google Scholar] [CrossRef]
- Mahe, M.M.; Sundaram, N.; Watson, C.L.; Shroyer, N.F.; Helmrath, M.A. Establishment of human epithelial enteroids and colonoids from whole tissue and biopsy. J. Vis. Exp. 2015, 97, e52483. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Aguilar-Rojas, A.; Olivo-Marin, J.-C.; Guillen, N. Human intestinal models to study interactions between intestine and microbes. Open Biol. 2020, 10, 200199. [Google Scholar] [CrossRef]
- Ho, B.X.; Pek, N.M.Q.; Soh, B.-S. Disease modeling using 3D organoids derived from human induced pluripotent stem cells. Int. J. Mol. Sci. 2018, 19, 936. [Google Scholar] [CrossRef] [Green Version]
- Jensen, C.; Teng, Y. Is it time to start transitioning from 2D to 3D cell culture? Front. Mol. Biosci. 2020, 7, 33. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Barker, N. Adult intestinal stem cells: Critical drivers of epithelial homeostasis and regeneration. Nat. Rev. Mol. Cell Biol. 2014, 15, 19–33. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Barker, N.; van Es, J.H.; Kuipers, J.; Kujala, P.; van den Born, M.; Cozijnsen, M.; Haegebarth, A.; Korving, J.; Begthel, H.; Peters, P.J.; et al. Identification of stem cells in small intestine and colon by marker gene Lgr5. Nature 2007, 449, 1003–1007. [Google Scholar] [CrossRef] [PubMed]
- Sato, T.; Vries, R.G.; Snippert, H.J.; van de Wetering, M.; Barker, N.; Stange, D.E.; van Es, J.H.; Abo, A.; Kujala, P.; Peters, P.J.; et al. Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche. Nature 2009, 459, 262–265. [Google Scholar] [CrossRef]
- Dutta, D.; Clevers, H. Organoid culture systems to study host–pathogen interactions. Curr. Op. Immunol. 2017, 48, 15–22. [Google Scholar] [CrossRef]
- Forbester, J.L.; Lees, E.A.; Goulding, D.; Forrest, S.; Yeung, A.; Speak, A.; Clare, S.; Coomber, E.L.; Mukhopadhyay, S.; Kraiczy, J.; et al. Interleukin-22 promotes phagolysosomal fusion to induce protection against Salmonella enterica Typhimurium in human epithelial cells. Proc. Natl. Acad. Sci. USA 2018, 115, 10118–10123. [Google Scholar] [CrossRef] [Green Version]
- DesRochers, T.M.; Kimmerling, E.P.; Jandhyala, D.M.; El-Jouni, W.; Zhou, J.; Thorpe, C.M.; Leong, J.M.; Kaplan, D.L. Effects of Shiga toxin type 2 on a bioengineered three-dimensional model of human renal tissue. Infect. Immun. 2015, 83, 28–38. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Marquez, L.B.; Araoz, A.; Repetto, H.A.; Ibarra, F.R.; Silberstein, C. Effects of shiga toxin 2 on cellular regeneration mechanisms in primary and three-dimensional cultures of human renal tubular epithelial cells. Microb. Pathog. 2016, 99, 87–94. [Google Scholar] [CrossRef]
- Rozman, J.; Krajnc, M.; Ziherl, P. Collective cell mechanics of epithelial shells with organoid-like morphologies. Nat. Commun. 2020, 11, 3805. [Google Scholar] [CrossRef] [PubMed]
- Co, J.Y.; Margalef-Català, M.; Li, X.; Mah, A.T.; Kuo, C.J.; Monack, D.M.; Amieva, M.R. Controlling epithelial polarity: A human enteroid model for host-pathogen interactions. Cell Rep. 2019, 26, 2509–2520.e4. [Google Scholar] [CrossRef] [Green Version]
- Wang, A.Z.; Ojakian, G.K.; Nelson, W.J. Steps in the morphogenesis of a polarized epithelium. II. Disassembly and assembly of plasma membrane domains during reversal of epithelial cell polarity in multicellular epithelial (MDCK) cysts. J. Cell Sci. 1990, 95 Pt 1, 153–165. [Google Scholar] [CrossRef]
- VanDussen, K.L.; Marinshaw, J.M.; Shaikh, N.; Miyoshi, H.; Moon, C.; Tarr, P.I.; Ciorba, M.A.; Stappenbeck, T.d.S. Development of an enhanced human gastrointestinal epithelial culture system to facilitate patient-based assays. Gut 2015, 64, 911–920. [Google Scholar] [CrossRef] [Green Version]
- Noel, G.; Baetz, N.W.; Staab, J.F.; Donowitz, M.; Kovbasnjuk, O.; Pasetti, M.F.; Zachos, N.C. A primary human macrophage-enteroid co-culture model to investigate mucosal gut physiology and host-pathogen interactions. Sci. Rep. 2017, 7, 45270. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yuki, K.; Cheng, N.; Nakano, M.; Kuo, C.J. Organoid models of tumor immunology. Trends Immunol. 2020, 41, 652–664. [Google Scholar] [CrossRef]
- Rogoz, A.; Reis, B.S.; Karssemeijer, R.A.; Mucida, D. A 3-D enteroid-based model to study T-cell and epithelial cell interaction. J. Immunol. Methods 2015, 421, 89–95. [Google Scholar] [CrossRef] [Green Version]
- Barrila, J.; Crabbé, A.; Yang, J.; Franco, K.; Nydam, S.D.; Forsyth, R.J.; Davis, R.R.; Gangaraju, S.; Ott, C.M.; Coyne, C.B.; et al. Modeling host-pathogen interactions in the context of the microenvironment: Three-dimensional cell culture comes of age. Infect. Immun. 2018, 86, e00282-18. [Google Scholar] [CrossRef] [Green Version]
- Bartfeld, S. Modeling infectious diseases and host-microbe interactions in gastrointestinal organoids. Dev. Biol. 2016, 420, 262–270. [Google Scholar] [CrossRef] [PubMed]
- Han, X.; Mslati, M.A.; Davies, E.G.; Chen, Y.; Allaire, J.M.; Vallance, B.A. Creating a more perfect union: Modeling intestinal bacteria-epithelial interactions using organoids. Cell. Mol. Gastroenterol. Hepatol. 2021, 12, 769–782. [Google Scholar] [CrossRef]
- Kim, J.; Koo, B.-K.; Knoblich, J.A. Human organoids: Model systems for human biology and medicine. Nat. Rev. Mol. Cell Biol. 2020, 21, 571–584. [Google Scholar] [CrossRef] [PubMed]
- Yu, H.; Hasan, N.M.; In, J.G.; Estes, R.K.; Kovbasnjuk, O.; Zachos, N.C.; Donowitz, M. The contributions of human mini-intestines to the study of intestinal physiology and pathophysiology. Annu. Rev. Physiol. 2017, 79, 291–312. [Google Scholar] [CrossRef] [Green Version]
- Holly, M.K.; Han, X.; Zhao, E.J.; Crowley, S.M.; Allaire, J.M.; Knodler, L.A.; Vallance, B.A.; Smith, J.G. Salmonella enterica infection of murine and human enteroid-derived monolayers elicits differential activation of epithelial-intrinsic inflammasomes. Infect. Immun. 2020, 88, e00017-20. [Google Scholar] [CrossRef]
- In, J.; Foulke-Abel, J.; Zachos, N.C.; Hansen, A.M.; Kaper, J.B.; Bernstein, H.D.; Halushka, M.; Blutt, S.; Estes, M.K.; Donowitz, M.; et al. Enterohemorrhagic Escherichia coli reduce mucus and intermicrovillar bridges in human stem cell-derived colonoids. Cell. Mol. Gastroenterol. Hepatol. 2016, 2, 48–62.e3. [Google Scholar] [CrossRef] [Green Version]
- Karve, S.S.; Pradhan, S.; Ward, D.V.; Weiss, A.A. Intestinal organoids model human responses to infection by commensal and Shiga toxin producing Escherichia coli. PLoS ONE 2017, 12, e0178966. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McCall, L.I.; Siqueira-Neto, J.L.; McKerrow, J.H. Location, location, location: Five facts about tissue tropism and pathogenesis. PLoS Pathog. 2016, 12, e1005519. [Google Scholar] [CrossRef] [Green Version]
- In, J.G.; Foulke-Abel, J.; Estes, M.K.; Zachos, N.C.; Kovbasnjuk, O.; Donowitz, M. Human mini-guts: New insights into intestinal physiology and host–pathogen interactions. Nat. Rev. Gastroenterol. Hepatol. 2016, 13, 633–642. [Google Scholar] [CrossRef] [Green Version]
- Harrington, S.M.; Dudley, E.G.; Nataro, J.P. Pathogenesis of enteroaggregative Escherichia coli infection. FEMS Microbiol. Lett. 2006, 254, 12–18. [Google Scholar] [CrossRef] [Green Version]
- Rajan, A.; Vela, L.; Zeng, X.-L.; Yu, X.; Shroyer, N.F.; Blutt, S.E.; Poole, N.M.; Carlin, L.G.; Nataro, J.P.; Estes, M.K.; et al. Novel segment- and host-specific patterns of enteroaggregative Escherichia coli adherence to human intestinal enteroids. mBio 2018, 9, e02419-17. [Google Scholar] [CrossRef] [Green Version]
- Koestler, B.J.; Ward, C.M.; Fisher, C.R.; Rajan, A.; Maresso, A.W.; Payne, S.M. Human intestinal enteroids as a model system of Shigella pathogenesis. Infect. Immun. 2019, 87, e00733-18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sansonetti, P.J. Rupture, invasion and inflammatory destruction of the intestinal barrier by Shigella, making sense of prokaryote-eukaryote cross-talks. FEMS Microbiol. Rev. 2001, 25, 3–14. [Google Scholar] [CrossRef]
- Hamon, M.; Bierne, H.; Cossart, P. Listeria monocytogenes: A multifaceted model. Nat. Rev. Microbiol. 2006, 4, 423–434. [Google Scholar] [CrossRef]
- Knodler, L.A.; Crowley, S.M.; Sham, H.P.; Yang, H.; Wrande, M.; Ma, C.; Ernst, R.K.; Steele-Mortimer, O.; Celli, J.; Vallance, B.A. Noncanonical inflammasome activation of Caspase-4/Caspase-11 mediates epithelial defenses against enteric bacterial pathogens. Cell Host Microb. 2014, 16, 249–256. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pentecost, M.; Kumaran, J.; Ghosh, P.; Amieva, M.R. Listeria monocytogenes Internalin B activates junctional endocytosis to accelerate intestinal invasion. PLoS Pathog. 2010, 6, e1000900. [Google Scholar] [CrossRef] [PubMed]
- Galán, J.E. Salmonella interactions with host cells: Type III secretion at work. Annu. Rev. Cell Dev. Biol. 2001, 17, 53–86. [Google Scholar] [CrossRef] [Green Version]
- Johansson, M.E.; Phillipson, M.; Petersson, J.; Velcich, A.; Holm, L.; Hansson, G.C. The inner of the two Muc2 mucin-dependent mucus layers in colon is devoid of bacteria. Proc. Natl. Acad. Sci. USA 2008, 105, 15064–15069. [Google Scholar] [CrossRef] [Green Version]
- Ranganathan, S.; Doucet, M.; Grassel, C.L.; Delaine-Elias, B.; Zachos, N.C.; Barry, E.M. Evaluating Shigella flexneri pathogenesis in the human enteroid model. Infect. Immun. 2019, 87, e00740-18. [Google Scholar] [CrossRef] [Green Version]
- Rey, C.; Chang, Y.-Y.; Latour-Lambert, P.; Varet, H.; Proux, C.; Legendre, R.; Coppée, J.-Y.; Enninga, J. Transcytosis subversion by M cell-to-enterocyte spread promotes Shigella flexneri and Listeria monocytogenes intracellular bacterial dissemination. PLoS Pathog. 2020, 16, e1008446. [Google Scholar] [CrossRef] [Green Version]
- Chiba, S.; Nagai, T.; Hayashi, T.; Baba, Y.; Nagai, S.; Koyasu, S. Listerial invasion protein internalin B promotes entry into ileal Peyer’s patches in vivo. Microbiol. Immunol. 2011, 55, 123–129. [Google Scholar] [CrossRef]
- Jones, B.D.; Ghori, N.; Falkow, S. Salmonella Typhimurium initiates murine infection by penetrating and destroying the specialized epithelial M cells of the Peyer’s patches. J. Exp. Med. 1994, 180, 15–23. [Google Scholar] [CrossRef] [Green Version]
- Mabbott, N.A.; Donaldson, D.S.; Ohno, H.; Williams, I.R.; Mahajan, A. Microfold (M) cells: Important immunosurveillance posts in the intestinal epithelium. Mucosal Immunol. 2013, 6, 666–677. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rouch, J.D.; Scott, A.; Lei, N.Y.; Solorzano-Vargas, R.S.; Wang, J.; Hanson, E.M.; Kobayashi, M.; Lewis, M.; Stelzner, M.G.; Dunn, J.C.Y.; et al. Development of functional Microfold (M) cells from intestinal stem cells in primary human enteroids. PLoS ONE 2016, 11, e0148216. [Google Scholar] [CrossRef] [Green Version]
- Tahoun, A.; Mahajan, S.; Paxton, E.; Malterer, G.; Donaldson, D.S.; Wang, D.; Tan, A.; Gillespie, T.L.; O’Shea, M.; Roe, A.J.; et al. Salmonella transforms follicle-associated epithelial cells into M cells to promote intestinal invasion. Cell Host Microb. 2012, 12, 645–656. [Google Scholar] [CrossRef] [Green Version]
- Wood, M.B.; Rios, D.; Williams, I.R. TNF-α augments RANKL-dependent intestinal M cell differentiation in enteroid cultures. Am. J. Physiol. Cell. Physiol. 2016, 311, C498–C507. [Google Scholar] [CrossRef] [Green Version]
- Scanu, T.; Spaapen, R.M.; Bakker, J.M.; Pratap, C.B.; Wu, L.-E.; Hofland, I.; Broeks, A.; Shukla, V.K.; Kumar, M.; Janssen, H.; et al. Salmonella manipulation of host signaling pathways provokes cellular transformation associated with gallbladder carcinoma. Cell Host Microb. 2015, 17, 763–774. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Forbester, J.L.; Goulding, D.; Vallier, L.; Hannan, N.; Hale, C.; Pickard, D.; Mukhopadhyay, S.; Dougan, G. Interaction of Salmonella enterica serovar Typhimurium with intestinal organoids derived from human induced pluripotent stem cells. Infect. Immun. 2015, 83, 2926–2934. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zheng, D.; Liwinski, T.; Elinav, E. Inflammasome activation and regulation: Toward a better understanding of complex mechanisms. Cell Discov. 2020, 6, 36. [Google Scholar] [CrossRef]
- Crowley, S.M.; Han, X.; Allaire, J.M.; Stahl, M.; Rauch, I.; Knodler, L.A.; Vallance, B.A. Intestinal restriction of Salmonella Typhimurium requires caspase-1 and caspase-11 epithelial intrinsic inflammasomes. PLoS Pathog. 2020, 16, e1008498. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Y.-G.; Wu, S.; Xia, Y.; Sun, J. Salmonella-infected crypt-derived intestinal organoid culture system for host-bacterial interactions. Physiol. Rep. 2014, 2, e12147. [Google Scholar] [CrossRef] [Green Version]
- Singer, M.; Sansonetti, P.J. IL-8 is a key chemokine regulating neutrophil recruitment in a new mouse model of Shigella-induced colitis. J. Immunol. 2004, 173, 4197–4206. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wilson, S.S.; Tocchi, A.; Holly, M.K.; Parks, W.C.; Smith, J.G. A small intestinal organoid model of non-invasive enteric pathogen–epithelial cell interactions. Mucosal Immunol. 2015, 8, 352–361. [Google Scholar] [CrossRef] [PubMed]
- Geiser, P.; Di Martino, M.L.; Ventayol, P.S.; Eriksson, J.; Sima, E.; Al-Saffar, A.K.; Ahl, D.; Phillipson, M.; Webb, D.-L.; Sundbom, M.; et al. Salmonella enterica serovar Typhimurium exploits cycling through epithelial cells to colonize human and murine enteroids. mBio 2021, 12, e02684-20. [Google Scholar] [CrossRef]
- Verma, S.; Prescott, R.A.; Ingano, L.; Nickerson, K.P.; Hill, E.; Faherty, C.S.; Fasano, A.; Senger, S.; Cherayil, B.J. The YrbE phospholipid transporter of Salmonella enterica serovar Typhi regulates the expression of flagellin and influences motility, adhesion and induction of epithelial inflammatory responses. Gut Microb. 2019, 11, 526–538. [Google Scholar] [CrossRef]
- Tse, C.; In, J.G.; Yin, J.; Donowitz, M.; Doucet, M.; Foulke-Abel, J.; Ruiz-Perez, F.; Nataro, J.P.; Zachos, N.C.; Kaper, J.B.; et al. Enterohemorrhagic E. coli (EHEC)—secreted serine protease EspP stimulates electrogenic ion transport in human colonoid monolayers. Toxins 2018, 10, 351. [Google Scholar] [CrossRef] [Green Version]
- Xue, R.-Y.; Liu, C.; Xiao, Q.-T.; Sun, S.; Zou, Q.-M.; Li, H.-B. HtrA family proteases of bacterial pathogens: Pros and cons for their therapeutic use. Clin. Microbiol. Infect. 2021, 27, 559–564. [Google Scholar] [CrossRef]
- Nickerson, K.P.; Senger, S.; Zhang, Y.; Lima, R.; Patel, S.; Ingano, L.; Flavahan, W.A.; Kumar, D.K.V.; Fraser, C.M.; Faherty, C.S.; et al. Salmonella Typhi colonization provokes extensive transcriptional changes aimed at evading host mucosal immune defense during early infection of human intestinal tissue. EBioMedicine 2018, 31, 92–109. [Google Scholar] [CrossRef] [Green Version]
- Roodsant, T.; Navis, M.; Aknouch, I.; Renes, I.B.; van Elburg, R.M.; Pajkrt, D.; Wolthers, K.C.; Schultsz, C.; van der Ark, K.C.H.; Sridhar, A.; et al. A human 2D primary organoid-derived epithelial monolayer model to study host-pathogen interaction in the small intestine. Front. Cell. Infect. Microbiol. 2020, 10, 272. [Google Scholar] [CrossRef] [PubMed]
- Huang, J.; Zhou, C.; Zhou, G.; Li, H.; Ye, K. Effect of Listeria monocytogenes on intestinal stem cells in the co-culture model of small intestinal organoids. Microb. Pathog. 2021, 153, 104776. [Google Scholar] [CrossRef]
- Allali, I.; Delgado, S.; Marron, P.I.; Astudillo, A.; Yeh, J.J.; Ghazal, H.; Amzazi, S.; Keku, T.; Azcarate-Peril, M.A. Gut microbiome compositional and functional differences between tumor and non-tumor adjacent tissues from cohorts from the US and Spain. Gut Microb. 2015, 6, 161–172. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gradel, K.O.; Nielsen, H.L.; Schønheyder, H.C.; Ejlertsen, T.; Kristensen, B.; Nielsen, H. Increased short- and long-term risk of inflammatory bowel disease after Salmonella or Campylobacter gastroenteritis. Gastroenterology 2009, 137, 495–501. [Google Scholar] [CrossRef]
- Newman, A.; Lambert, J.R. Campylobacter jejuni causing flare-up in inflammatory bowel disease. Lancet 1980, 2, 919. [Google Scholar] [CrossRef]
- Warren, R.L.; Freeman, D.J.; Pleasance, S.; Watson, P.; Moore, R.A.; Cochrane, K.; Allen-Vercoe, E.; Holt, R.A. Co-occurrence of anaerobic bacteria in colorectal carcinomas. Microbiome 2013, 1, 16. [Google Scholar] [CrossRef] [Green Version]
- He, Z.; Gharaibeh, R.Z.; Newsome, R.C.; Pope, J.L.; Dougherty, M.W.; Tomkovich, S.; Pons, B.; Mirey, G.; Vignard, J.; Hendrixson, D.R.; et al. Campylobacter jejuni promotes colorectal tumorigenesis through the action of cytolethal distending toxin. Gut 2019, 68, 289–300. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vejborg, R.M.; Friis, C.; Hancock, V.; Schembri, M.A.; Klemm, P. A virulent parent with probiotic progeny: Comparative genomics of Escherichia coli strains CFT073, Nissle 1917 and ABU 83972. Mol. Genet. Genom. 2010, 283, 469–484. [Google Scholar] [CrossRef]
- Pradhan, S.; Weiss, A.A. Probiotic properties of Escherichia coli Nissle in human intestinal organoids. mBio 2020, 11, e01470-20. [Google Scholar] [CrossRef] [PubMed]
- Lu, X.; Xie, S.; Ye, L.; Zhu, L.; Yu, Q. Lactobacillus protects against S. Typhimurium–induced intestinal inflammation by determining the fate of epithelial proliferation and differentiation. Mol. Nutr. Food Res. 2020, 64, 1900655. [Google Scholar] [CrossRef]
- Lu, R.; Shang, M.; Zhang, Y.G.; Jiao, Y.; Xia, Y.; Garrett, S.; Bakke, D.; Bäuerl, C.; Martinez, G.P.; Kim, C.H.; et al. Lactic acid bacteria isolated from korean kimchi activate the vitamin D receptor-autophagy signaling pathways. Inflamm. Bowel. Dis. 2020, 26, 1199–1211. [Google Scholar] [CrossRef]
- Beaumont, M.; Blanc, F.; Cherbuy, C.; Egidy, G.; Giuffra, E.; Lacroix-Lamandé, E.; Wiedemann, A. Intestinal organoids in farm animals. Vet. Res. 2021, 52, 33. [Google Scholar] [CrossRef] [PubMed]
- Derricott, H.; Luu, L.; Fong, W.Y.; Hartley, C.S.; Johnston, L.J.; Armstrong, S.D.; Randle, N.; Duckworth, C.A.; Campbell, B.J.; Wastling, J.M.; et al. Developing a 3D intestinal epithelium model for livestock species. Cell Tissue Res. 2019, 375, 409–424. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Williamson, I.A.; Arnold, J.W.; Samsa, L.A.; Gaynor, L.; DiSalvo, M.; Cocchiaro, J.L.; Carroll, I.; Azcarate-Peril, M.A.; Rawls, J.F.; Allbritton, N.L.; et al. A high-throughput organoid microinjection platform to study gastrointestinal microbiota and luminal physiology. Cell. Mol. Gastroenterol. Hepatol. 2018, 6, 301–319. [Google Scholar] [CrossRef] [Green Version]
- Perrone, F.; Zilbauer, M. Biobanking of human gut organoids for translational research. Exp. Mol. Med. 2021, 53, 1451–1458. [Google Scholar] [CrossRef]
- Pasch, C.A.; Favreau, P.F.; Yueh, A.E.; Babiarz, C.P.; Gillette, A.A.; Sharick, J.T.; Karim, M.R.; Nickel, K.P.; DeZeeuw, A.K.; Sprackling, C.M.; et al. Patient-derived cancer organoid cultures to predict sensitivity to chemotherapy and radiation. Clin. Cancer Res. 2019, 25, 5376–5387. [Google Scholar] [CrossRef]
- Vlachogiannis, G.; Hedayat, S.; Vatsiou, A.; Jamin, Y.; Fernández-Mateos, J.; Khan, K.; Lampis, A.; Eason, K.; Huntingford, I.; Burke, R.; et al. Patient-derived organoids model treatment response of metastatic gastrointestinal cancers. Science 2018, 359, 920–926. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yan, H.H.N.; Siu, H.C.; Law, S.; Ho, S.L.; Yue, S.S.K.; Tsui, W.Y.; Chan, D.; Chan, A.S.; Ma, S.; Lam, K.O.; et al. A comprehensive human gastric cancer organoid biobank captures tumor subtype heterogeneity and enables therapeutic screening. Cell. Stem Cell 2018, 23, 882–897. [Google Scholar] [CrossRef] [Green Version]
| Comparison | 2D Monolayer Cell Culture | 3D Cell Culture |
|---|---|---|
| Cell differentiation into enterocyte or goblet cell | ✓ | ✓ |
| Cell differentiation into Paneth cell and enteroendocrine lineages | - | ✓ |
| Easily accessible to the apical side of cells | ✓ | - |
| Include immune, nerve, or vascular cells | - | - |
| Cell polarisation | ✓ | ✓ |
| Formation of cell–cell tight junctions | ✓ | ✓ |
| Development of villus-like and crypt-like structures—three-dimensional architecture | - | ✓ |
| Expanded indefinitely | ✓ (if derived from tumour cells) | ✓ |
| Cryopreservation for long-term storage | ✓ (if derived from tumour cells) | ✓ |
| Reproducibility | +++ | + |
| Cost | + | +++ |
| Advantages | Disadvantages |
|---|---|
| Better mimic endogenous tissues, including organization and spontaneous differentiation of multiple cell types into physiologically relevant 3-D structures, expression and localization of tight junctions, mucus production, polarity, gene expression, cell viability and proliferation, cytokine production | Heterogeneity in size, shape, and viability of organoids within a culture and across different samples, owing to the diversity of individuals and protocols. Protocols for organoid establishment and quality control are not globally standardized. |
| Contain highly polarized cells that differentiate into the cell lineages of the tissue of origin, i.e., intestinal organoids contain fully mature goblet cells, enterocytes, Paneth cells, and enteroendocrine cells. | Lack of neural innervation, immune cells, vasculature, and amicrobiome → coculture systems with other cell types are not firmly established. Lack of mechanical stress (peristalsis) and luminal and basolateral flow → towards organoid on chip. |
| Personalization: induced pluripotent stem cells and organoids can be obtained from individuals | Infection experiments: closed system that represents a nonphysiological route for pathogens that infect via the apical/luminal side, i.e., the luminal side is inaccessible without microinjection or disruption of organoid polarization. Microinjection remains a technical challenge. |
| Genetic engineering: most modern genetic engineering tools can be applied to induced pluripotent stem cells or directly to organoid systems | Relatively costly: organoids cost less than animal models, but they are relatively expensive compared to traditional cell lines (mainly due to medium composition with growth factors and volume required for culturing large numbers of cells). |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Aguirre Garcia, M.; Hillion, K.; Cappelier, J.-M.; Neunlist, M.; Mahe, M.M.; Haddad, N. Intestinal Organoids: New Tools to Comprehend the Virulence of Bacterial Foodborne Pathogens. Foods 2022, 11, 108. https://doi.org/10.3390/foods11010108
Aguirre Garcia M, Hillion K, Cappelier J-M, Neunlist M, Mahe MM, Haddad N. Intestinal Organoids: New Tools to Comprehend the Virulence of Bacterial Foodborne Pathogens. Foods. 2022; 11(1):108. https://doi.org/10.3390/foods11010108
Chicago/Turabian StyleAguirre Garcia, Mayra, Killian Hillion, Jean-Michel Cappelier, Michel Neunlist, Maxime M. Mahe, and Nabila Haddad. 2022. "Intestinal Organoids: New Tools to Comprehend the Virulence of Bacterial Foodborne Pathogens" Foods 11, no. 1: 108. https://doi.org/10.3390/foods11010108
APA StyleAguirre Garcia, M., Hillion, K., Cappelier, J.-M., Neunlist, M., Mahe, M. M., & Haddad, N. (2022). Intestinal Organoids: New Tools to Comprehend the Virulence of Bacterial Foodborne Pathogens. Foods, 11(1), 108. https://doi.org/10.3390/foods11010108