Quantitative Determination of Nitrofurazone Metabolites in Animal-Derived Foods Based on a Background Fluorescence Quenching Immunochromatographic Assay
Abstract
:1. Introduction
2. Materials and Methods
2.1. Chemicals and Equipment
2.2. Preparation and Characterization of AuNPs-Labeled Antibody Immunoprobe
2.3. Preparation of bFQICA Strip
2.4. The Procedure of bFQICA for SEM
2.5. Standard Curves and Generation of QR-Code
2.6. Sample Pretreatment
2.7. Validation of bFQICA
3. Results and Discussion
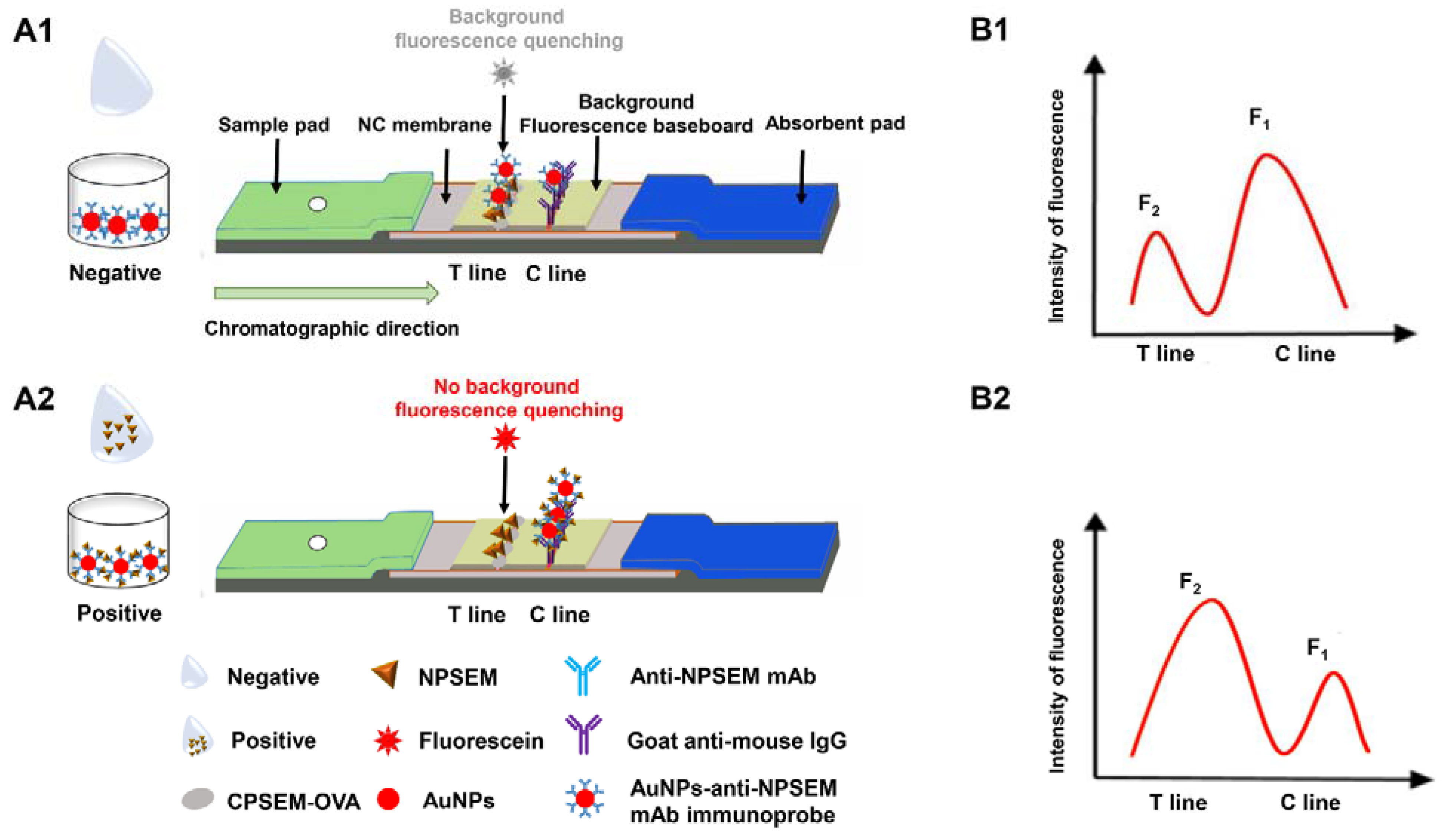
3.1. Principle of bFQICA for Quantification of SEM
3.2. Characterization of AuNPs
3.3. Optimization and Identification of AuNPs-Labeled Antibody Immunoprobe
3.4. Optimization of the bFQICA
3.5. Detectability
3.6. Specificity
3.7. Validation of bFQICA
3.7.1. Limit of Detection
3.7.2. Accuracy and Precision
3.8. Application in Field Samples
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
References
- Vass, M.; Hruska, K.; Franek, M. Nitrofuran antibiotics: A review on the application, prohibition and residual analysis. Vet. Med. 2008, 53, 469–500. [Google Scholar] [CrossRef] [Green Version]
- Liu, H.; Liang, D.P.; Hua, T.G.; She, Z.Y.; Deng, X.F.; Wu, S.Z.; Wu, B. Progress in analytical methods of nitrofuran antibiotics and their metabolites in food: A review. J. Food Saf. Qual. 2013, 4, 383–388. [Google Scholar]
- Kwon, J. Semicarbazide: Natural occurrence and uncertain evidence of its formation from food processing. Food Control. 2017, 72, 268–275. [Google Scholar] [CrossRef]
- Yang, W.Y.; Dong, J.X.; Shen, Y.D.; Yang, J.Y.; Wang, H.; Xu, Z.L.; Yang, X.X.; Sun, Y.M. Indirect competitive chemiluminescence enzyme immunoassay for furaltadone metabolite in Metapenaeus Ensis. Chin. J. Anal. Chem. 2012, 40, 1816–1821. [Google Scholar] [CrossRef]
- Zhao, H.X.; Guo, W.X.; Quan, W.N.; Jiang, J.Q.; Qu, B.C. Occurrence and levels of nitrofuran metabolites in sea cucumber from Dalian, China. Food Addit. Contam. A 2016, 33, 1672–1677. [Google Scholar] [CrossRef]
- European Commission. Regulation (EC) No 470/2009 of the European Parliament and of the council of 6 may 2009. Off. J. Eur. Union. 2009, L152, 11–22. [Google Scholar]
- Federal, R. Topical nitrofurans, extralabel animal drug use, order of prohibition. Fed. Reg. 2002, 67, 5470–5471. [Google Scholar]
- Announcement No.250, Ministry of Agriculture and Rural Affairs of the People’s Republic of China. 2019. Available online: http://extwprlegs1.fao.org/docs/pdf/chn192931.pdf (accessed on 8 July 2021). (In Chinese).
- European Commission. 2003/181/EC Commission decision of 13 march amending decision 2002/657/EC. Off. J. Eur. Union. 2003, L71, 17–18. [Google Scholar]
- Tang, T.; Wei, F.D.; Wang, X.; Ma, Y.J.; Song, Y.Y.; Ma, Y.S.; Song, Q.; Xu, G.H.; Cen, Y.; Hu, Q. Determination of semicarbazide in fish by molecularly imprinted stir bar sorptive extraction coupled with high performance liquid chromatography. J. Chromatog. B 2018, 1076, 8–14. [Google Scholar] [CrossRef] [PubMed]
- Sheng, L.Q.; Chen, M.M.; Chen, S.S.; Du, N.N.; Liu, Z.D.; Song, C.F.; Qiao, R. High-performance liquid chromatography with fluorescence detection for the determination of nitrofuran metabolites in pork muscle. Food Addit. Contam. A 2013, 30, 2114–2122. [Google Scholar] [CrossRef] [PubMed]
- Aldeek, F.; Hsieh, K.C.; Ugochukwu, O.N.; Gerard, G.; Hammack, W. Accurate quantitation and analysis of nitrofuran metabolites, chloramphenicol, and florfenicol in seafood by ultrahigh-performance liquid chromatography-tandem mass spectrometry: Method validation and regulatory samples. J. Agric. Food Chem. 2018, 66, 5018–5030. [Google Scholar] [CrossRef]
- Zhang, Y.; Qiao, H.; Chen, C.; Wang, Z.; Xia, X. Determination of nitrofurans metabolites residues in aquatic products by ultra-performance liquid chromatography-tandem mass spectrometry. Food Chem. 2016, 192, 612–617. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Z.W.; Wu, Y.P.; Li, X.W.; Wang, Y.Y.; Li, H.; Fu, Q.; Shan, Y.W.; Liu, T.H.; Xia, X. Multi-class method for the determination of nitroimidazoles, nitrofurans, and chloramphenicol in chicken muscle and egg by dispersive-solid phase extraction and ultra-high performance liquid chromatography-tandem mass spectrometry. Food Chem. 2017, 217, 182–190. [Google Scholar] [CrossRef] [PubMed]
- Valera-Tarifa, N.M.; Plaza-Bolaños, P.; Romero-González, R.; Martínez-Vidal, J.L.; Garrido-Frenich, A. Determination of nitrofuran metabolites in seafood by ultra-high performance liquid chromatography coupled to triple quadrupole tandem mass spectrometry. J. Food Compos. Anal. 2013, 30, 86–93. [Google Scholar] [CrossRef]
- Kulikovskii, A.V.; Gorlov, I.F.; Slozhenkina, M.I.; Vostrikova, N.L.; Kuznetsova, O.A. Determination of nitrofuran metabolites in muscular tissue by high-performance liquid chromatography with mass spectrometric detection. J. Anal. Chem. 2019, 74, 906–912. [Google Scholar] [CrossRef]
- Jiang, W.X.; Luo, P.J.; Wang, X.; Chen, X.; Zhao, Y.F.; Shi, W.; Wu, X.P.; Wu, Y.N.; Shen, J.Z. Development of an enzyme-linked immunosorbent assay for the detection of nitrofurantoin metabolite, 1-amino-hydantoin, in animal tissues. Food Control. 2012, 23, 20–25. [Google Scholar] [CrossRef]
- Sun, Q.; Luo, J.H.; Zhang, L.; Zhang, Z.H.; Le, T. Development of monoclonal antibody-based ultrasensitive enzyme-linked immunosorbent assay and fluorescence-linked immunosorbent assay for 1-aminohydantoin detection in aquatic animals. J. Pharmaceut. Biomed. 2018, 147, 417–424. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Liu, Y.; Wang, M.; Chen, Y.; Jiang, W. A multiplex immunochromatographic test using gold nanoparticles for the rapid and simultaneous detection of four nitrofuran metabolites in fish samples. Anal. Bioanal. Chem. 2018, 410, 223–233. [Google Scholar] [CrossRef]
- Sharma, D.; Lee, J.; Seo, J.; Shin, H.J.S. Development of a sensitive electrochemical enzymatic reaction-based cholesterol biosensor using nano-sized carbon interdigitated electrodes decorated with gold nanoparticles. Sensors 2017, 17, 2128. [Google Scholar] [CrossRef] [Green Version]
- Dykmana, L.; Khlebtsov, N. Gold Nanoparticles in Biomedical Applications: Recent Advances and Perspectives. Chem. Soc. Rev. 2012, 41, 2256–2282. [Google Scholar] [CrossRef]
- Dreaden, E.; Alkilany, A.; Huang, X.; Murphy, C.; El-Sayed, M. The Golden Age: Gold Nanoparticles for Biomedicine. Chem. Soc. Rev. 2012, 41, 2740–2779. [Google Scholar] [CrossRef] [Green Version]
- Grzelczak, M.; Liz-Marzan, L.; Klajn, R. Stimuli-Responsive Self-Assembly of Nanoparticles. Chem. Soc. Rev. 2019, 48, 1342–1361. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, X.J.; Xu, Y.Y.; Yu, J.S.; Li, J.T.; Zhou, X.L.; Wu, C.Y.; Ji, Q.L.; Ren, Y.; Wang, L.Q.; Huang, Z.Y.; et al. Antigen detection based on background fluorescence quenching immunochromatographic assay. Anal. Chim. Acta. 2014, 841, 44–50. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.; Tian, X.; Xu, L.; Li, J.; Li, X.; Wang, Y. Determination of aflatoxin m1 and chloramphenicol in milk based on background fluorescence quenching immunochromatographic assay. BioMed Res. Int. 2017, 2017, 8649314. [Google Scholar] [CrossRef] [PubMed]
- Wan, X.L.; Wan, X.; Tao, X.Q. Determination of 3-methyl-quinoxaline-2-carboxylic acid and quinoxaline-2-carboxylic acid in pork based on a background fluorescence quenching immunochromatographic assay. Anal. Sci. 2020, 36, 783–785. [Google Scholar] [CrossRef]
- Zhou, Y.; Pan, F.; Li, Y.; Zhang, Y.; Zhang, J.; Lu, S.; Ren, H.; Liu, Z. Colloidal Gold Probe-Based Immunochromatographic Assay for the Rapid Detection of Brevetoxins in Fishery Product Samples. Biosens. Bioelectron. 2009, 24, 2744–2747. [Google Scholar] [CrossRef]
- Wu, W.; Li, J.; Pan, D.; Li, J.; Song, S.; Rong, M.; Li, Z.; Gao, J.; Lu, J. Gold Nanoparticle-Based Enzyme-Linked Antibody-Aptamer Sandwich Assay for Detection of Salmonella Typhimurium. ACS Appl. Mater. Inter. 2014, 6, 16974–16981. [Google Scholar] [CrossRef] [PubMed]
- Sun, X.; Zhao, X.; Jian, T.; Zhou, J.; Chu, F.S. Preparation of gold-labeled antibody probe and its use in immunochromatography assay for detection of aflatoxin B1. Int. J. Food Microbiol. 2005, 99, 185–194. [Google Scholar]
- Peng, J.C.; Liu, X.D.; Ding, X.P.; Fu, Z.J.; Wang, Q.L. Evaluation of the particle diameter of colloidal gold and its distribution through visible spectroscopy. Bull. Acad. Mil. Med. Sci. 2000, 24, 211–212. [Google Scholar]
- Wang, K.; Kou, Y.; Wang, M.; Ma, X.; Wang, J. Determination of nitrofuran metabolites in fish by ultraperformance liquid chromatography-photodiode array detection with thermostatic ultrasound-assisted derivatization. ACS Omega 2020, 5, 18887–18893. [Google Scholar] [CrossRef]
- Points, J.; Burns, D.T.; Walker, M.J. Forensic issues in the analysis of trace nitrofuran veterinary residues in food of animal origin. Food Control. 2015, 50, 92–103. [Google Scholar] [CrossRef]
- Berlina, A.N.; Taranova, N.A.; Zherdev, A.V. Quantum dot-based lateral flow immunoassay for detection of chloramphenicol in milk. Anal. Bioanal. Chem. 2013, 405, 4997–5000. [Google Scholar] [CrossRef]
- Wang, L.; Wang, S.; Zhang, J.Y.; Liu, J.W.; Zhang, Y. Enzyme-linked immunosorbent assay and colloidal gold immunoassay for sulphamethazine residues in edible animal foods: Investigation of the effects of the analytical conditions and the sample matrix on assay performance. Anal. Bioanal. Chem. 2008, 390, 1619–1627. [Google Scholar] [CrossRef] [PubMed]
- Mitchell, J.S.; Lowe, T.E. Matrix effects on an antigen immobilized format for competitive enzyme immunoassay of salivary testosterone. J. Immunol. Methods 2009, 349, 61–66. [Google Scholar] [CrossRef]
- Cooper, K.M.; Caddell, A.; Elliott, C.T.; Kennedy, D.G. Production and characterisation of polyclonal antibodies to a derivative of 3-amino-2-oxazolidinone, a metabolite of the nitrofuran furazolidone. Anal. Chim. Acta 2004, 520, 79–86. [Google Scholar] [CrossRef]
- Lu, X.W.; Liang, X.L.; Dong, J.H.; Fang, Z.Y.; Zeng, L.W. Lateral flow biosensor for multiplex detection of nitrofuran metabolites based on functionalized magnetic beads. Anal. Bioanal. Chem. 2016, 408, 6703–6709. [Google Scholar] [CrossRef]
- Tao, Y.F.; Chen, D.M.; Wei, H.M.; Pan, Y.H.; Liu, Z.L.; Huang, L.L.; Wang, Y.L.; Xie, S.Y.; Yuan, Z.H. Development of an accelerated solvent extraction, ultrasonic derivatisation LC-MS/MS method for the determination of the marker residues of nitrofurans in freshwater fish. Food Addit. Contam. Part A 2012, 29, 736–745. [Google Scholar] [CrossRef] [PubMed]



| Characterization | Results | |
|---|---|---|
| AuNPs-anti-NPSEM mAb probes (1mL reaction system) | The particle size of AuNPs | 30 nm |
| pH | 8 (0.8% v/v K2CO3) | |
| Anti-NPSEM mAb (μg mL−1) | 2.55 | |
| Storage buffer | 0.02 M PB (0.5% BSA, 0.5% Triton X-100, 5% sucrose, 0.03% NaN3, pH 7.4) | |
| Optimum parameters of the established bFQICA (50 μL reaction system) | The dosage of AuNPs-anti-NPSEM mAb probe | 4 μL per well |
| AuNPs-anti-NPSEM mAb probe (μg mL−1) | 2.55 | |
| CPSEM-OVA (mg mL−1) | 0.17 | |
| Concentration of goat anti-mouse IgG (mg mL−1) | 0.33 | |
| rehydrated solution (μL) | 46 (0.02 PB) | |
| Analytical parameters of NPSEM standard curve | IC50 (μg L−1) | 0.19 |
| 20–80% inhibition (μg L−1) | 0.08–0.41 | |
| LODs (μg kg−1) | 0.09 (egg), 0.10 (chicken), 0.12 (fish), 0.15 (shrimp) |
| Samples | Spiked Concentration (μg kg−1) | Intra-Assay a | Inter-Assay b | ||
|---|---|---|---|---|---|
| Measure ± SD c (μg kg−1) | Recovery ± CV d (%) | Measure ± SD (μg kg−1) | Recovery ± CV (%) | ||
| Egg | 0.09 | 0.074 ± 0.007 | 82.2 ± 8.9 | 0.077 ± 0.008 | 85.8 ± 9.9 |
| 0.18 | 0.181 ± 0.012 | 100.6 ± 6.4 | 0.168 ± 0.017 | 93.4 ± 10.3 | |
| 0.36 | 0.302 ± 0.011 | 83.8 ± 3.8 | 0.323 ± 0.016 | 89.6 ± 5.1 | |
| 1.00 | 0.981 ± 0.037 | 98.1 ± 3.8 | 0.932 ± 0.049 | 93.2 ± 5.3 | |
| Chicken | 0.10 | 0.090 ± 0.006 | 90.1 ± 7.2 | 0.078 ± 0.007 | 78.1 ± 8.5 |
| 0.20 | 0.167 ± 0.015 | 83.5 ± 9.1 | 0.187 ± 0.012 | 93.6 ± 6.2 | |
| 0.40 | 0.418 ± 0.015 | 104.5 ± 3.6 | 0.420 ± 0.016 | 105.1 ± 3.9 | |
| 1.00 | 1.012 ± 0.050 | 101.2 ± 4.9 | 0.946 ± 0.069 | 94.6 ± 7.3 | |
| Fish | 0.12 | 0.091 ± 0.006 | 75.9 ± 6.9 | 0.101 ± 0.006 | 83.9 ± 6.0 |
| 0.24 | 0.212 ± 0.012 | 88.4 ± 5.8 | 0.211 ± 0.018 | 88.1 ± 8.6 | |
| 0.48 | 0.461 ± 0.034 | 96.0 ± 7.4 | 0.478 ± 0.045 | 99.6 ± 9.4 | |
| 1.00 | 0.858 ± 0.040 | 85.8 ± 4.7 | 0.916 ± 0.057 | 91.6 ± 6.2 | |
| Shrimp | 0.15 | 0.118 ± 0.012 | 78.4 ± 10.2 | 0.114 ± 0.013 | 75.7 ± 11.5 |
| 0.30 | 0.269 ± 0.017 | 89.8 ± 6.3 | 0.279 ± 0.025 | 92.9 ± 8.9 | |
| 0.60 | 0.555 ± 0.049 | 92.5 ± 8.8 | 0.609 ± 0.033 | 101.5 ± 5.4 | |
| 1.00 | 0.880 ± 0.032 | 88.0 ± 3.6 | 0.900 ± 0.060 | 90.0 ± 6.7 | |
| Sample | No. | bFQICA, Mean ± SD (μg kg−1) | LC-MS/MS, Mean ± SD (μg kg−1) |
|---|---|---|---|
| Egg | 1–9 | ND a | ND |
| 10 | ND | ND | |
| 11–20 | ND | ND | |
| Chicken | 1–6 | ND | ND |
| 7 | ND | ND | |
| 8–20 | ND | ND | |
| Fish | 1 | 0.88 ± 0.04 | 0.92 ± 0.03 |
| 2–20 | ND | ND | |
| Shrimp | 1–12 | ND | ND |
| 13 | ND | ND | |
| 14–20 | ND | ND |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wu, Y.; Wang, J.; Zhou, Y.; Qi, Y.; Ma, L.; Wang, X.; Tao, X. Quantitative Determination of Nitrofurazone Metabolites in Animal-Derived Foods Based on a Background Fluorescence Quenching Immunochromatographic Assay. Foods 2021, 10, 1668. https://doi.org/10.3390/foods10071668
Wu Y, Wang J, Zhou Y, Qi Y, Ma L, Wang X, Tao X. Quantitative Determination of Nitrofurazone Metabolites in Animal-Derived Foods Based on a Background Fluorescence Quenching Immunochromatographic Assay. Foods. 2021; 10(7):1668. https://doi.org/10.3390/foods10071668
Chicago/Turabian StyleWu, Yuping, Jia Wang, Yong Zhou, Yonghua Qi, Licai Ma, Xuannian Wang, and Xiaoqi Tao. 2021. "Quantitative Determination of Nitrofurazone Metabolites in Animal-Derived Foods Based on a Background Fluorescence Quenching Immunochromatographic Assay" Foods 10, no. 7: 1668. https://doi.org/10.3390/foods10071668
APA StyleWu, Y., Wang, J., Zhou, Y., Qi, Y., Ma, L., Wang, X., & Tao, X. (2021). Quantitative Determination of Nitrofurazone Metabolites in Animal-Derived Foods Based on a Background Fluorescence Quenching Immunochromatographic Assay. Foods, 10(7), 1668. https://doi.org/10.3390/foods10071668






