A Proteomic Study for the Discovery of Beef Tenderness Biomarkers and Prediction of Warner–Bratzler Shear Force Measured on Longissimus thoracis Muscles of Young Limousin-Sired Bulls
Abstract
1. Introduction
2. Materials and Methods
2.1. Meat Sample Collection
2.2. Warner–Bratzler Shear Force Measurement
2.3. Muscle Protein Extraction
2.4. Shotgun Proteomics
2.4.1. One Dimensional SDS-PAGE and Protein Bands Preparation
2.4.2. LC-MS/MS
2.4.3. LC-MS/MS Data Processing and Protein Identification
2.5. Bioinformatics Analyses
2.5.1. Protein-Protein Interactions (PPI)
2.5.2. Gene Ontology and Pathway and Process Enrichment Analyses
2.6. Statistical Analyses
3. Results
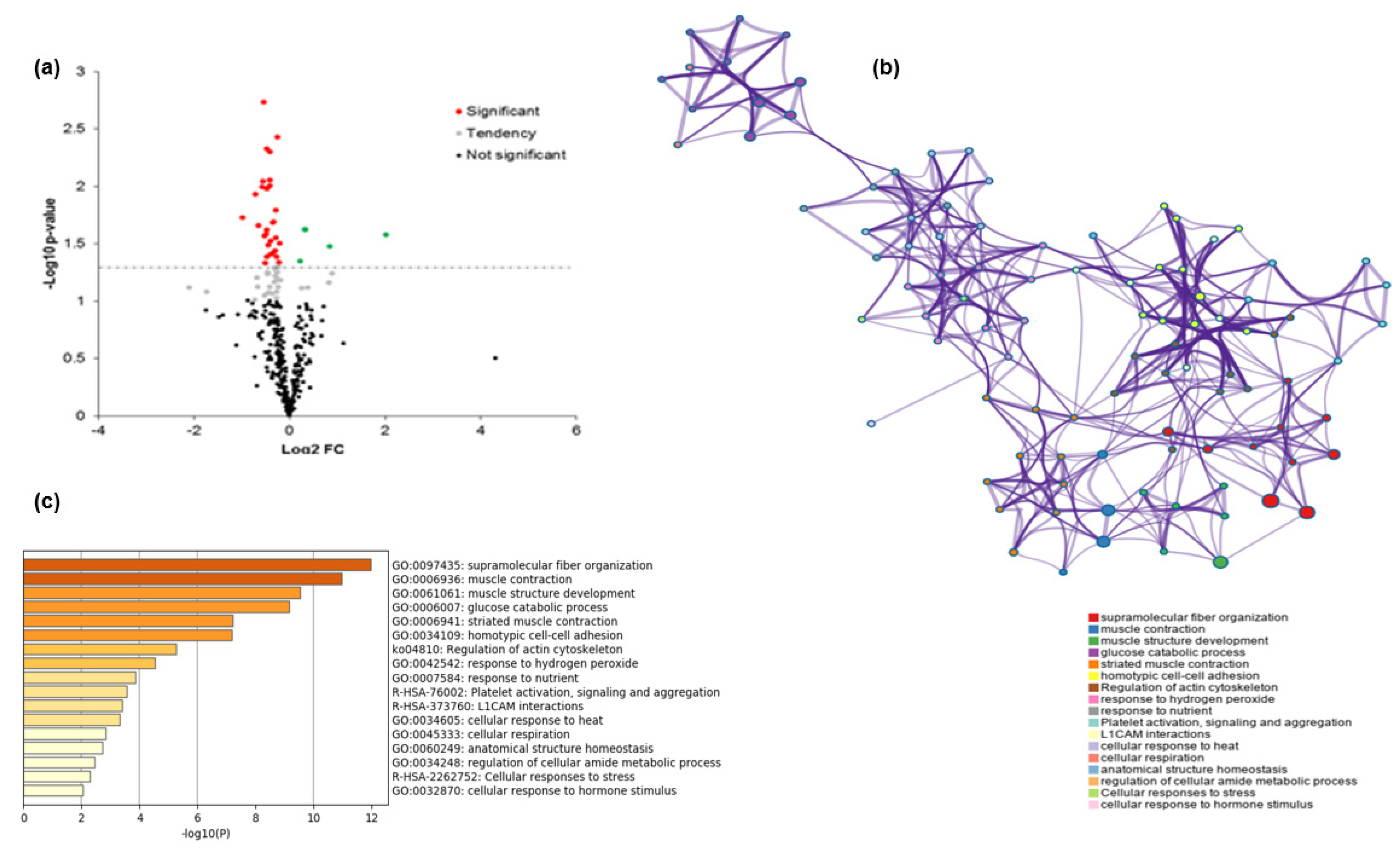
3.1. Differential Proteins between Extreme Groups of High and Low WBSF Values
3.2. Partial Least Squares to Explain the Variability of WBSF Values
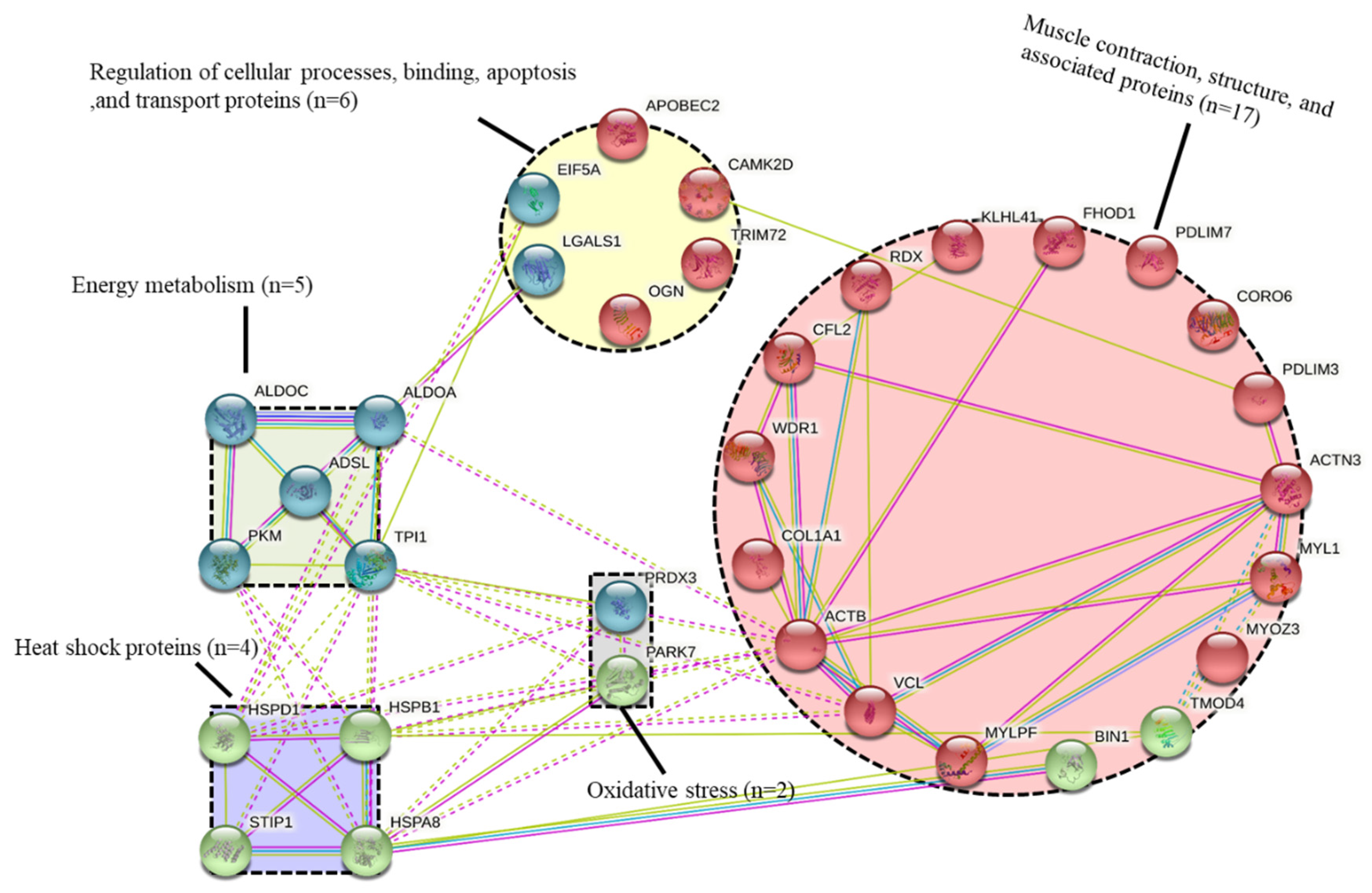
3.3. Protein-Protein Interactions (PPI)
3.4. Pathway and Process Enrichment Analysis
4. Discussion
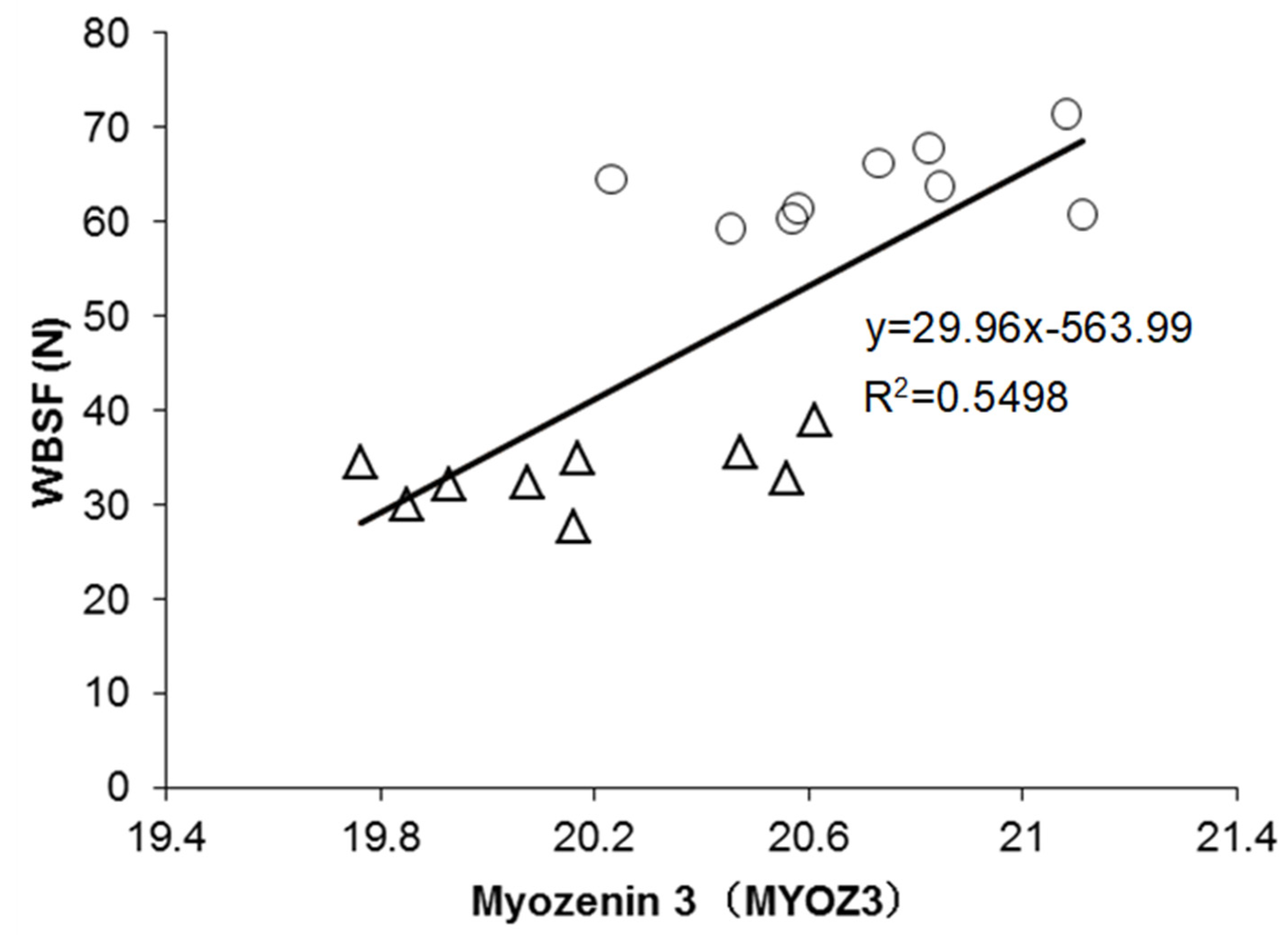
4.1. The Best Explanatory Proteins in the Regression Model of WBSF
4.2. Dominant Pathway Related to WBSF of Young Limousin-Sired Bulls
4.3. Candidate Protein Biomarkers of WBSF from the Energy Metabolism Pathway
4.4. Heat Shock Proteins (HSPs) as Important Indicators of WBSF
4.5. Putative Biomarkers of Tenderness Related to Oxidative Stress
4.6. Proteins from Other Pathways
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kerth, C.R.; Miller, R.K. Beef Flavor: A Review from Chemistry to Consumer. J. Sci. Food Agric. 2015, 95, 2783–2798. [Google Scholar] [CrossRef]
- Purslow, P.P.; Gagaoua, M.; Warner, R.D. Insights on Meat Quality from Combining Traditional Studies and Proteomics. Meat Sci. 2021, 174, 108423. [Google Scholar] [CrossRef] [PubMed]
- Henchion, M.; McCarthy, M.; Resconi, V.C.; Troy, D. Meat Consumption: Trends and Quality Matters. Meat Sci. 2014, 98, 561–568. [Google Scholar] [CrossRef] [PubMed]
- Cuvelier, C.; Clinquart, A.; Hocquette, J.F.; Cabaraux, J.F.; Dufrasne, I.; Istasse, L.; Hornick, J.L. Comparison of Composition and Quality Traits of Meat from Young Finishing Bulls from Belgian Blue, Limousin and Aberdeen Angus Breeds. Meat Sci. 2006, 74, 522–531. [Google Scholar] [CrossRef] [PubMed]
- Gagaoua, M.; Terlouw, C.; Richardson, I.; Hocquette, J.-F.; Picard, B. The Associations between Proteomic Biomarkers and Beef Tenderness Depend on the End-Point Cooking Temperature, the Country Origin of the Panelists and Breed. Meat Sci. 2019, 157, 107871. [Google Scholar] [CrossRef] [PubMed]
- Cafferky, J.; Hamill, R.M.; Allen, P.; O’Doherty, J.V.; Cromie, A.; Sweeney, T. Effect of Breed and Gender on Meat Quality of M. Longissimus Thoracis et Lumborum Muscle from Crossbred Beef Bulls and Steers. Foods 2019, 8, 173. [Google Scholar] [CrossRef]
- Nian, Y.; Kerry, J.P.; Prendiville, R.; Allen, P. The Eating Quality of Beef from Young Dairy Bulls Derived from Two Breed Types at Three Ages from Two Different Production Systems. Ir. J. Agric. Food Res. 2017, 56, 31–44. [Google Scholar] [CrossRef]
- Picard, B.; Gagaoua, M. Muscle Fiber Properties in Cattle and Their Relationships with Meat Qualities: An Overview. J. Agric. Food Chem. 2020, 68, 6021–6039. [Google Scholar] [CrossRef] [PubMed]
- Gagaoua, M.; Monteils, V.; Picard, B. Data from the Farmgate-to-Meat Continuum Including Omics-Based Biomarkers to Better Understand the Variability of Beef Tenderness: An Integromics Approach. J. Agric. Food Chem. 2018, 66, 13552–13563. [Google Scholar] [CrossRef]
- Terlouw, E.M.C.; Picard, B.; Deiss, V.; Berri, C.; Hocquette, J.-F.; Lebret, B.; Lefèvre, F.; Hamill, R.; Gagaoua, M. Understanding the Determination of Meat Quality Using Biochemical Characteristics of the Muscle: Stress at Slaughter and Other Missing Keys. Foods 2021, 10, 84. [Google Scholar] [CrossRef]
- Gagaoua, M.; Monteils, V.; Couvreur, S.; Picard, B. Identification of Biomarkers Associated with the Rearing Practices, Carcass Characteristics, and Beef Quality: An Integrative Approach. J. Agric. Food Chem. 2017, 65, 8264–8278. [Google Scholar] [CrossRef] [PubMed]
- Picard, B.; Gagaoua, M. Meta-Proteomics for the Discovery of Protein Biomarkers of Beef Tenderness: An Overview of Integrated Studies. Food Res. Int. 2020, 127, 108739. [Google Scholar] [CrossRef] [PubMed]
- Gagaoua, M.; Terlouw, E.M.C.; Mullen, A.M.; Franco, D.; Warner, R.D.; Lorenzo, J.M.; Purslow, P.P.; Gerrard, D.; Hopkins, D.L.; Troy, D.; et al. Molecular Signatures of Beef Tenderness: Underlying Mechanisms Based on Integromics of Protein Biomarkers from Multi-Platform Proteomics Studies. Meat Sci. 2021, 172, 108311. [Google Scholar] [CrossRef]
- Gagaoua, M.; Bonnet, M.; Picard, B. Protein Array-Based Approach to Evaluate Biomarkers of Beef Tenderness and Marbling in Cows: Understanding of the Underlying Mechanisms and Prediction. Foods 2020, 9, 1180. [Google Scholar] [CrossRef]
- Picard, B.; Gagaoua, M. Chapter 11—Proteomic Investigations of Beef Tenderness. In Proteomics in Food Science; Colgrave, M.L., Ed.; Academic Press: Cambridge, MA, USA, 2017; pp. 177–197. ISBN 978-0-12-804007-2. [Google Scholar]
- Munekata, P.E.; Pateiro, M.; López-Pedrouso, M.; Gagaoua, M.; Lorenzo, J.M. Foodomics in Meat Quality. Curr. Opin. Food Sci. 2021, 38, 79–85. [Google Scholar] [CrossRef]
- Gagaoua, M.; Hughes, J.; Terlouw, E.M.C.; Warner, R.D.; Purslow, P.P.; Lorenzo, J.M.; Picard, B. Proteomic Biomarkers of Beef Colour. Trends Food Sci. Technol. 2020, 101, 234–252. [Google Scholar] [CrossRef]
- Zhu, Y.; Mullen, A.M.; Rai, D.K.; Kelly, A.L.; Sheehan, D.; Cafferky, J.; Hamill, R.M. Assessment of RNAlater® as a Potential Method to Preserve Bovine Muscle Proteins Compared with Dry Ice in a Proteomic Study. Foods 2019, 8, 60. [Google Scholar] [CrossRef]
- Bouley, J.; Chambon, C.; Picard, B. Mapping of Bovine Skeletal Muscle Proteins Using Two-Dimensional Gel Electrophoresis and Mass Spectrometry. Proteomics 2004, 4, 1811–1824. [Google Scholar] [CrossRef]
- Bradford, M.M. A Rapid and Sensitive Method for the Quantitation of Microgram Quantities of Protein Utilizing the Principle of Protein-Dye Binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Zhu, Y.; Gagaoua, M.; Mullen, A.M.; Viala, D.; Rai, D.K.; Kelly, A.L.; Sheehan, D.; Hamill, R.M. Shotgun Proteomics for the Preliminary Identification of Biomarkers of Beef Sensory Tenderness, Juiciness and Chewiness from Plasma and Muscle of Young Limousin-Sired Bulls. Meat Sci. 2021, 176, 108488. [Google Scholar] [CrossRef] [PubMed]
- Gagaoua, M.; Troy, D.; Mullen, A.M. The Extent and Rate of the Appearance of the Major 110 and 30 KDa Proteolytic Fragments during Post-Mortem Aging of Beef Depend on the Glycolysing Rate of the Muscle and Aging Time: An LC–MS/MS Approach to Decipher Their Proteome and Associated Pathways. J. Agric. Food Chem. 2021, 69, 602–614. [Google Scholar] [CrossRef] [PubMed]
- Kaspric, N.; Picard, B.; Reichstadt, M.; Tournayre, J.; Bonnet, M. ProteINSIDE to Easily Investigate Proteomics Data from Ruminants: Application to Mine Proteome of Adipose and Muscle Tissues in Bovine Foetuses. PLoS ONE 2015, 10, e0128086. [Google Scholar] [CrossRef]
- Zhou, Y.; Zhou, B.; Pache, L.; Chang, M.; Khodabakhshi, A.H.; Tanaseichuk, O.; Benner, C.; Chanda, S.K. Metascape Provides a Biologist-Oriented Resource for the Analysis of Systems-Level Datasets. Nat. Commun. 2019, 10, 1523. [Google Scholar] [CrossRef]
- Gagaoua, M.; Terlouw, E.M.C.; Picard, B. The Study of Protein Biomarkers to Understand the Biochemical Processes Underlying Beef Color Development in Young Bulls. Meat Sci. 2017, 134, 18–27. [Google Scholar] [CrossRef] [PubMed]
- Huffman, K.L.; Miller, M.F.; Hoover, L.C.; Wu, C.K.; Brittin, H.C.; Ramsey, C.B. Effect of Beef Tenderness on Consumer Satisfaction with Steaks Consumed in the Home and Restaurant. J. Anim Sci. 1996, 74, 91–97. [Google Scholar] [CrossRef]
- Ye, M.; Ye, F.; He, L.; Luo, B.; Yang, F.; Cui, C.; Zhao, X.; Yin, H.; Li, D.; Xu, H.; et al. Transcriptomic Analysis of Chicken Myozenin 3 Regulation Reveals Its Potential Role in Cell Proliferation. PLoS ONE 2017, 12, e0189476. [Google Scholar] [CrossRef]
- Ye, M.; Ye, F.; He, L.; Liu, Y.; Zhao, X.; Yin, H.; Li, D.; Xu, H.; Zhu, Q.; Wang, Y. Molecular Cloning, Expression Profiling, and Marker Validation of the Chicken Myoz3 Gene. Biomed. Res. Int. 2017, 2017, 5930918. [Google Scholar] [CrossRef] [PubMed]
- Boudon, S.; Ounaissi, D.; Viala, D.; Monteils, V.; Picard, B.; Cassar-Malek, I. Label Free Shotgun Proteomics for the Identification of Protein Biomarkers for Beef Tenderness in Muscle and Plasma of Heifers. J. Proteom. 2020, 217, 103685. [Google Scholar] [CrossRef] [PubMed]
- Elliott, K.; Sakamuro, D.; Basu, A.; Du, W.; Wunner, W.; Staller, P.; Gaubatz, S.; Zhang, H.; Prochownik, E.; Eilers, M.; et al. Bin1 Functionally Interacts with Myc and Inhibits Cell Proliferation via Multiple Mechanisms. Oncogene 1999, 18, 3564–3573. [Google Scholar] [CrossRef]
- Karisa, B.K.; Thomson, J.; Wang, Z.; Stothard, P.; Moore, S.S.; Plastow, G.S. Candidate Genes and Single Nucleotide Polymorphisms Associated with Variation in Residual Feed Intake in Beef Cattle1. J. Anim. Sci. 2013, 91, 3502–3513. [Google Scholar] [CrossRef] [PubMed]
- Hu, S.-M.; Li, F.; Yu, H.-M.; Li, R.-Y.; Ma, Q.-Y.; Ye, T.-J.; Lu, Z.-Y.; Chen, J.-L.; Song, H.-D. The Mimecan Gene Expressed in Human Pituitary and Regulated by Pituitary Transcription Factor-1 as a Marker for Diagnosing Pituitary Tumors. J. Clin. Endocrinol. Metab. 2005, 90, 6657–6664. [Google Scholar] [CrossRef] [PubMed]
- Ohsaki, H.; Okada, M.; Sasazaki, S.; Hinenoya, T.; Sawa, T.; Iwanaga, S.; Tsuruta, H.; Mukai, F.; Mannen, H. Proteomic Comparison between Japanese Black and Holstein Cattle by Two-Dimensional Gel Electrophoresis and Identification of Proteins. Asian-Australas. J. Anim. Sci. 2007, 20, 638–644. [Google Scholar] [CrossRef]
- Tasheva, E.S.; Koester, A.; Paulsen, A.Q.; Garrett, A.S.; Boyle, D.L.; Davidson, H.J.; Song, M.; Fox, N.; Conrad, G.W. Mimecan/Osteoglycin-Deficient Mice Have Collagen Fibril Abnormalities. Mol. Vis. 2002, 8, 407–415. [Google Scholar] [PubMed]
- Lowey, S.; Waller, G.S.; Trybus, K.M. Skeletal Muscle Myosin Light Chains Are Essential for Physiological Speeds of Shortening. Nature 1993, 365, 454–456. [Google Scholar] [CrossRef] [PubMed]
- Picard, B.; Gagaoua, M.; Micol, D.; Cassar-Malek, I.; Hocquette, J.-F.; Terlouw, C.E.M. Inverse Relationships between Biomarkers and Beef Tenderness According to Contractile and Metabolic Properties of the Muscle. J. Agric. Food Chem. 2014, 62, 9808–9818. [Google Scholar] [CrossRef]
- Mato, A.; Rodríguez-Vázquez, R.; López-Pedrouso, M.; Bravo, S.; Franco, D.; Zapata, C. The First Evidence of Global Meat Phosphoproteome Changes in Response to Pre-Slaughter Stress. Bmc Genom. 2019, 20, 590. [Google Scholar] [CrossRef] [PubMed]
- De Rodrigues, R.T.S.; Chizzotti, M.L.; Vital, C.E.; Baracat-Pereira, M.C.; Barros, E.; Busato, K.C.; Gomes, R.A.; Ladeira, M.M.; Martins, T.D.S. Differences in Beef Quality between Angus (Bos Taurus Taurus) and Nellore (Bos Taurus Indicus) Cattle through a Proteomic and Phosphoproteomic Approach. PLoS ONE 2017, 12, e0170294. [Google Scholar] [CrossRef]
- Ouali, A.; Gagaoua, M.; Boudida, Y.; Becila, S.; Boudjellal, A.; Herrera-Mendez, C.H.; Sentandreu, M.A. Biomarkers of Meat Tenderness: Present Knowledge and Perspectives in Regards to Our Current Understanding of the Mechanisms Involved. Meat Sci. 2013, 95, 854–870. [Google Scholar] [CrossRef] [PubMed]
- Weston, A.R.; Rogers, R.W.; Althen, T.G. Review: The Role of Collagen in Meat Tenderness. Prof. Anim. Sci. 2002, 18, 107–111. [Google Scholar] [CrossRef]
- Lepetit, J. Collagen Contribution to Meat Toughness: Theoretical Aspects. Meat Sci. 2008, 80, 960–967. [Google Scholar] [CrossRef]
- Bjarnadóttir, S.G.; Hollung, K.; Høy, M.; Bendixen, E.; Codrea, M.C.; Veiseth-Kent, E. Changes in Protein Abundance between Tender and Tough Meat from Bovine Longissimus Thoracis Muscle Assessed by Isobaric Tag for Relative and Absolute Quantitation (ITRAQ) and 2-Dimensional Gel Electrophoresis Analysis1. J. Anim. Sci. 2012, 90, 2035–2043. [Google Scholar] [CrossRef]
- Liao, H.; Zhang, X.H.; Qi, Y.X.; Wang, Y.Q.; Pang, Y.Z.; Liu, Z.B.Z.P. The Relationships of Collagen and ADAMTS2 Expressionlevels with Meat Quality Traits in Cattle. Indian J. Anim. Res. 2016, 52, 167–172. [Google Scholar] [CrossRef]
- Cui, L.; Cheng, Z.; Hu, K.; Pang, Y.; Liu, Y.; Qian, T.; Quan, L.; Dai, Y.; Pang, Y.; Ye, X.; et al. Prognostic Value of the PDLIM Family in Acute Myeloid Leukemia. Am. J. Transl Res. 2019, 11, 6124–6131. [Google Scholar]
- Ríos, H.; Paganelli, A.R.; Fosser, N.S. The Role of PDLIM1, a PDZ-LIM Domain Protein, at the Ribbon Synapses in the Chicken Retina. J. Comp. Neurol. 2020, 528, 1820–1832. [Google Scholar] [CrossRef]
- Schönichen, A.; Mannherz, H.G.; Behrmann, E.; Mazur, A.J.; Kühn, S.; Silván, U.; Schoenenberger, C.-A.; Fackler, O.T.; Raunser, S.; Dehmelt, L.; et al. FHOD1 Is a Combined Actin Filament Capping and Bundling Factor That Selectively Associates with Actin Arcs and Stress Fibers. J. Cell Sci. 2013, 126, 1891–1901. [Google Scholar] [CrossRef]
- Hemerich, D.; Pei, J.; Harakalova, M.; van Setten, J.; Boymans, S.; Boukens, B.J.; Efimov, I.R.; Michels, M.; van der Velden, J.; Vink, A.; et al. Integrative Functional Annotation of 52 Genetic Loci Influencing Myocardial Mass Identifies Candidate Regulatory Variants and Target Genes. Circ. Genom Precis Med. 2019, 12, e002328. [Google Scholar] [CrossRef]
- Hansen, M.D.H.; Kwiatkowski, A.V. Chapter One—Control of Actin Dynamics by Allosteric Regulation of Actin Binding Proteins. In International Review of Cell and Molecular Biology; Jeon, K.W., Ed.; Academic Press: Cambridge, MA, USA, 2013; Volume 303, pp. 1–25. [Google Scholar]
- Ferguson, D.M.; Gerrard, D.E. Regulation of Post-Mortem Glycolysis in Ruminant Muscle. Anim. Prod. Sci. 2014, 54, 464–481. [Google Scholar] [CrossRef]
- Orosz, F.; Oláh, J.; Ovádi, J. Triosephosphate Isomerase Deficiency: New Insights into an Enigmatic Disease. Biochim. Biophys. Acta (BBA) Mol. Basis Dis. 2009, 1792, 1168–1174. [Google Scholar] [CrossRef]
- Esposito, G.; Vitagliano, L.; Costanzo, P.; Borrelli, L.; Barone, R.; Pavone, L.; Izzo, P.; Zagari, A.; Salvatore, F. Human Aldolase A Natural Mutants: Relationship between Flexibility of the C-Terminal Region and Enzyme Function. Biochem. J. 2004, 380, 51–56. [Google Scholar] [CrossRef]
- Hughes, J.; Clarke, F.; Li, Y.; Purslow, P.; Warner, R. Differences in Light Scattering between Pale and Dark Beef Longissimus Thoracis Muscles Are Primarily Caused by Differences in the Myofilament Lattice, Myofibril and Muscle Fibre Transverse Spacings. Meat Sci. 2019, 149, 96–106. [Google Scholar] [CrossRef]
- Israelsen, W.J.; Vander Heiden, M.G. Pyruvate Kinase: Function, Regulation and Role in Cancer. Semin. Cell Dev. Biol. 2015, 43, 43–51. [Google Scholar] [CrossRef]
- Daugaard, M.; Rohde, M.; Jäättelä, M. The Heat Shock Protein 70 Family: Highly Homologous Proteins with Overlapping and Distinct Functions. Febs Lett. 2007, 581, 3702–3710. [Google Scholar] [CrossRef]
- Jiang, B.; Liang, P.; Deng, G.; Tu, Z.; Liu, M.; Xiao, X. Increased Stability of Bcl-2 in HSP70-Mediated Protection against Apoptosis Induced by Oxidative Stress. Cell Stress Chaperones 2011, 16, 143–152. [Google Scholar] [CrossRef]
- Mayer, M.P. Hsp70 Chaperone Dynamics and Molecular Mechanism. Trends Biochem. Sci. 2013, 38, 507–514. [Google Scholar] [CrossRef]
- Morzel, M.; Terlouw, C.; Chambon, C.; Micol, D.; Picard, B. Muscle Proteome and Meat Eating Qualities of Longissimus Thoracis of “Blonde d’Aquitaine” Young Bulls: A Central Role of HSP27 Isoforms. Meat Sci. 2008, 78, 297–304. [Google Scholar] [CrossRef]
- Caruso Bavisotto, C.; Alberti, G.; Vitale, A.M.; Paladino, L.; Campanella, C.; Rappa, F.; Gorska, M.; Conway de Macario, E.; Cappello, F.; Macario, A.J.L.; et al. Hsp60 Post-Translational Modifications: Functional and Pathological Consequences. Front. Mol. Biosci. 2020, 7. [Google Scholar] [CrossRef]
- McDonagh, B.; Sheehan, D. Redox Proteomics in the Blue Mussel Mytilus Edulis: Carbonylation Is Not a Pre-Requisite for Ubiquitination in Acute Free Radical-Mediated Oxidative Stress. Aquat. Toxicol. 2006, 79, 325–333. [Google Scholar] [CrossRef]
- Sierra, V.; Oliván, M. Role of Mitochondria on Muscle Cell Death and Meat Tenderization. Recent Pat. Endocr. Metab. Immune Drug Discov. 2013. [Google Scholar] [CrossRef]
- Lana, A.; Zolla, L. Apoptosis or Autophagy, That Is the Question: Two Ways for Muscle Sacrifice towards Meat. Trends Food Sci. Technol. 2015, 46, 231–241. [Google Scholar] [CrossRef]
- Thomas, K.J.; McCoy, M.K.; Blackinton, J.; Beilina, A.; van der Brug, M.; Sandebring, A.; Miller, D.; Maric, D.; Cedazo-Minguez, A.; Cookson, M.R. DJ-1 Acts in Parallel to the PINK1/Parkin Pathway to Control Mitochondrial Function and Autophagy. Hum. Mol. Genet. 2011, 20, 40–50. [Google Scholar] [CrossRef]
- Gagaoua, M.; Bonnet, M.; Ellies-Oury, M.-P.; De Koning, L.; Picard, B. Reverse Phase Protein Arrays for the Identification/Validation of Biomarkers of Beef Texture and Their Use for Early Classification of Carcasses. Food Chem. 2018, 250, 245–252. [Google Scholar] [CrossRef]
- Jia, X.; Veiseth-Kent, E.; Grove, H.; Kuziora, P.; Aass, L.; Hildrum, K.I.; Hollung, K. Peroxiredoxin-6--a Potential Protein Marker for Meat Tenderness in Bovine Longissimus Thoracis Muscle. J. Anim. Sci. 2009, 87, 2391–2399. [Google Scholar] [CrossRef]
- Fan, J.; Ren, H.; Jia, N.; Fei, E.; Zhou, T.; Jiang, P.; Wu, M.; Wang, G. DJ-1 Decreases Bax Expression through Repressing P53 Transcriptional Activity. J. Biol. Chem. 2008, 283, 4022–4030. [Google Scholar] [CrossRef] [PubMed]
- Fisher, A.B. Peroxiredoxin 6: A Bifunctional Enzyme with Glutathione Peroxidase and Phospholipase A2 Activities. Antioxid. Redox Signal. 2010, 15, 831–844. [Google Scholar] [CrossRef]
- Cao, Z.; Bhella, D.; Lindsay, J.G. Reconstitution of the Mitochondrial PrxIII Antioxidant Defence Pathway: General Properties and Factors Affecting PrxIII Activity and Oligomeric State. J. Mol. Biol. 2007, 372, 1022–1033. [Google Scholar] [CrossRef] [PubMed]
- Grabež, V.; Kathri, M.; Phung, V.; Moe, K.M.; Slinde, E.; Skaugen, M.; Saarem, K.; Egelandsdal, B. Protein Expression and Oxygen Consumption Rate of Early Postmortem Mitochondria Relate to Meat Tenderness. J. Anim. Sci. 2015, 93, 1967–1979. [Google Scholar] [CrossRef]
- Polati, R.; Menini, M.; Robotti, E.; Millioni, R.; Marengo, E.; Novelli, E.; Balzan, S.; Cecconi, D. Proteomic Changes Involved in Tenderization of Bovine Longissimus Dorsi Muscle during Prolonged Ageing. Food Chem. 2012, 135, 2052–2069. [Google Scholar] [CrossRef]
- Malheiros, J.M.; Braga, C.P.; Grove, R.A.; Ribeiro, F.A.; Calkins, C.R.; Adamec, J.; Chardulo, L.A.L. Influence of Oxidative Damage to Proteins on Meat Tenderness Using a Proteomics Approach. Meat Sci. 2019, 148, 64–71. [Google Scholar] [CrossRef]
- Gagaoua, M.; Claudia Terlouw, E.M.; Boudjellal, A.; Picard, B. Coherent Correlation Networks among Protein Biomarkers of Beef Tenderness: What They Reveal. J. Proteom. 2015, 128, 365–374. [Google Scholar] [CrossRef] [PubMed]
- Cai, C.; Masumiya, H.; Weisleder, N.; Matsuda, N.; Nishi, M.; Hwang, M.; Ko, J.-K.; Lin, P.; Thornton, A.; Zhao, X.; et al. MG53 Nucleates Assembly of Cell Membrane Repair Machinery. Nat. Cell Biol. 2009, 11, 56–64. [Google Scholar] [CrossRef] [PubMed]
| Quality Traits | Min | Max | Mean | SD | CV (%) |
|---|---|---|---|---|---|
| WBSF (N) (n = 9) | 27.70 | 38.85 | 33.21 | 3.24 | 9.75 |
| WBSF (N) (n = 9) | 59.25 | 71.40 | 63.96 | 3.98 | 6.22 |
| Uniprot ID | Gene Name | Full Protein Name | Differences | Pearson Correlations a | Overlap with Gagaoua et al. Database [13] | |
|---|---|---|---|---|---|---|
| Fold Change (Log2) | p-Value | WBSF | ||||
| Muscle contraction, structure and associated proteins (n = 17) | ||||||
| Q08DI7 | MYOZ3 b | Myozenin 3 | −0.53 | 0.002 | 0.741 *** | ✓ |
| E1BNG8 | BIN1 | Bridging Integrator-1 | −0.25 | 0.003 | 0.616 ** | |
| Q148F1 | CFL2 | Cofilin-2 | −0.47 | 0.003 | 0.670 ** | |
| E1BIN0 | FHOD1 | Formin homology 2 domain containing 1 | −0.40 | 0.005 | 0.714 *** | |
| A6QLZ8 | CORO6 | Coronin | −0.57 | 0.007 | 0.607 ** | |
| Q0P571 | MYLPF | Myosin regulatory light chain 2 | −0.71 | 0.009 | 0.613 ** | ✓ |
| Q3SX40 | PDLIM7 b | PDZ and LIM domain protein 7 | −0.47 | 0.010 | 0.642 ** | ✓ |
| Q0VC48 | TMOD4 | Tropomodulin-4 | −0.42 | 0.010 | 0.640 ** | TMOD1 |
| Q2KJH4 | WDR1 | WD repeat-containing protein 1 | −0.49 | 0.019 | 0.542 * | ✓ |
| P60712 | ACTB | Actin, cytoplasmic 1 | −0.98 | 0.023 | 0.524 * | ✓ |
| A0JNJ5 | MYL1 | Myosin light chain 1/3, skeletal muscle isoform | −0.47 | 0.024 | 0.554 * | ✓ |
| P02453 | COL1A1 | Collagen alpha-1(I) chain | 0.89 | 0.025 | −0.536 * | ✓ |
| Q3SYZ8 | PDLIM3 | PDZ and LIM domain protein 3 | −0.51 | 0.029 | 0.512 * | PDLIM7/PDLIM1 |
| Q0III9 | ACTN3 c | Alpha-actinin-3 | −0.34 | 0.034 | 0.529 * | ✓ |
| A4FV78 | KLHL41 | KBTBD10 protein | −0.38 | 0.037 | ✓ | |
| F1N789 | VCL | Vinculin | −0.21 | 0.040 | ✓ | |
| Q32LP2 | RDX | Radixin | −0.26 | 0.043 | 0.476 * | |
| Energy metabolism (n = 5) | ||||||
| Q5E956 | TPI1 | Triosephosphate isomerase | −0.35 | 0.018 | 0.631 ** | ✓ |
| Q3ZBY4 | ALDOC | Fructose-bisphosphate aldolase | −0.33 | 0.018 | 0.621 ** | ✓ |
| A5D984 | PKM | Pyruvate kinase | −0.39 | 0.029 | 0.540 * | ✓ |
| A6QLL8 | ALDOA | Fructose-bisphosphate aldolase | −0.29 | 0.029 | 0.554 * | ✓ |
| A3KN12 | ADSL | Adenylosuccinate lyase | 0.27 | 0.019 | −0.629 ** | |
| Heat shock proteins (n = 4) | ||||||
| P19120 | HSPA8 b | Heat shock cognate 71 kDa protein | −0.41 | 0.006 | 0.590 ** | ✓ |
| P31081 | HSPD1 | 60 kDa heat shock protein, mitochondrial | 0.84 | 0.016 | −0.567 * | |
| Q3T149 | HSPB1 | Heat shock protein beta-1 | −0.44 | 0.038 | 0.531 * | ✓ |
| Q3ZBZ8 | STIP1 b,c | Stress-induced-phosphoprotein 1 | −0.29 | 0.040 | 0.484 * | ✓ |
| Oxidative stress (n = 2) | ||||||
| P35705 | PRDX3 | Thioredoxin-dependent peroxide reductase | 0.32 | 0.038 | −0.488 * | PRDX6/PRDX1/PRDX2 |
| Q5E946 | PARK7 | Protein/nucleic acid deglycase DJ-1 | −0.47 | 0.046 | 0.501 * | ✓ |
| Other pathways (n = 6) | ||||||
| P11116 | LGALS1 | Galectin-1 | −0.56 | 0.006 | 0.639 ** | ✓ |
| E1BE77 | TRIM72 | Tripartite motif containing 72 | −0.45 | 0.008 | 0.620 ** | ✓ |
| P19879 | OGN b | Mimecan | −0.64 | 0.017 | 0.589 * | |
| Q2HJF7 | CAMK2D | Calcium/calmodulin-dependent protein kinase | −0.20 | 0.038 | 0.510 * | |
| Q6EWQ7 | EIF5A | Eukaryotic translation initiation factor 5A-1 | −0.45 | 0.046 | 0.574 * | |
| Q3SYR3 | APOBEC2 | Probable C->U-editing enzyme APOBEC-2 | −0.66 | 0.047 | 0.530 * | |
| R-Squared a | S.E | Entered Independent Variable b | Partial R-Squared | Regression Coefficient | t-Value | p-Value |
|---|---|---|---|---|---|---|
| 0.79 ** | 0.125 | MYOZ3 | 0.52 | 0.486 | 3.875 | 0.002 |
| 0.116 | BIN1 | 0.17 | 0.454 | 3.907 | 0.002 | |
| 0.121 | OGN | 0.1 | 0.347 | 2.868 | 0.012 |
| Proteins | VIP | Direction (+ or −) |
|---|---|---|
| MYOZ3: Myozenin 3 | 1.291 | − |
| FHOD1: Formin homology 2 domain containing 1 | 1.243 | − |
| CFL2: Cofilin-2 | 1.168 | − |
| PDLIM7: PDZ and LIM domain protein 7 | 1.119 | − |
| TMOD4: Tropomodulin-4 | 1.115 | − |
| LGALS1: Galectin-1 | 1.113 | − |
| TPI1: Triosephosphate isomerase | 1.099 | − |
| ADSL: Adenylosuccinate lyase | 1.095 | + |
| ALDOC: Fructose-bisphosphate aldolase | 1.082 | − |
| TRIM72: Tripartite motif containing 72 | 1.080 | − |
| BIN1: Bridging Integrator-1 | 1.073 | − |
| MYLPF: Myosin regulatory light chain 2, skeletal muscle isoform | 1.067 | − |
| CORO6: Coronin | 1.056 | − |
| HSPA8: Heat shock cognate 71 kDa protein | 1.027 | − |
| OGN: Mimecan | 1.025 | − |
| EIF5A: Eukaryotic translation initiation factor 5A-1 | 1.000 | − |
| HSPD1: 60 kDa heat shock protein, mitochondrial | 0.987 | + |
| MYL1: Myosin light chain 1/3, skeletal muscle isoform | 0.965 | − |
| ALDOA: Fructose-bisphosphate aldolase | 0.965 | − |
| WDR1: WD repeat-containing protein 1 | 0.943 | − |
| PKM: Pyruvate kinase | 0.940 | − |
| COL1A1: Collagen alpha-1(I) chain | 0.934 | + |
| HSPB1: Heat shock protein beta-1 | 0.924 | − |
| APOBEC2: Probable C->U-editing enzyme APOBEC-2 | 0.923 | − |
| ACTN3: Alpha-actinin-3 | 0.922 | − |
| ACTB: Actin, cytoplasmic 1 | 0.913 | − |
| PDLIM3: PDZ and LIM domain protein 3 | 0.892 | − |
| CAMK2D: Calcium/calmodulin-dependent protein kinase type II subunit delta | 0.889 | − |
| PARK7: Protein/nucleic acid deglycase DJ-1 | 0.873 | − |
| PRDX3: Thioredoxin-dependent peroxide reductase, mitochondrial | 0.850 | + |
| STIP1: Stress-induced-phosphoprotein 1 | 0.843 | − |
| RDX: Radixin | 0.830 | − |
| KLHL41: KBTBD10 protein | 0.771 | − |
| VCL: Vinculin | 0.708 | − |
| GO | Function | Gene Name | GO Frequency within the Dataset (%) | GO Frequency within the Genome (%) | p-Values |
|---|---|---|---|---|---|
| Biological Process (BP) | |||||
| GO:0061621 | canonical glycolysis | ALDOC TPI1 PKM ALDOA | 11.76 | 14.81 | 2.13 × 10−9 |
| GO:0006096 | glycolytic process | ALDOC ALDOA PKM TPI1 | 11.76 | 10.26 | 5.4 × 10−9 |
| GO:0006936 | muscle contraction | TRIM72 MYL1 VCL MYLPF TMOD4 | 14.71 | 2.35 | 2.86 × 10−8 |
| GO:0043312 | neutrophil degranulation | VCL HSPA8 ALDOA ALDOC PKM | 14.71 | 1.03 | 1.28 × 10−6 |
| GO:0070527 | platelet aggregation | ACTB VCL HSPB1 | 8.82 | 7.14 | 2.06 × 10−6 |
| GO:0006094 | gluconeogenesis | ALDOC TPI1 ALDOA | 8.82 | 6.82 | 2.27 × 10−6 |
| GO:0006986 | response to unfolded protein | HSPB1 HSPA8 HSPD1 | 8.82 | 6.25 | 2.62 × 10−6 |
| GO:0035633 | maintenance of blood-brain barrier | VCL ACTB | 5.88 | 66.67 | 5.99 × 10−6 |
| GO:0030388 | fructose 1,6-bisphosphate metabolic process | ALDOA ALDOC | 5.88 | 28.57 | 1.98 × 10−5 |
| GO:0030042 | actin filament depolymerisation | WDR1 CFL2 | 5.88 | 25 | 2.34 × 10−5 |
| GO:0043297 | apical junction assembly | VCL WDR1 | 5.88 | 25 | 2.34 × 10−5 |
| GO:0002576 | platelet degranulation | ALDOA WDR1 VCL | 8.82 | 2.44 | 3.11 × 10−5 |
| GO:0030836 | positive regulation of actin filament depolymerisation | CFL2 WDR1 | 5.88 | 15.38 | 4.82 × 10−5 |
| GO:0006000 | fructose metabolic process | ALDOA ALDOC | 5.88 | 13.33 | 5.97 × 10−5 |
| GO:0007015 | actin filament organisation | ALDOA CORO6 TMOD4 | 8.82 | 1.54 | 9.6 × 10−5 |
| GO:0042026 | protein refolding | HSPD1 HSPA8 | 5.88 | 9.52 | 9.98 × 10−5 |
| GO:0030239 | myofibril assembly | KLHL41 TMOD4 | 5.88 | 7.14 | 0.000162 |
| GO:0034333 | adherens junction assembly | ACTB VCL | 5.88 | 5.88 | 0.000221 |
| GO:0043066 | negative regulation of apoptotic process | HSPD1 HSPB1 PRDX3 PARK7 | 11.76 | 0.49 | 0.000221 |
| GO:0086091 | regulation of heart rate by cardiac conduction | BIN1 CAMK2D | 5.88 | 5.56 | 0.000239 |
| Cellular Component (CC) | |||||
| GO:0005829 | cytosol | VCL TPI1 MYLPF HSPA8 STIP1 EIF5A CAMK2D ADSL MYL1 ACTN3 ALDOC HSPD1 BIN1 PRDX3 PARK7 PDLIM3 ALDOA HSPB1 WDR1 PKM FHOD1 PDLIM7 ACTB KLHL41 | 70.59 | 0.5 | 3.03 × 10−21 |
| GO:0070062 | extracellular exosome | ALDOC HSPD1 ACTN3 ACTB PARK7 TPI1 PKM VCL RDX CFL2 LGALS1 OGN ALDOA HSPA8 HSPB1 WDR1 | 47.06 | 0.58 | 1.67 × 10−15 |
| GO:0005737 | cytoplasm | PARK7 FHOD1 COL1A1 EIF5A PKM BIN1 HSPD1 HSPA8 KLHL41 PRDX3 TRIM72 APOBEC2 CAMK2D ACTB HSPB1 CFL2 LGALS1 | 50 | 0.41 | 3 × 10−14 |
| GO:0005615 | extracellular space | HSPD1 RDX ALDOA CFL2 HSPA8 TPI1 OGN COL1A1 HSPB1 LGALS1 ACTB | 32.35 | 0.77 | 3 × 10−12 |
| GO:0005925 | focal adhesion | VCL HSPB1 RDX HSPA8 ACTN3 ACTB PDLIM7 | 20.59 | 1.82 | 2.33 × 10−10 |
| GO:0015629 | actin cytoskeleton | CFL2 MYOZ3 BIN1 PDLIM7 ACTB ALDOA | 17.65 | 3.14 | 2.54 × 10−10 |
| GO:0005856 | cytoskeleton | VCL KLHL41 HSPB1 BIN1 TMOD4 ACTB FHOD1 ALDOC | 23.53 | 1.08 | 3.05 × 10−10 |
| GO:0030018 | Z disc | PDLIM7 MYOZ3 BIN1 CFL2 PDLIM3 | 14.71 | 4.2 | 2.57 × 10−9 |
| GO:1904813 | ficolin-1-rich granule lumen | ALDOC HSPA8 PKM ALDOA VCL | 14.71 | 4.03 | 2.92 × 10−9 |
| GO:0005634 | nucleus | STIP1 CAMK2D EIF5A BIN1 FHOD1 ALDOA PKM HSPB1 APOBEC2 ACTB HSPA8 PARK7 TPI1 | 38.24 | 0.26 | 4.61 × 10−9 |
| GO:0005576 | extracellular region | PKM ALDOC WDR1 LGALS1 ALDOA HSPA8 VCL COL1A1 OGN | 26.47 | 0.49 | 7.91 × 10−9 |
| GO:0034774 | secretory granule lumen | ALDOC PKM VCL ALDOA HSPA8 | 14.71 | 1.56 | 2 × 10−7 |
| GO:0031674 | I band | ALDOA CFL2 BIN1 | 8.82 | 13.64 | 3.96 × 10−7 |
| GO:0005912 | adherens junction | PARK7 ACTB PDLIM3 PDLIM7 VCL | 14.71 | 1.04 | 1.28 × 10−6 |
| GO:0030864 | cortical actin cytoskeleton | RDX CFL2 WDR1 | 8.82 | 8.57 | 1.28 × 10−6 |
| GO:0001725 | stress fibre | PDLIM3 PDLIM7 FHOD1 | 8.82 | 6.25 | 2.62 × 10−6 |
| GO:0030424 | axon | PARK7 HSPA8 BIN1 ACTB | 11.76 | 1.65 | 3.52 × 10−6 |
| GO:0005654 | nucleoplasm | PARK7 CAMK2D ACTB KLHL41 HSPA8 FHOD1 PDLIM7 | 20.59 | 0.24 | 3.81 × 10−5 |
| GO:0101031 | chaperone complex | STIP1 HSPA8 | 5.88 | 15.38 | 4.82 × 10−5 |
| GO:0005886 | plasma membrane | VCL HSPA8 BIN1 HSPD1 ACTB RDX WDR1 KLHL41 | 23.53 | 0.18 | 5.47 × 10−5 |
| Molecular Function (MF) | |||||
| GO:0005515 | protein binding | PRDX3 PARK7 ALDOA OGN MYOZ3 ACTN3 FHOD1 CAMK2D PKM HSPD1 TRIM72 COL1A1 CFL2 TMOD4 STIP1 ALDOC CORO6 BIN1 LGALS1 VCL EIF5A HSPA8 HSPB1 PDLIM3 TPI1 KLHL41 RDX ACTB PDLIM7 | 85.29 | 0.45 | 9.5 × 10−24 |
| GO:0042802 | identical protein binding | ALDOA PRDX3 TRIM72 PARK7 ACTB CAMK2D ADSL BIN1 HSPB1 APOBEC2 ACTN3 FHOD1 COL1A1 | 38.24 | 0.92 | 2.98 × 10−15 |
| GO:0003723 | RNA binding | HSPB1 HSPD1 LGALS1 EIF5A APOBEC2 HSPA8 PKM ALDOA STIP1 RDX | 29.41 | 0.62 | 2.33 × 10−10 |
| GO:0045296 | cadherin binding | PARK7 HSPA8 VCL RDX ALDOA PKM | 17.65 | 2.03 | 2.44 × 10−9 |
| GO:0051015 | actin filament binding | BIN1 FHOD1 WDR1 CFL2 CORO6 | 14.71 | 3.57 | 4.61 × 10−9 |
| GO:0003779 | actin binding | PDLIM3 VCL ALDOA PDLIM7 MYOZ3 RDX | 17.65 | 1.48 | 1.01 × 10−8 |
| GO:0008307 | structural constituent of muscle | ACTN3 MYL1 MYLPF | 8.82 | 6.52 | 2.48 × 10−6 |
| GO:0004332 | fructose-bisphosphate aldolase activity | ALDOA ALDOC | 5.88 | 66.67 | 5.99 × 10−6 |
| GO:0031625 | ubiquitin protein ligase binding | HSPA8 HSPD1 VCL TPI1 | 11.76 | 1.36 | 6.77 × 10−6 |
| GO:0051087 | chaperone binding | BIN1 HSPD1 HSPA8 | 8.82 | 3.3 | 1.44 × 10−5 |
| GO:0048156 | tau protein binding | BIN1 ACTB | 5.88 | 20 | 3.26 × 10−5 |
| GO:0023026 | MHC class II protein complex binding | PKM HSPA8 | 5.88 | 11.76 | 7.34 × 10−5 |
| GO:0051371 | muscle alpha-actinin binding | PDLIM3 PDLIM7 | 5.88 | 11.11 | 7.98 × 10−5 |
| GO:0044183 | protein folding chaperone | HSPA8 HSPB1 | 5.88 | 7.14 | 0.000162 |
| GO:0042803 | protein homodimerisation activity | PARK7 CAMK2D TPI1 HSPB1 | 11.76 | 0.53 | 0.000173 |
| GO:0003697 | single-stranded DNA binding | HSPD1 PARK7 | 5.88 | 2.06 | 0.001335 |
| GO:0019901 | protein kinase binding | HSPB1 ACTB PRDX3 | 8.82 | 0.54 | 0.001393 |
| GO:0044325 | ion channel binding | ACTN3 CAMK2D | 5.88 | 1.82 | 0.001617 |
| GO:0002020 | protease binding | COL1A1 BIN1 | 5.88 | 1.61 | 0.001708 |
| GO:0070626 | (S)-2-(5-amino-1-(5-phospho-D-ribosyl)imidazole-4-carboxamido)succinate AMP-lyase (fumarate-forming) activity | ADSL | 2.94 | 100 | 0.001708 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhu, Y.; Gagaoua, M.; Mullen, A.M.; Kelly, A.L.; Sweeney, T.; Cafferky, J.; Viala, D.; Hamill, R.M. A Proteomic Study for the Discovery of Beef Tenderness Biomarkers and Prediction of Warner–Bratzler Shear Force Measured on Longissimus thoracis Muscles of Young Limousin-Sired Bulls. Foods 2021, 10, 952. https://doi.org/10.3390/foods10050952
Zhu Y, Gagaoua M, Mullen AM, Kelly AL, Sweeney T, Cafferky J, Viala D, Hamill RM. A Proteomic Study for the Discovery of Beef Tenderness Biomarkers and Prediction of Warner–Bratzler Shear Force Measured on Longissimus thoracis Muscles of Young Limousin-Sired Bulls. Foods. 2021; 10(5):952. https://doi.org/10.3390/foods10050952
Chicago/Turabian StyleZhu, Yao, Mohammed Gagaoua, Anne Maria Mullen, Alan L. Kelly, Torres Sweeney, Jamie Cafferky, Didier Viala, and Ruth M. Hamill. 2021. "A Proteomic Study for the Discovery of Beef Tenderness Biomarkers and Prediction of Warner–Bratzler Shear Force Measured on Longissimus thoracis Muscles of Young Limousin-Sired Bulls" Foods 10, no. 5: 952. https://doi.org/10.3390/foods10050952
APA StyleZhu, Y., Gagaoua, M., Mullen, A. M., Kelly, A. L., Sweeney, T., Cafferky, J., Viala, D., & Hamill, R. M. (2021). A Proteomic Study for the Discovery of Beef Tenderness Biomarkers and Prediction of Warner–Bratzler Shear Force Measured on Longissimus thoracis Muscles of Young Limousin-Sired Bulls. Foods, 10(5), 952. https://doi.org/10.3390/foods10050952








