Comparative Study of Several Machine Learning Algorithms for Classification of Unifloral Honeys
Abstract
:1. Introduction
2. Materials and Methods
2.1. Honey Samples
2.2. Microscopical Analysis
2.3. Electrical Conductivity
2.4. Water Content
2.5. pH Measurement
2.6. Color
2.7. Sugars
2.8. Classification Using Statistical Multivariate and Machine Learning Algorithms
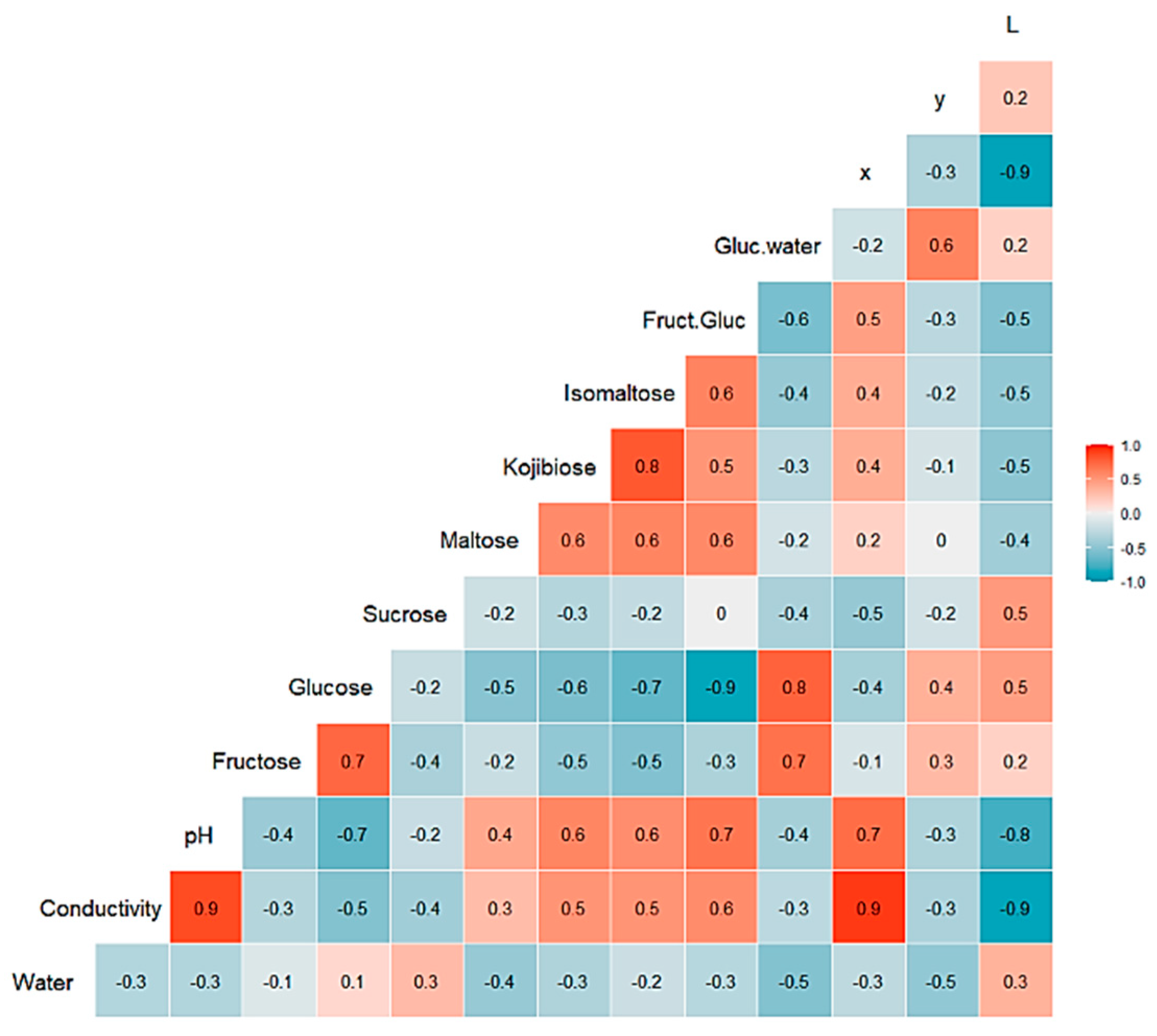
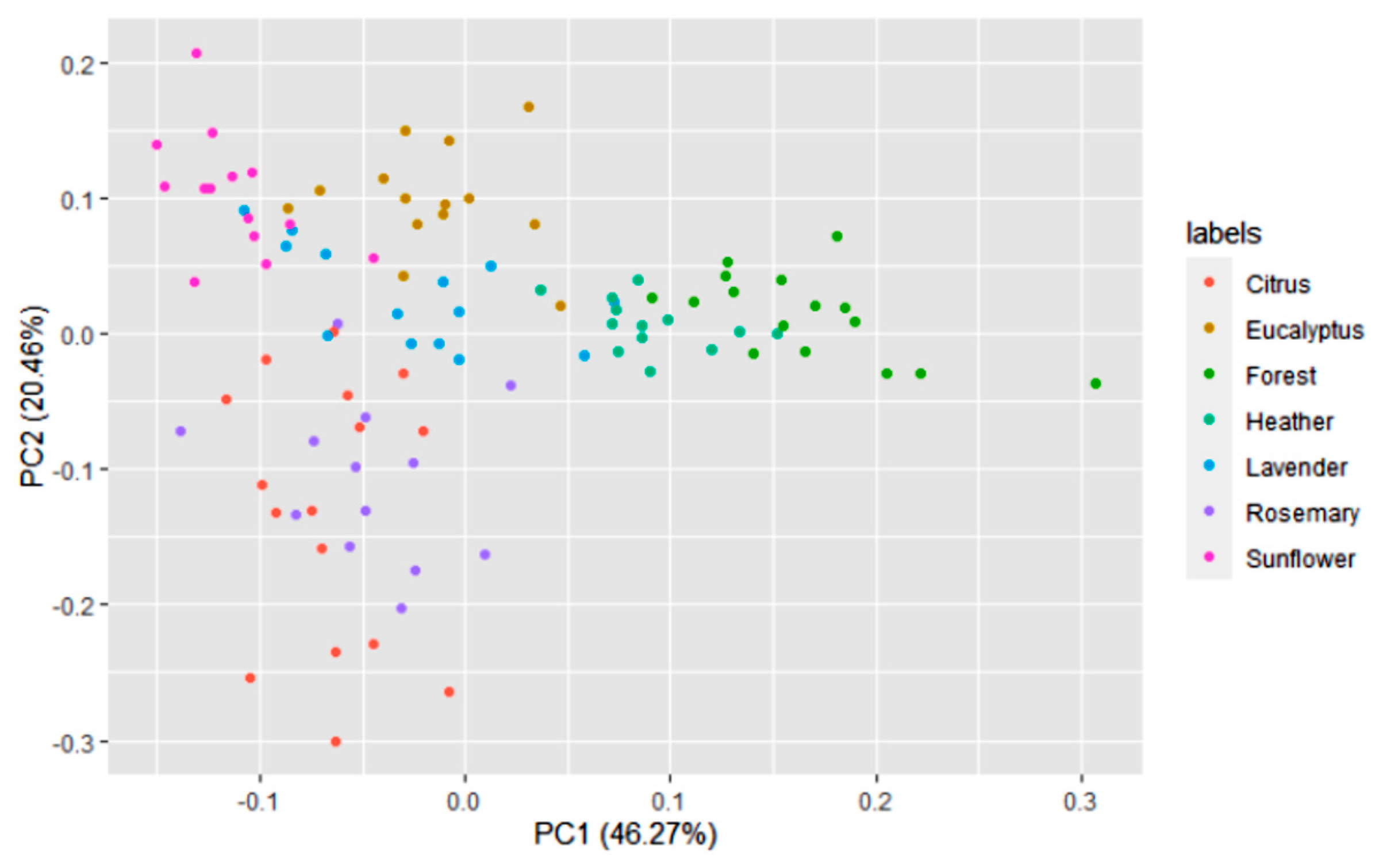
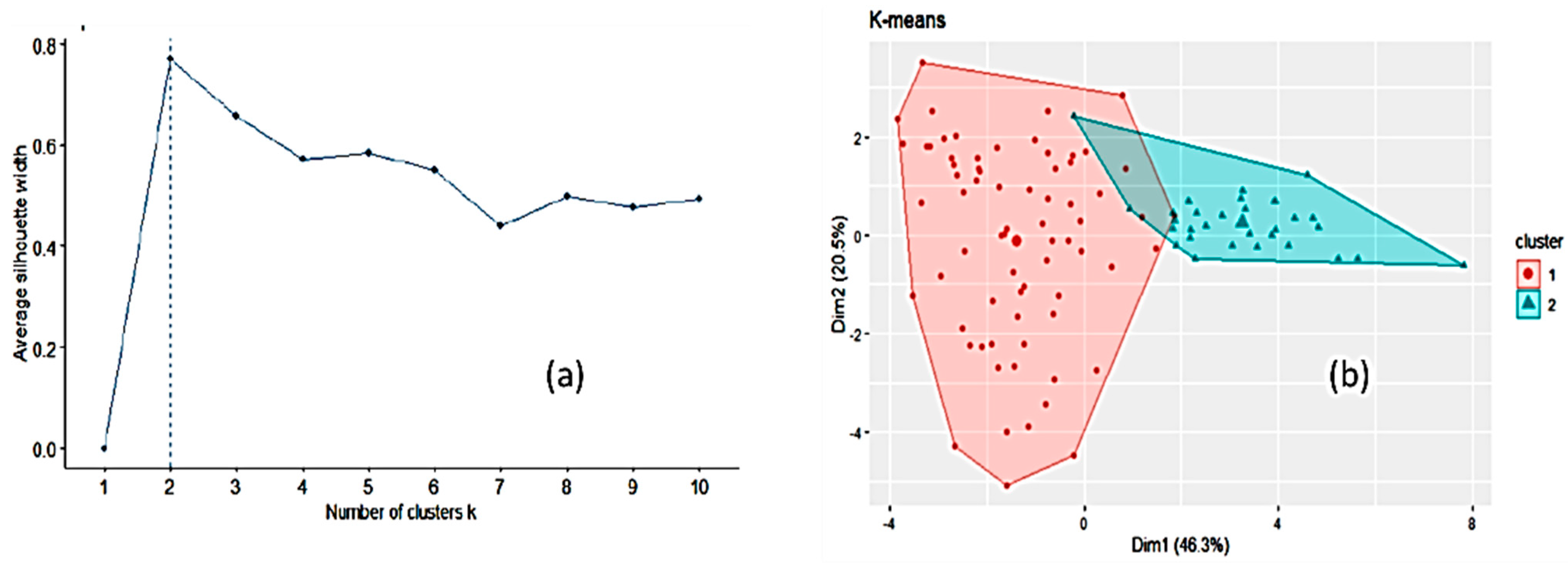
3. Results
3.1. Honey Dataset
3.2. Statistical and ML Algorithms
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Cianciosi, D.; Forbes-Hernández, T.Y.; Afrin, S.; Gasparrini, M.; Reboredo-Rodriguez, P.; Manna, P.P.; Zhang, J.; Bravo Lamas, L.; Martínez Flórez, S.; Agudo Toyos, P.; et al. Phenolic compounds in honey and their associated health benefits: A review. Molecules 2018, 23, 2322. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Afrin, S.; Haneefa, S.M.; Fernandez-Cabezudo, M.J.; Giampieri, F.; al-Ramadi, B.K.; Battino, M. Therapeutic and preventive properties of honey and its bioactive compounds in cancer: An evidence-based review. Nutr. Res. Rev. 2020, 33, 50–76. [Google Scholar] [CrossRef] [PubMed]
- European Commission. Regulation (EC) No 178/2002 of the European Parliament and of the council of 28 January 2002 laying down the general principles and requirements of food law, establishing the European food safety authority and laying down procedures in matters of food safety. Off. J. Eur. Commun. 2002, L 31, 1–24. [Google Scholar]
- Council Directive 2001/110/EC of 20 December 2001 relating to honey. Off. J. Eur. Comm. 2001, L 10, 47–52.
- Directive 2014/63/EU of the European Parliament and of the Council of 15 May 2014 amending Council Directive 2001/110/EC relating to honey. Off. J. Eur. Union 2014, L 164, 1–5.
- Codex Alimentarius Standard for honey CXS 12-1981 Adopted in 1981. Revised in 1987, 2001. Amended in 2019. 2001. Available online: http://www.fao.org/fao-who-codexalimentarius/sh-proxy/en/?lnk=1&url=https%253A%252F%252Fworkspace.fao.org%252Fsites%252Fcodex%252FStandards%252FCXS%2B12-1981%252FCXS_012e.pdf (accessed on 21 June 2021).
- Ampuero, S.; Bogdanov, S.; Bosset, J.O. Classification of unifloral honeys with an MS-based electronic nose using different sampling modes: SHS, SPME and INDEX. Eur. Food Res. Technol. 2004, 218, 198–207. [Google Scholar] [CrossRef]
- Cavaco, A.M.; Miguel, G.; Antunes, D.; Guerra, R. Determination of geographical and botanical origin of honey: From sensory evaluation to the state of the art of non-invasive technology. In Honey: Production, Consumption and Health Benefits; Bondurand, G., Bosch, H., Eds.; Nova Science Publishers: Hauppauge, NY, USA, 2012; pp. 1–40. [Google Scholar]
- Maurizio, A. Microscopy of honey. In Honey: A Comprehensive Survey; Crane, E., Ed.; Heinemann in Cooperation with the International Bee Research Association: London, UK, 1975; pp. 240–257. [Google Scholar]
- Louveaux, J.; Maurizio, A.; Vorwohl, G. Methods of melissopalynology. Bee World 1978, 59, 139–157. [Google Scholar] [CrossRef]
- Mateo, R.; Bosch-Reig, F. Classification of Spanish unifloral honeys by discriminant analysis of electrical conductivity, color, water content, sugars, and pH. J. Agric. Food Chem. 1998, 46, 393–400. [Google Scholar] [CrossRef] [PubMed]
- White, J.W.; Bryant, V.M., Jr. Assessing citrus honey quality: Pollen and methyl anthranilate content. J. Agric. Food Chem. 1996, 44, 3423–3425. [Google Scholar] [CrossRef]
- Persano-Oddo, L.; Piro, R. Main European unifloral honeys: Descriptive sheets1. Apidologie 2004, 35, S38–S81. [Google Scholar] [CrossRef]
- Mateo Castro, R.; Jiménez Escamilla, M.; Bosch Reig, F. Evaluation of the color of some Spanish unifloral honey types as a characterization parameter. J. AOAC Int. 1992, 75, 537–542. [Google Scholar] [CrossRef]
- Mateo, R.; Bosch-Reig, F. Sugar profiles of Spanish unifloral honeys. Food Chem. 1997, 60, 33–41. [Google Scholar] [CrossRef]
- de la Fuente, E.; Ruiz-Matute, A.I.; Valencia-Barrera, R.M.; Sanz, J.; Martínez Castro, I. Carbohydrate composition of Spanish unifloral honeys. Food Chem. 2011, 129, 1483–1489. [Google Scholar] [CrossRef] [Green Version]
- Weston, R.J.; Brocklebank, L.K. The oligosaccharide composition of some New Zealand honeys. Food Chem. 1999, 64, 33–37. [Google Scholar] [CrossRef]
- Bouseta, A.; Scheirman, V.; Collin, S. Flavor and free amino acid composition of lavender and eucalyptus honeys. J. Food Sci. 1996, 61, 683–687, 694. [Google Scholar] [CrossRef]
- Baroni, M.V.; Nores, M.L.; Díaz, M.D.P.; Chiabrando, G.A.; Fassano, J.P.; Costa, C.; Wunderlin, D.A. Determination of volatile organic compound patterns characteristic of five unifloral honey by solid-phase microextraction−gas chromatography−mass spectrometry coupled to chemometrics. J. Agric. Food Chem. 2006, 54, 7235–7241. [Google Scholar] [CrossRef]
- Revell, L.E.; Morris, B.; Manley-Harris, M. Analysis of volatile compounds in New Zealand unifloral honeys by SPME–GC–MS and chemometric-based classification of floral source. Food Meas. 2014, 8, 81–91. [Google Scholar] [CrossRef]
- Castro-Vázquez, L.; Díaz-Maroto, M.C.; González-Viñas, M.A.; Pérez-Coello, M.S. Differentiation of monofloral citrus, rosemary, eucalyptus, lavender, thyme and heather honeys based on volatile composition and sensory descriptive analysis. Food Chem. 2009, 112, 1022–1030. [Google Scholar] [CrossRef]
- Machado, A.M.; Miguel, M.G.; Vilas-Boas, M.; Figueiredo, A.C. Honey volatiles as a fingerprint for botanical origin—A review on their occurrence on monofloral honeys. Molecules 2020, 25, 374. [Google Scholar] [CrossRef] [Green Version]
- Sun, Z.; Zhao, L.; Cheng, N.; Xue, X.; Wu, L.; Zheng, J.; Cao, W. Identification of botanical origin of Chinese unifloral honeys by free amino acid profiles and chemometric methods. J. Pharm. Anal. 2017, 7, 317–323. [Google Scholar] [CrossRef]
- Oroian, M.; Sorina, R. Honey authentication based on physicochemical parameters and phenolic compounds. Comput. Electron. Agric. 2017, 138, 148–156. [Google Scholar] [CrossRef]
- Karabagias, I.K.; Louppis, A.P.; Kontakos, S.; Drouza, C.; Papastephanou, C. Characterization and botanical differentiation of monofloral and multifloral honeys produced in Cyprus, Greece, and Egypt using physicochemical parameter analysis and mineral content in conjunction with supervised statistical techniques. J. Anal. Meth. Chem. 2018, 7698251. [Google Scholar] [CrossRef] [PubMed]
- Ruoff, K.; Luginbühl, W.; Kilchenmann, V.; Bosset, J.O.; von der Ohec, K.; von der Ohec, W.; Amad, R. Authentication of the botanical origin of honey using profiles of classical measurands and discriminant analysis. Apidologie 2007, 38, 438–452. [Google Scholar] [CrossRef]
- Lenhardt, L.; Zeković, I.; Dramićanin, T.; Tešić, Ž.; Milojković-Opsenica, D.; Dramićanin, M.D. Authentication of the botanical origin of unifloral honey by infrared spectroscopy coupled with support vector machine algorithm. Phys. Scr. 2014, T162, 014042. [Google Scholar] [CrossRef]
- Minaei, S.; Shafiee, S.; Polder, G.; Moghadam-Charkari, N.; van Ruth, S.; Barzegar, M.; Zahiri, J.; Alewijn, M.; Kus, P.M. VIS/NIR imaging application for honey floral origin determination. Infrared Phys. Technol. 2017, 86, 218–225. [Google Scholar] [CrossRef] [Green Version]
- Corvucci, F.; Nobili, L.; Melucci, D.; Grillenzoni, F.V. The discrimination of honey origin using melissopalynology and Raman spectroscopy techniques coupled with multivariate analysis. Food Chem. 2015, 169, 297–304. [Google Scholar] [CrossRef]
- Oroian, M.; Ropciuc, S. Botanical authentication of honeys based on Raman spectra. Food Meas. 2018, 12, 545–554. [Google Scholar] [CrossRef]
- Xagoraris, M.; Lazarou, E.; Kaparakou, E.H.; Alissandrakis, E.; Tarantilisa, P.A.; Pappas, C.S. Botanical origin discrimination of Greek honeys: Physicochemical parameters versus Raman spectroscopy. J. Sci. Food Agric. 2021, 101, 3319–3327. [Google Scholar] [CrossRef]
- Melucci, D.; Cocchi, M.; Corvucci, F.; Boi, M.; Tositti, L.; de Laurentiis, F.; Zappi, A.; Locatelli, C.; Locatelli, M. Chemometrics for the direct analysis of solid samples by spectroscopic and chromatographic techniques. In Chemometrics: Methods, Applications and New Research; Luna, A.S., Ed.; Nova Science Publishers: Hauppauge, NY, USA, 2017; pp. 173–204. [Google Scholar]
- Siddiqui, A.J.; Musharraf, S.G.; Choudhary, M.I. Application of analytical methods in authentication and adulteration of honey. Food Chem. 2017, 217, 687–698. [Google Scholar] [CrossRef]
- Zahed, N.; Najib, M.S.; Tajuddin, S.N. Categorization of gelam, acacia and tualang honey odor-profile using k-nearest neighbors. Int. J. Soft. Eng. Comput Syst. 2018, 4, 15–28. [Google Scholar] [CrossRef]
- Major, N.; Marković, K.; Krpan, M.; Šarić, G.; Hrusˇkar, M.; Vahčić, N. Rapid honey characterization and botanical classification by an electronic tongue. Talanta 2011, 85, 569–574. [Google Scholar] [CrossRef]
- Anjos, O.; Iglesias, C.; Peres, F.; Martínez, J.; García, Á.; Taboada, J. Neural networks applied to discriminate botanical origin of honeys. Food Chem. 2015, 175, 128–136. [Google Scholar] [CrossRef]
- Popek, S.; Halagarda, M.; Kursa, K. A new model to identify botanical origin of Polish honeys based on the physicochemical parameters and chemometric analysis. LWT Food Sci. Technol. 2017, 77, 482–487. [Google Scholar] [CrossRef]
- Maione, C.; Barbosa, F., Jr.; Barbosa, R.M. Predicting the botanical and geographical origin of honey with multivariate data analysis and machine learning techniques: A review. Comput. Electron. Agric. 2019, 157, 436–446. [Google Scholar] [CrossRef]
- Escuredo, O.; Fernández-González, M.; Seijo, M.C. Differentiation of blossom honey and honeydew honey from Northwest Spain. Agriculture 2012, 2, 25–37. [Google Scholar]
- Seijo, M.C.; Escuredo, O.; Rodríguez-Flores, M.S. Physicochemical properties and pollen profile of oak honeydew and evergreen oak honeydew honeys from Spain: A comparative study. Foods 2019, 8, 126. [Google Scholar] [CrossRef] [Green Version]
- Orden de 12 de junio de 1986 por la que se aprueban los métodos oficiales de análisis para la miel. (Order of 12 June 1986 approving the official methods of analysis for honey). BOE 1986, 145, 22195–22202. Available online: https://www.boe.es/eli/es/o/1986/06/12/(3)/dof/spa/pdf (accessed on 22 June 2021).
- AOAC 969. 38B MAFF Validated method V21 for moisture in honey. J. Assoc. Public Anal. 1992, 28, 183–187. [Google Scholar]
- CIE (Commission Internationale de l’Eclairage). In Proceedings of the Eighth Session, Cambridge, UK, September 1931. Available online: http://classify.oclc.org/classify2/ClassifyDemo?owi=25128274 (accessed on 22 June 2021).
- Hastie, T.; Bujas, A.; Tibsihrani, R. Penalized discriminant analysis. Ann. Stat. 1995, 23, 73–102. [Google Scholar] [CrossRef]
- Hechenbichler, K.; Schliep, K. Weighted k-Nearest-Neighbor Techniques and Ordinal Classification; Sonderforschungsbereich 386, Paper 399; Ludwig-Maximilians-Universität: München, Germany, 2004; pp. 1–16. Available online: https://epub.ub.uni-muenchen.de/1769/1/paper_399.pdf (accessed on 4 May 2021).
- Kuhn, M.; Wing, J.; Weston, S.; Williams, A.; Keefer, C.; Engelhardt, A.; Cooper, T.; Mayer, Z.; Kenkel, B.; The R Core Team; et al. Classification and Regression Training. R Package Version 2016, 6.0–71. Available online: https://CRAN.R-project.org/package=caret (accessed on 5 April 2021).
- Bouveyron, C.; Girard, S.; Schmid, C. High-dimensional discriminant analysis. Comm. Stat. Theor. Meth. 2007, 36, 2607–2623. [Google Scholar] [CrossRef] [Green Version]
- Tibshirani, R.; Hastie, T.; Narasimhan, B.; Chu, G. Diagnosis of multiple cancer types by shrunken centroids of gene expression. Proc. Natl. Acad. Sci. USA 2002, 99, 6567–6572. [Google Scholar] [CrossRef] [Green Version]
- Tibshirani, R.; Hastie, T.; Narasimhan, B.; Chu, G. Class prediction by nearest shrunken centroids, with applications to DNA microarrays. Stat. Sci. 2003, 18, 104–117. [Google Scholar] [CrossRef]
- Geurst, P.; Louis, D.; Wehenke, L. Extremely randomized trees. Mach. Learn. 2006, 63, 3–42. [Google Scholar]
- Ahdesmäki, M.; Strimmer, K. Feature selection in omics prediction problems using cat scores and false non discovery rate control. Ann. Appl. Stat. 2010, 4, 503–519. [Google Scholar] [CrossRef]
- Günther, F.; Fritsch, S. Neuralnet: Training of neural networks. R J 2010, 2, 30–38. [Google Scholar] [CrossRef] [Green Version]
- Cortes, C.; Vapnik, V. Support-vector networks. Mach. Learn. 1995, 20, 273–297. [Google Scholar] [CrossRef]
- Vapnik, V.N. Statistical Learning Theory; Wiley: New York, NY, USA, 1998. [Google Scholar]
- Liaw, A.; Wiener, M.C. Classification and regression by random forest. R News 2002, 2, 18–22. [Google Scholar]
- Chen, T.; Guestrin, C. XGBoost: A scalable tree boosting system. In Proceedings of the 22nd ACM SIGKDD International Conference on knowledge discovery and data mining, San Francisco, CA, USA, 13–17 August 2016; pp. 785–794. [Google Scholar] [CrossRef] [Green Version]
- Bishop, C.M. Neural Networks for Pattern Recognition; Oxford University Press: Oxford, UK, 1995. [Google Scholar]
- Tapas Kanungo, D.M. A local search approximation algorithm for k-means clustering. In Proceedings of the 18th Annual Symposium On Computational Geometry, Barcelona, Spain, 5–7 June 2002; ACM Press: New York, NY, USA, 2002; pp. 10–18. [Google Scholar]
- Kursa, M.B.; Rudnicki, W.R. Feature selection with the boruta package. J. Stat. Soft. 2010, 36, 1–13. Available online: http://www.jstatsoft.org/v36/i11/ (accessed on 12 May 2021). [CrossRef] [Green Version]
- Stevinho, L.M.; Chambó, E.D.; Pereira, A.P.R.; Carvalho, C.A.L.D.; de Toledo, V.D.A.A. Characterization of Lavandula spp. honey using multivariate techniques. PLoS ONE 2016, 11, e016220. [Google Scholar]
- Escriche, I.; Sobrino-Gregorio, L.; Conchado, A.; Juan-Borrás, M. Volatile profile in the accurate labelling of monofloral honey. The case of lavender and thyme honey. Food Chem. 2017, 226, 61–68. [Google Scholar] [CrossRef]
- Ciulu, M.; Oertel, E.; Serra, R.; Farre, R.; Spano, N.; Caredda, M.; Malfatti, L.; Sanna, G. Classification of unifloral honeys from SARDINIA (Italy) by ATR-FTIR spectroscopy and random forest. Molecules 2021, 26, 88. [Google Scholar] [CrossRef]
- Bisutti, V.; Merlanti, R.; Serva, L.; Lucatello, L.; Mirisola, M.; Balzan, S.; Tenti, S.; Fontana, F.; Trevisan, G.; Montanucci, L.; et al. Multivariate and machine learning approaches for honey botanical origin authentication using near infrared spectroscopy. J. Near Infrared Spectrosc. 2019, 27, 65–74. [Google Scholar] [CrossRef]
- Chen, L.; Wang, J.; Ye, Z.; Zhao, J.; Xue, X.; Vander Heyden, Y.; Sun, Q. Classification of Chinese honeys according to their floral origin by near infrared spectroscopy. Food Chem. 2012, 135, 338–342. [Google Scholar] [CrossRef] [PubMed]
- Benedetti, S.; Mannino, S.; Sabatini, A.G.; Marcazzan, G.L. Electronic nose and neural network use for the classification of honey. Apidologie 2004, 35, 397–402. [Google Scholar] [CrossRef] [Green Version]
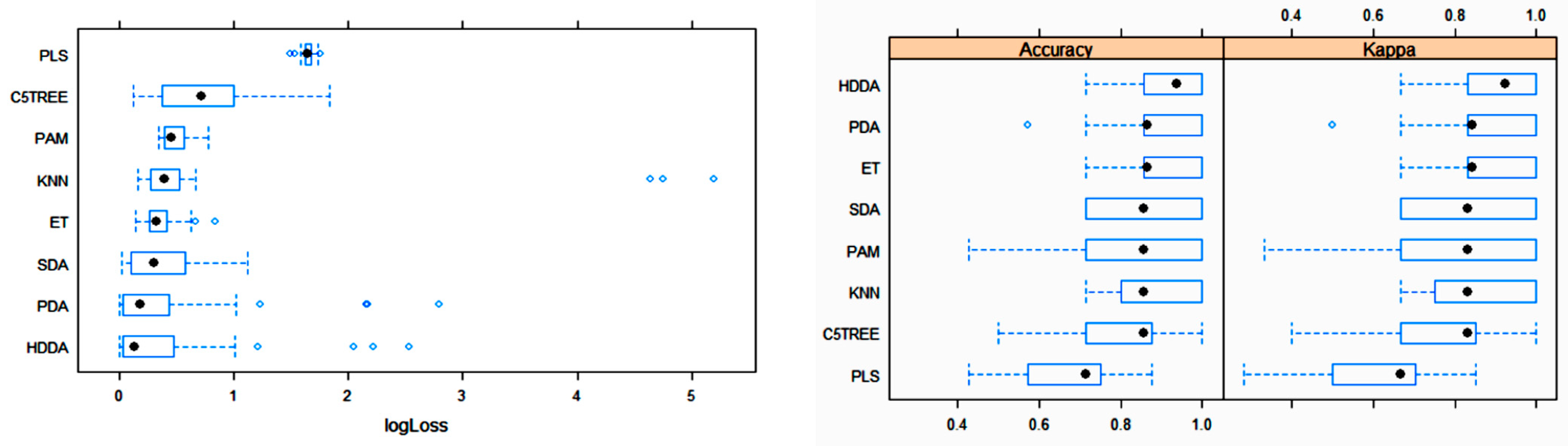
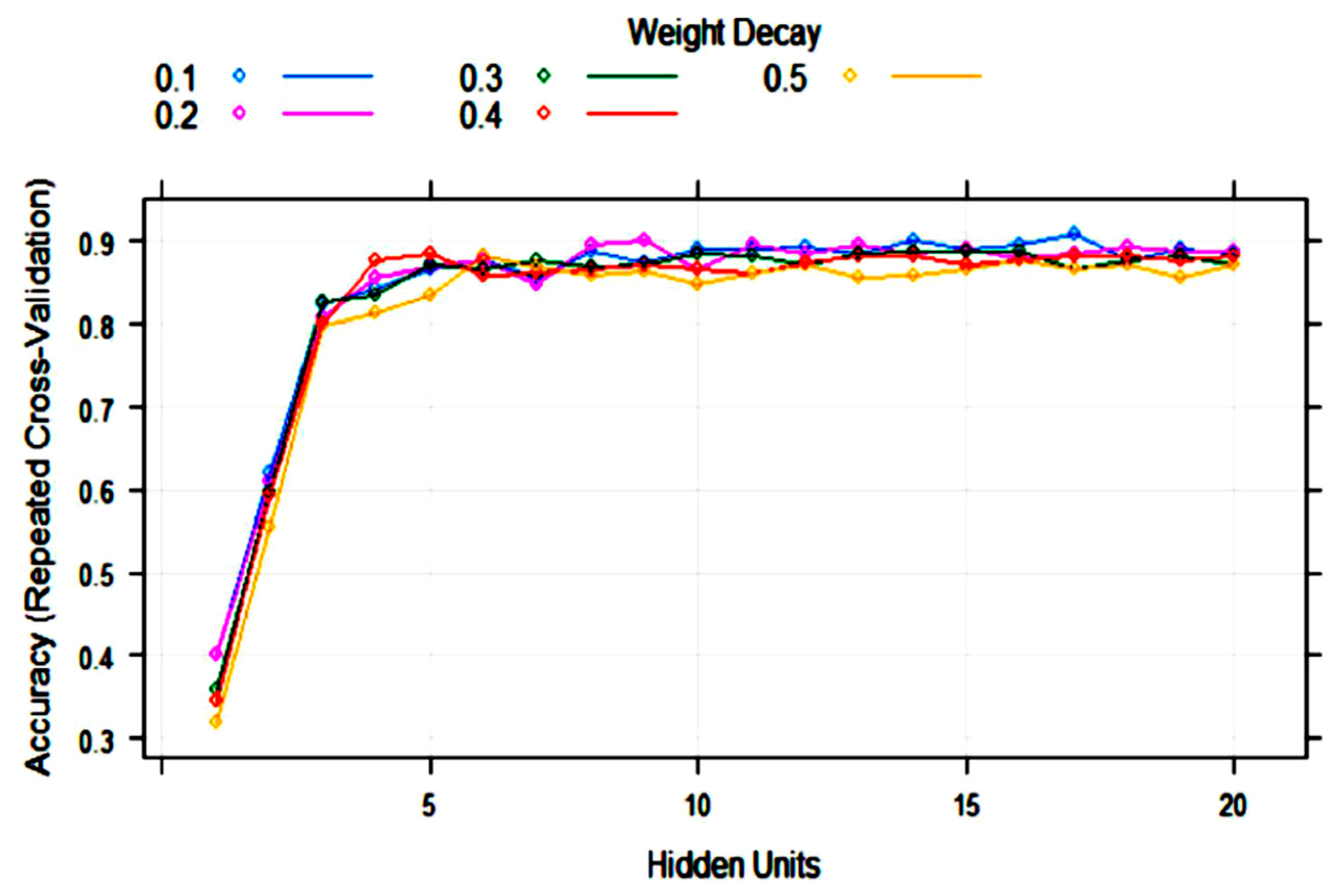
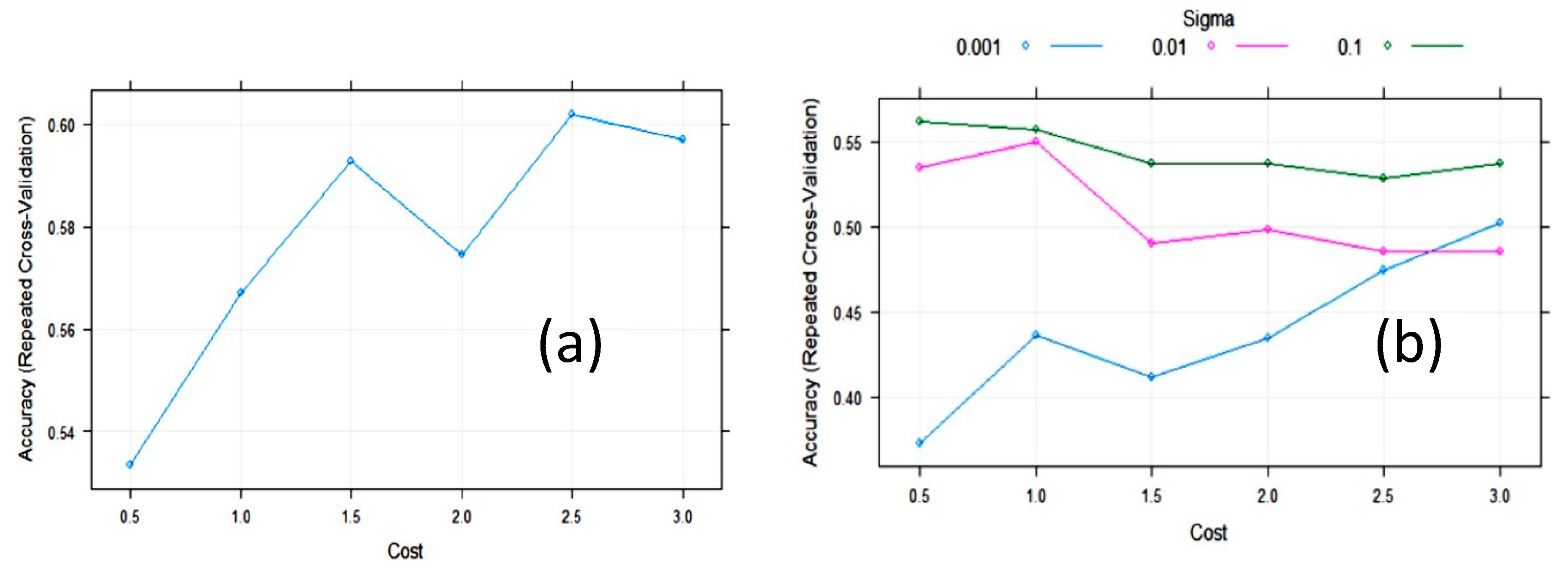
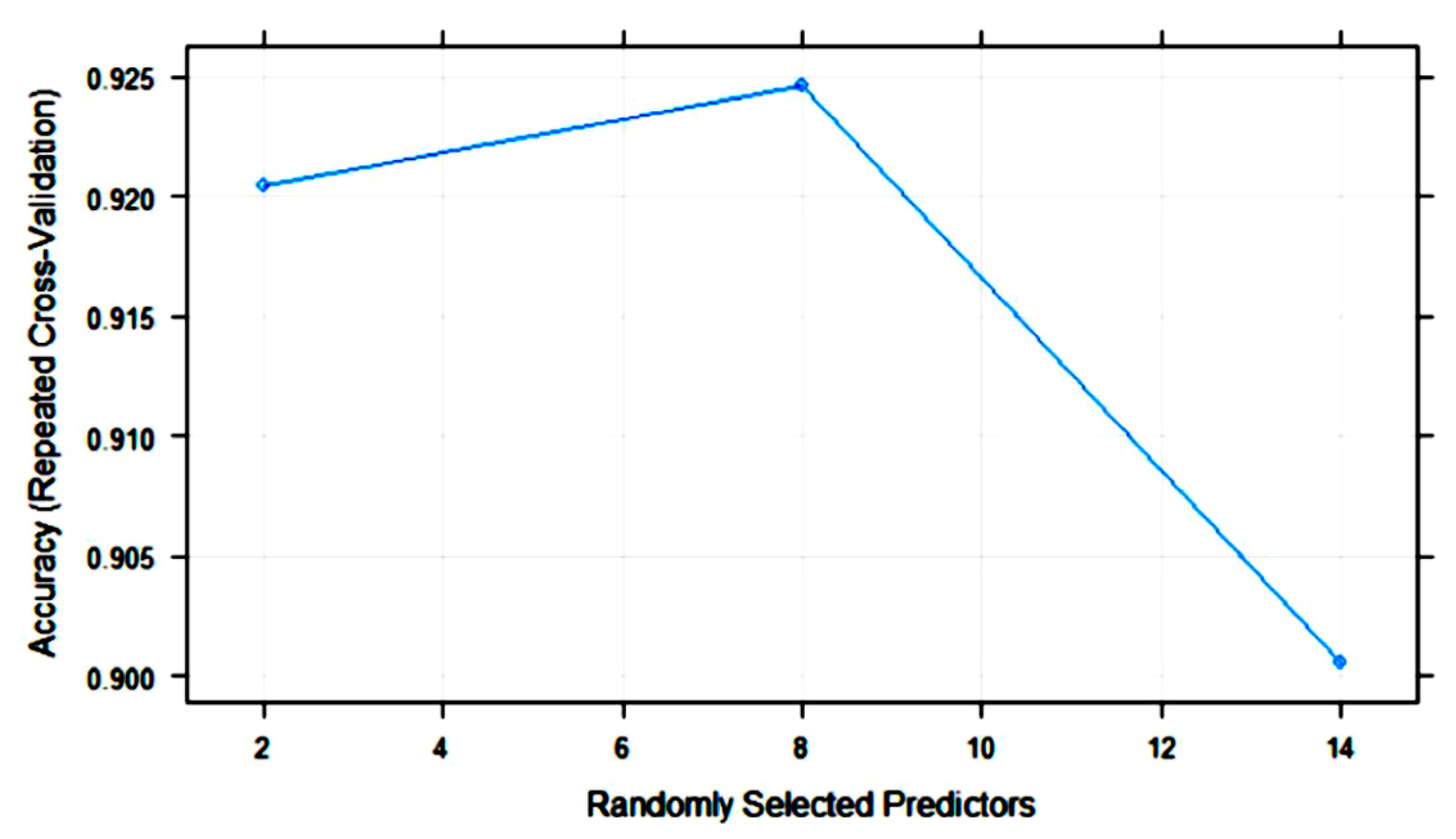
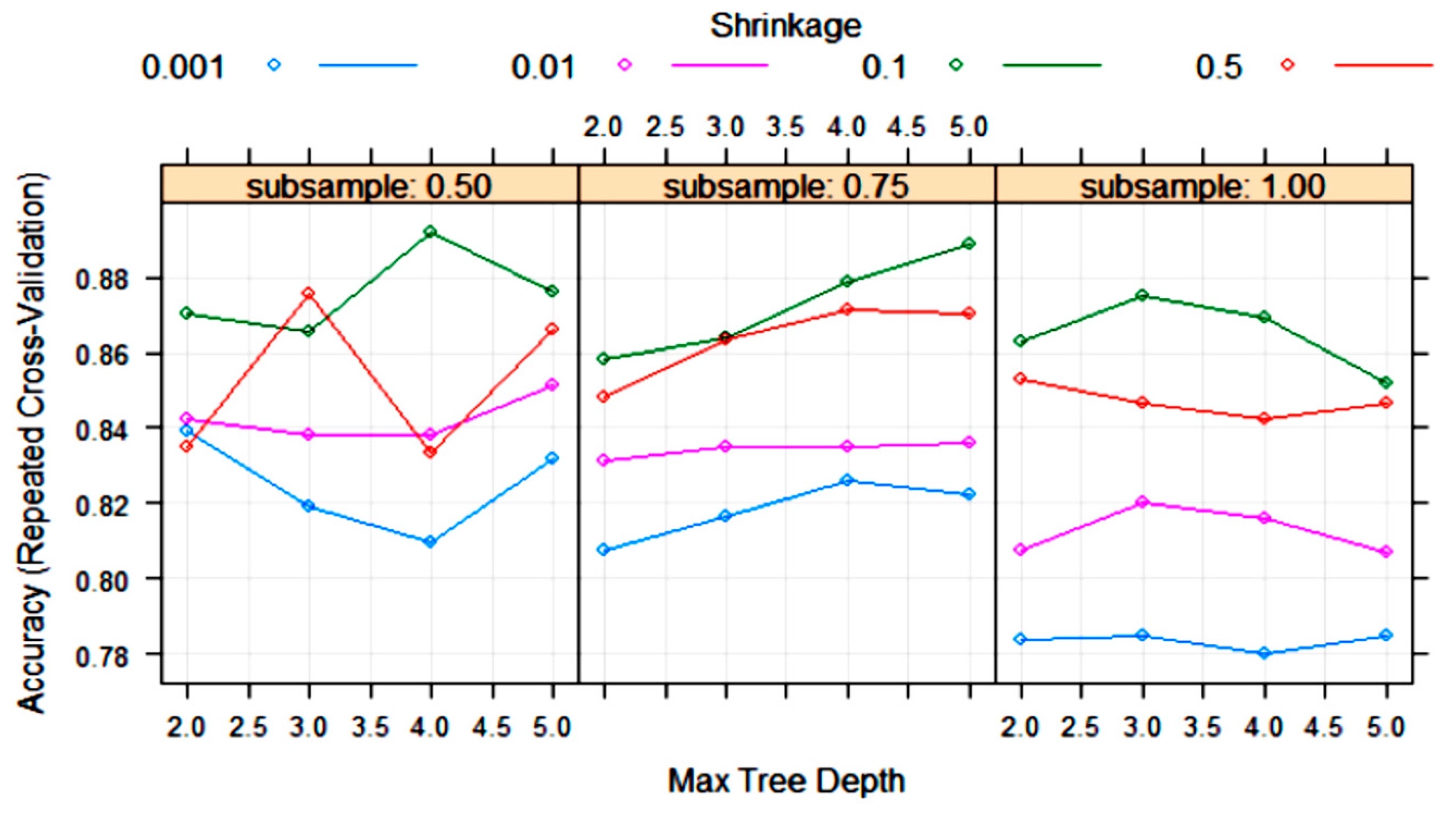
| Algorithm | Tuning Parameter | Mean Log Loss Values |
|---|---|---|
| KKNN | Kmax = 5 | 0.8339319 |
| Kmax = 7 | 0.9017721 | |
| Kmax = 9 | 0.9808674 | |
| PDA | Lambda = 1 | 0.5689435 |
| Lambda = 0.0001 | 0.5687306 | |
| Lambda = 0.1 | 0.4611719 | |
| HDDA | Thershold = 0.05 | 0.4360396 |
| Thershold = 0.175 | 1.3732500 | |
| Thershold = 0.300 | 1.0080708 | |
| SDA | Lambda = 0.0 | 0.6320813 |
| Lambda = 0.5 | 0.3968958 | |
| Lambda = 1.0 | 0.4908678 | |
| PAM | Threshold = 0.7608929 | 0.4986565 |
| Threshold = 11.0329476 | 1.9483062 | |
| Threshold = 21.3050022 | 1.9483062 | |
| PLS | Ncomp = 1 | 1.826913 |
| Ncomp = 2 | 1.733439 | |
| Ncomp = 3 | 1.643669 | |
| C5.0 tree | 0.7482527 | |
| ET | 0.3590714 |
| Reference | ||||||||
|---|---|---|---|---|---|---|---|---|
| Prediction | Citrus | Eucalyptus | Forest | Heather | Lavender | Rosemary | Sunflower | |
| PDA | Citrus | 5 | 0 | 0 | 0 | 0 | 1 | 0 |
| Eucalyptus | 0 | 4 | 0 | 0 | 1 | 0 | 0 | |
| Forest | 0 | 0 | 5 | 0 | 0 | 0 | 0 | |
| Heather | 0 | 0 | 0 | 4 | 0 | 0 | 0 | |
| Lavender | 0 | 0 | 0 | 0 | 2 | 0 | 0 | |
| Rosemary | 0 | 0 | 0 | 0 | 0 | 3 | 0 | |
| Sunflower | 0 | 0 | 0 | 0 | 1 | 0 | 4 | |
| SDA | Citrus | 4 | 0 | 0 | 0 | 0 | 1 | 0 |
| Eucalyptus | 0 | 4 | 0 | 0 | 1 | 0 | 0 | |
| Forest | 0 | 0 | 5 | 0 | 0 | 0 | 0 | |
| Heather | 0 | 0 | 0 | 4 | 0 | 0 | 0 | |
| Lavender | 0 | 0 | 0 | 0 | 3 | 1 | 0 | |
| Rosemary | 1 | 0 | 0 | 0 | 0 | 2 | 0 | |
| Sunflower | 0 | 0 | 0 | 0 | 0 | 0 | 4 | |
| ET | Citrus | 4 | 0 | 0 | 0 | 0 | 0 | 0 |
| Eucalyptus | 0 | 4 | 0 | 0 | 2 | 0 | 0 | |
| Forest | 0 | 0 | 5 | 0 | 0 | 0 | 0 | |
| Heather | 0 | 0 | 0 | 4 | 0 | 0 | 0 | |
| Lavender | 0 | 0 | 0 | 0 | 2 | 1 | 0 | |
| Rosemary | 1 | 0 | 0 | 0 | 0 | 3 | 0 | |
| Sunflower | 0 | 0 | 0 | 0 | 0 | 0 | 4 | |
| PLS | Citrus | 5 | 0 | 0 | 0 | 0 | 3 | 0 |
| Eucalyptus | 0 | 3 | 0 | 0 | 0 | 0 | 0 | |
| Forest | 0 | 0 | 5 | 0 | 0 | 1 | 0 | |
| Heather | 0 | 0 | 0 | 4 | 0 | 0 | 0 | |
| Lavender | 0 | 0 | 0 | 0 | 1 | 0 | 0 | |
| Rosemary | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| Sunflower | 0 | 1 | 0 | 0 | 3 | 0 | 4 | |
| C5.0 tree | Citrus | 4 | 1 | 0 | 0 | 0 | 1 | 0 |
| Eucalyptus | 0 | 2 | 0 | 0 | 2 | 0 | 0 | |
| Forest | 0 | 0 | 5 | 0 | 0 | 0 | 0 | |
| Heather | 0 | 0 | 0 | 4 | 0 | 0 | 0 | |
| Lavender | 0 | 1 | 0 | 0 | 1 | 0 | 0 | |
| Rosemary | 0 | 0 | 0 | 0 | 0 | 3 | 0 | |
| Sunflower | 1 | 0 | 0 | 0 | 1 | 0 | 4 | |
| Reference | ||||||||
|---|---|---|---|---|---|---|---|---|
| Prediction | Citrus | Eucalyptus | Forest | Heather | Lavender | Rosemary | Sunflower | |
| KKNN | Citrus | 4 | 0 | 0 | 0 | 0 | 1 | 0 |
| Eucalyptus | 0 | 4 | 0 | 0 | 0 | 0 | 0 | |
| Forest | 0 | 0 | 5 | 0 | 0 | 0 | 0 | |
| Heather | 0 | 0 | 0 | 4 | 0 | 0 | 0 | |
| Lavender | 0 | 0 | 0 | 0 | 1 | 0 | 0 | |
| Rosemary | 1 | 0 | 0 | 0 | 0 | 3 | 0 | |
| Sunflower | 0 | 0 | 0 | 0 | 3 | 0 | 4 | |
| PAM | Citrus | 5 | 0 | 0 | 0 | 0 | 1 | 0 |
| Eucalyptus | 0 | 3 | 0 | 0 | 1 | 0 | 0 | |
| Forest | 0 | 0 | 5 | 0 | 0 | 0 | 0 | |
| Heather | 0 | 0 | 0 | 4 | 0 | 0 | 0 | |
| Lavender | 0 | 0 | 0 | 0 | 2 | 0 | 0 | |
| Rosemary | 0 | 0 | 0 | 0 | 0 | 3 | 0 | |
| Sunflower | 0 | 1 | 0 | 0 | 1 | 0 | 4 | |
| HDDA | Citrus | 4 | 0 | 0 | 0 | 0 | 1 | 0 |
| Eucalyptus | 0 | 4 | 0 | 0 | 1 | 0 | 0 | |
| Forest | 0 | 0 | 4 | 0 | 0 | 0 | 0 | |
| Heather | 0 | 0 | 1 | 4 | 0 | 0 | 0 | |
| Lavender | 0 | 0 | 0 | 0 | 2 | 0 | 0 | |
| Rosemary | 0 | 0 | 0 | 0 | 0 | 3 | 0 | |
| Sunflower | 1 | 0 | 0 | 0 | 1 | 0 | 4 | |
| ANN | Citrus | 5 | 0 | 0 | 0 | 0 | 1 | 0 |
| Eucalyptus | 0 | 4 | 0 | 0 | 0 | 0 | 0 | |
| Forest | 0 | 0 | 5 | 0 | 0 | 0 | 0 | |
| Heather | 0 | 0 | 0 | 4 | 0 | 0 | 0 | |
| Lavender | 0 | 0 | 0 | 0 | 1 | 0 | 0 | |
| Rosemary | 0 | 0 | 0 | 0 | 0 | 3 | 0 | |
| Sunflower | 0 | 0 | 0 | 0 | 3 | 0 | 4 | |
| RF | Citrus | 5 | 1 | 0 | 0 | 0 | 1 | 0 |
| Eucalyptus | 0 | 2 | 0 | 0 | 1 | 0 | 0 | |
| Forest | 0 | 0 | 5 | 1 | 0 | 0 | 0 | |
| Heather | 0 | 0 | 0 | 3 | 0 | 0 | 0 | |
| Lavender | 0 | 1 | 0 | 0 | 2 | 0 | 0 | |
| Rosemary | 0 | 0 | 0 | 0 | 0 | 3 | 0 | |
| Sunflower | 0 | 0 | 0 | 0 | 1 | 0 | 4 | |
| Reference | ||||||||
|---|---|---|---|---|---|---|---|---|
| Prediction | Citrus | Eucalyptus | Forest | Heather | Lavender | Rosemary | Sunflower | |
| SVML | Citrus | 3 | 0 | 0 | 0 | 0 | 1 | 0 |
| Eucalyptus | 0 | 4 | 0 | 1 | 1 | 0 | 0 | |
| Forest | 0 | 0 | 5 | 2 | 0 | 0 | 0 | |
| Heather | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| Lavender | 0 | 0 | 0 | 1 | 0 | 0 | 0 | |
| Rosemary | 2 | 0 | 0 | 0 | 1 | 3 | 0 | |
| Sunflower | 0 | 0 | 0 | 0 | 2 | 0 | 4 | |
| SVMR | Citrus | 5 | 0 | 0 | 1 | 0 | 4 | 0 |
| Eucalyptus | 0 | 4 | 0 | 0 | 2 | 0 | 0 | |
| Forest | 0 | 0 | 5 | 0 | 0 | 0 | 0 | |
| Heather | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| Lavender | 0 | 0 | 0 | 3 | 0 | 0 | 0 | |
| Rosemary | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| Sunflower | 0 | 0 | 0 | 0 | 2 | 0 | 4 | |
| XGB | Citrus | 5 | 0 | 0 | 0 | 0 | 0 | 0 |
| Eucalyptus | 0 | 4 | 0 | 0 | 0 | 0 | 0 | |
| Forest | 0 | 0 | 4 | 0 | 0 | 0 | 0 | |
| Heather | 0 | 0 | 1 | 4 | 0 | 0 | 0 | |
| Lavender | 0 | 0 | 0 | 0 | 2 | 0 | 0 | |
| Rosemary | 0 | 0 | 0 | 0 | 0 | 4 | 0 | |
| Sunflower | 0 | 0 | 0 | 0 | 2 | 0 | 4 | |
| ML Algorithm | Overall Accuracy per Test | Mean Overall Accuracy | |||
|---|---|---|---|---|---|
| Test 1 | Test 2 | Test 3 | Test 4 | ||
| PLS | 0.7333 | 0.6667 | 0.6333 | 0.7000 | 0.6833 |
| C5.0 tree | 0.7667 | 0.7667 | 0.7667 | 0.8000 | 0.7750 |
| KKNN | 0.8333 | 0.8333 | 0.7000 | 0.8000 | 0.7916 |
| PAM | 0.8333 | 0.8333 | 0.6667 | 0.8667 | 0.8000 |
| PDA | 0.9000 | 0.9333 | 0.7667 | 0.8667 | 0.8667 |
| SDA | 0.8667 | 0.8667 | 0.7667 | 0.8333 | 0.8333 |
| ET | 0.8333 | 0.8667 | 0.7667 | 0.9000 | 0.8417 |
| HDDA | 0.8333 | 0.8667 | 0.7667 | 0.9000 | 0.8417 |
| ANN | 0.8667 | 0.9333 | 0.7667 | 0.8667 | 0.8584 |
| RF | 0.8000 | 0.8333 | 0.8667 | 0.8667 | 0.8417 |
| SVML | 0.6333 | 0.4667 | 0.5000 | 0.6667 | 0.5667 |
| SVMR | 0.6000 | 0.6667 | 0.5333 | 0.5667 | 0.5917 |
| XGBoost | 0.9000 | 0.8333 | 0.7000 | 0.9333 | 0.8417 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mateo, F.; Tarazona, A.; Mateo, E.M. Comparative Study of Several Machine Learning Algorithms for Classification of Unifloral Honeys. Foods 2021, 10, 1543. https://doi.org/10.3390/foods10071543
Mateo F, Tarazona A, Mateo EM. Comparative Study of Several Machine Learning Algorithms for Classification of Unifloral Honeys. Foods. 2021; 10(7):1543. https://doi.org/10.3390/foods10071543
Chicago/Turabian StyleMateo, Fernando, Andrea Tarazona, and Eva María Mateo. 2021. "Comparative Study of Several Machine Learning Algorithms for Classification of Unifloral Honeys" Foods 10, no. 7: 1543. https://doi.org/10.3390/foods10071543
APA StyleMateo, F., Tarazona, A., & Mateo, E. M. (2021). Comparative Study of Several Machine Learning Algorithms for Classification of Unifloral Honeys. Foods, 10(7), 1543. https://doi.org/10.3390/foods10071543







