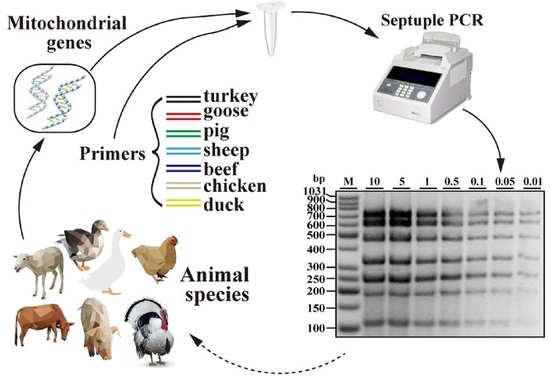
A Simple and Reliable Single Tube Septuple PCR Assay for Simultaneous Identification of Seven Meat Species
Abstract
1. Introduction
2. Materials and Methods
2.1. Preparation of Meat Samples and DNA Extraction
2.2. Design of Species-Specific Primers
2.3. Simplex and Multiplex PCR
2.4. Test of Primers’ Specificity, Sensitivity and Reproducibility
2.5. Commercial Samples
3. Results
3.1. Specificity Assays of Simplex PCR
3.2. Sensitivity Assays of Septuple PCR
3.3. Validation of Reproducibility Assay in Thermally Processed Meat
3.4. Application of Multiplex PCR Assay on Commercially Processed Meat Products
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Li, Y.; Liu, S.; Meng, F.; Liu, D.; Zhang, Y.; Wang, W.; Zhang, J. Comparative review and the recent progress in detection technologies of meat product adulteration. Compr. Rev. Food Sci. Food Saf. 2020, 19, 2256–2296. [Google Scholar] [CrossRef] [PubMed]
- Chung, S.M.; Hellberg, R.S. Effects of poor sanitation procedures on cross-contamination of animal species in ground meat products. Food Control. 2020, 109, 106927. [Google Scholar] [CrossRef]
- Da Costa Filho, P.A.; Cobuccio, L.; Mainali, D.; Rault, M.; Cavin, C. Rapid analysis of food raw materials adulteration using laser direct infrared spectroscopy and imaging. Food Control. 2020, 113, 107114. [Google Scholar] [CrossRef]
- Mansouri, M.; Khalilzadeh, B.; Barzegari, A.; Shoeibi, S.; Isildak, S.; Bargahi, N.; Omidi, Y.; Dastmalchi, S.; Rashidi, M.-R. Design a highly specific sequence for electrochemical evaluation of meat adulteration in cooked sausages. Biosens. Bioelectron. 2020, 150, 111916. [Google Scholar] [CrossRef] [PubMed]
- Mokhtar, N.F.K.; El Sheikha, A.F.; Azmi, N.I.; Mustafa, S. Potential authentication of various meat-based products using simple and efficient DNA extraction method. J. Sci. Food Agric. 2019, 100, 1687–1693. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Wang, X.; Zhang, Q.; Liu, Z.; Zhou, X.; Liu, B. A multiplex PCR method for detection of five animal species in processed meat products using novel species-specific nuclear DNA sequences. Eur. Food Res. Technol. 2020, 246, 1351–1360. [Google Scholar] [CrossRef]
- Barbin, D.F.; Badaró, A.T.; Honorato, D.C.; Ida, E.Y.; Shimokomaki, M. Identification of turkey meat and processed products using near infrared spectroscopy. Food Control. 2020, 107, 106816. [Google Scholar] [CrossRef]
- Safdar, M.; Junejo, Y.; Arman, K.; Abasıyanık, M. A highly sensitive and specific tetraplex PCR assay for soybean, poultry, horse and pork species identification in sausages: Development and validation. Meat Sci. 2014, 98, 296–300. [Google Scholar] [CrossRef]
- Tanabe, S.; Miyauchi, E.; Muneshige, A.; Mio, K.; Sato, C.; Sato, M. PCR method of detecting pork in foods for verifying allergen labeling and for identifying hidden pork ingredients in processed foods. Biosci. Biotechnol. Biochem. 2007, 71, 1663–1667. [Google Scholar] [CrossRef][Green Version]
- Li, T.T.; Jalbani, Y.M.; Zhang, G.L.; Zhao, Z.Y.; Wang, Z.Y.; Zhao, X.Y.; Chen, A.L. Detection of goat meat adulteration by real-time PCR based on a reference primer. Food Chem. 2019, 277, 554–557. [Google Scholar] [CrossRef]
- Li, J.; Li, J.; Xu, S.; Xiong, S.; Yang, J.; Chen, X.; Wang, S.; Qiao, X.; Zhou, T. A rapid and reliable multiplex PCR assay for simultaneous detection of fourteen animal species in two tubes. Food Chem. 2019, 295, 395–402. [Google Scholar] [CrossRef]
- Leng, T.; Li, F.; Xiong, L.; Xiong, Q.; Zhu, M.; Chen, Y. Quantitative detection of binary and ternary adulteration of minced beef meat with pork and duck meat by NIR combined with chemometrics. Food Control. 2020, 113, 107203. [Google Scholar] [CrossRef]
- Asensio, L.; González, I.; García, T.; Martín, R. Determination of food authenticity by enzyme-linked immunosorbent assay (ELISA). Food Control. 2008, 19, 1–8. [Google Scholar] [CrossRef]
- Sentandreu, M.Á.; Sentandreu, E. Authenticity of meat products: Tools against fraud. Food Res. Int. 2014, 60, 19–29. [Google Scholar] [CrossRef]
- Liu, G.; Luo, J.; Xu, W.; Li, C.; Guo, Y.; Guo, L. Improved triplex real-time PCR with endogenous control for synchronous identification of DNA from chicken, duck, and goose meat. Food Sci. Nutr. 2021. [Google Scholar] [CrossRef]
- Li, J.; Li, J.; Liu, R.; Wei, Y.; Wang, S. Identification of eleven meat species in foodstuff by a hexaplex real-time PCR with melting curve analysis. Food Control. 2021, 121, 107599. [Google Scholar] [CrossRef]
- Dolch, K.; Andrée, S.; Schwägele, F. Comparison of real-time PCR quantification methods in the identification of poultry species in meat products. Foods 2020, 9, 1049. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Wei, Y.; Li, J.; Liu, R.; Xu, S.; Xiong, S.; Guo, Y.; Qiao, X.; Wang, S. A novel duplex SYBR green real-time PCR with melting curve analysis method for beef adulteration detection. Food Chem. 2021, 338, 127932. [Google Scholar] [CrossRef]
- Zhu, T.; Zhou, X.; Zhang, W.; Wu, Y.; Yang, J.; Xu, L.; Chen, M.; Dong, W.; Xu, H. Multiplex and real-time PCR for qualitative and quantitative donkey meat adulteration. J. Food Meas. Charact. 2021, 15, 1161–1168. [Google Scholar] [CrossRef]
- Li, T.; Wang, J.; Wang, Z.; Qiao, L.; Liu, R.; Li, S.; Chen, A. Quantitative determination of mutton adulteration with single-copy nuclear genes by real-time PCR. Food Chem. 2020, 344, 128622. [Google Scholar] [CrossRef]
- Thanakiatkrai, P.; Dechnakarin, J.; Ngasaman, R.; Kitpipit, T. Direct pentaplex PCR assay: An adjunct panel for meat species identification in Asian food products. Food Chem. 2019, 271, 767–772. [Google Scholar] [CrossRef]
- Fajardo, V.; González, I.; Rojas, M.; García, T.; Martín, R. A review of current PCR-based methodologies for the authentication of meats from game animal species. Trends Food Sci. Technol. 2010, 21, 408–421. [Google Scholar] [CrossRef]
- Kumar, A.; Kumar, R.R.; Sharma, B.D.; Gokulakrishnan, P.; Mendiratta, S.K.; Sharma, D. Identification of species origin of meat and meat products on the dna basis: A review. Crit. Rev. Food Sci. Nutr. 2013, 55, 1340–1351. [Google Scholar] [CrossRef]
- Ali, M.E.; Razzak, M.A.; Hamid, S.B.A.; Rahman, M.M.; Amin, M.A.; Rashid, N.R.A. Multiplex PCR assay for the detection of five meat species forbidden in Islamic foods. Food Chem. 2015, 177, 214–224. [Google Scholar] [CrossRef] [PubMed]
- Galal-Khallaf, A. Multiplex PCR and 12S rRNA gene sequencing for detection of meat adulteration: A case study in the Egyptian markets. Gene 2021, 764, 145062. [Google Scholar] [CrossRef]
- Vaithiyanathan, S.; Vishnuraj, M.R.; Reddy, G.N.; Kulkarni, V.V. Application of DNA technology to check misrepresentation of animal species in illegally sold meat. Biocatal. Agric. Biotechnol. 2018, 16, 564–568. [Google Scholar] [CrossRef]
- Liu, W.; Tao, J.; Xue, M.; Ji, J.; Zhang, Y.; Zhang, L.; Sun, W. A multiplex PCR method mediated by universal primers for the identification of eight meat ingredients in food products. Eur. Food Res. Technol. 2019, 245, 2385–2392. [Google Scholar] [CrossRef]
- Martín, I.; García, T.; Fajardo, V.; Rojas, M.; Pegels, N.; Hernández, P.E.; González, I.; Martín, R. SYBR-green real-time PCR approach for the detection and quantification of pig DNA in feedstuffs. Meat Sci. 2009, 82, 252–259. [Google Scholar] [CrossRef]
- Pegels, N.; González, I.; Garcia, T.; Martín, R. Avian-specific real-time PCR assay for authenticity control in farm animal feeds and pet foods. Food Chem. 2014, 142, 39–47. [Google Scholar] [CrossRef]
- Cheng, X.; He, W.; Huang, F.; Huang, M.; Zhou, G. Multiplex real-time PCR for the identification and quantification of DNA from duck, pig and chicken in Chinese blood curds. Food Res. Int. 2014, 60, 30–37. [Google Scholar] [CrossRef]
- Iqbal, M.; Saleem, M.S.; Imran, M.; Khan, W.A.; Ashraf, K.; Zahoor, M.Y.; Rashid, I.; Rehman, H.-U.; Nadeem, A.; Ali, S.; et al. Single tube multiplex PCR assay for the identification of banned meat species. Food Addit. Contam. Part. B 2020, 13, 284–291. [Google Scholar] [CrossRef]
- Mafra, I.; Ferreira, I.M.P.L.V.O.; Oliveira, M.B.P.P. Food authentication by PCR-based methods. Eur. Food Res. Technol. 2008, 227, 649–665. [Google Scholar] [CrossRef]
- Prusakova, O.V.; Glukhova, X.A.; Afanas’Eva, G.V.; Trizna, Y.A.; Nazarova, L.F.; Beletsky, I.P. A simple and sensitive two-tube multiplex PCR assay for simultaneous detection of ten meat species. Meat Sci. 2018, 137, 34–40. [Google Scholar] [CrossRef] [PubMed]
- Dantas, V.V.; Cardoso, G.V.F.; Araújo, W.S.C.; De Oliveira, A.C.D.S.; Da Silva, A.S.; Da Silva, J.B.; Pedroso, S.C.D.S.; Roos, T.B.; De Moraes, C.M.; Lourenço, L.D.F.H. Application of a multiplex polymerase chain reaction (mPCR) assay to detect fraud by substitution of bovine meat cuts with water buffalo meat in Northern Brazil. CyTA J. Food 2019, 17, 790–795. [Google Scholar] [CrossRef]
- Li, X.; Guan, Y. Specific identification of the adulterated components in beef or mutton meats using multiplex PCR. J. AOAC Int. 2019, 102, 1181–1185. [Google Scholar] [CrossRef]
- Balakrishna, K.; Sreerohini, S.; Parida, M. Ready-to-use single tube quadruplex PCR for differential identification of mutton, chicken, pork and beef in processed meat samples. Food Addit. Contam. Part A 2019, 36, 1435–1444. [Google Scholar] [CrossRef] [PubMed]
- Sultana, S.; Hossain, M.; Zaidul, I.; Ali, E. Multiplex PCR to discriminate bovine, porcine, and fish DNA in gelatin and confectionery products. LWT 2018, 92, 169–176. [Google Scholar] [CrossRef]
- Safdar, M.; Junejo, Y. The development of a hexaplex-conventional PCR for identification of six animal and plant species in foodstuffs. Food Chem. 2016, 192, 745–749. [Google Scholar] [CrossRef]
- Qin, P.; Hong, Y.; Kim, H.-Y. Multiplex-PCR assay for simultaneous identification of lamb, beef and duck in raw and heat-treated meat mixtures. J. Food Saf. 2015, 36, 367–374. [Google Scholar] [CrossRef]
- He, H.; Hong, X.; Feng, Y.; Wang, Y.; Ying, J.; Liu, Q.; Qian, Y.; Zhou, X.; Wang, D. Application of quadruple multiplex PCR detection for beef, duck, mutton and pork in mixed meat. J. Food Nutr. Res. 2015, 3, 392–398. [Google Scholar] [CrossRef]
- Mane, B.; Mendiratta, S.; Tiwari, A. Polymerase chain reaction assay for identification of chicken in meat and meat products. Food Chem. 2009, 116, 806–810. [Google Scholar] [CrossRef]
- Hou, B.; Meng, X.; Zhang, L.; Guo, J.; Li, S.; Jin, H. Development of a sensitive and specific multiplex PCR method for the simultaneous detection of chicken, duck and goose DNA in meat products. Meat Sci. 2015, 101, 90–94. [Google Scholar] [CrossRef] [PubMed]
- Yacoub, H.A.; Sadek, M.A. Identification of fraud (with pig stuffs) in chicken-processed meat through information of mitochondrial cytochrome b. Mitochondrial DNA Part A 2016, 28, 855–859. [Google Scholar] [CrossRef] [PubMed]
- Kim, M.-J.; Kim, H.-Y. Species identification of commercial jerky products in food and feed using direct pentaplex PCR assay. Food Control. 2017, 78, 1–6. [Google Scholar] [CrossRef]
- Nejad, F.P.; Tafvizi, F.; Ebrahimi, M.T.; Hosseni, S.E. Optimization of multiplex PCR for the identification of animal species using mitochondrial genes in sausages. Eur. Food Res. Technol. 2014, 239, 533–541. [Google Scholar] [CrossRef]




| Primers | Genes | Sequence (5′–3′ Direction) | Amplicons (bp) | Reference or Source |
|---|---|---|---|---|
| Turkey | 16S rRNA | CTCTAGCCCAACCACCCAT | 110 | this study |
| GCGCCTAAGGTCTTTTCTATCAC | ||||
| Goose | 16S rRNA | TTAGACGCGATAGAGACCCCA | 194 | this study |
| GTTCGCTCTCTTTAACTGCTTG | ||||
| Pig | cytochrome c oxidase subunit I | CAGCCCGGAACCCTACTTG | 254 | this study |
| GTTCATCCAGTACCCGCTCC | ||||
| Sheep | cytochrome c oxidase subunit I | AGATATCGGCACCCTTTACCTTC | 329 | this study |
| CTGCTCCGGCCTCAACCAT | ||||
| Beef | 16S rRNA | GTGCCTGATAATACTCTGACCAC | 473 | this study |
| CACCCCAACCGAAACTACCAA | ||||
| Chicken | cytochrome b | TTTCGGCTCCCTATTAGCAGTC | 612 | this study |
| AGTATGAGAGTTAAGCCCAGA | ||||
| Duck | 12S rRNA | TGCCCTCAATAGCCTTCACC | 718 | this study |
| CATACTTCTTTCCGTGTTGCC | ||||
| Eukaryotes | 12S rRNA | CAACTGGGATTAGATACCCCACTAT | 456 | [26] |
| GAGGGTGACGGGCGGTGTGT | ||||
| Eukaryotes | 16S rRNA | AAGACGAGAAGACCCTATGGA | 240 | [27] |
| GATTGCGCTGTTATCCCTAGGGTA | ||||
| Eukaryotes | 18S rRNA | AGGATCCATTGGAGGGCAAGT | 99 | [28] |
| TCCAACTACGAGCTTTTTAACTGCA |
| Products | No | Labelled | Detected Species | Adulteration | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Turkey | Goose | Pig | Sheep | Beef | Chicken | Duck | ||||
| Beef | 15 | 5 (33.3%) | ||||||||
| meat balls | 5 | beef | 1/5 b | — | 1/5 a, 1/5 b | — | 5/5 | 1/5 a | — | |
| meat slices | 5 | beef | — | — | 1/5 | — | 5/5 | — | — | |
| kebab | 5 | beef | — | 2/5 | — | 5/5 | — | — | ||
| Mutton | 15 | 6 (40.0%) | ||||||||
| meat balls | 5 | mutton | — | — | 1/5 a, 1/5 b | 5/5 | — | 1/5 a | 1/5 b | |
| meat slices | 5 | mutton | — | — | 2/5 | 5/5 | — | — | — | |
| kebab | 5 | mutton | — | — | 2/5 | 5/5 | — | — | — | |
| Pork | 15 | 4 (26.7%) | ||||||||
| meat balls | 5 | pig | — | 1/5 b | 5/5 | — | — | 1/5 a, 1/5 b | 1/5 a | |
| sausages | 5 | pig | — | — | 5/5 | — | — | 1/5 a | 1/5 b | |
| cutlets | 5 | pig | — | — | 5/5 | — | — | — | — | |
| Turkey | 15 | |||||||||
| cutlets | 5 | turkey | 5/5 | — | — | — | — | 1/5 | — | 1 (6.7%) |
| meat slices | 3 | turkey | 3/3 | — | — | — | — | — | — | |
| breast | 5 | turkey | 5/5 | — | — | — | — | — | — | |
| jerky | 2 | turkey | 2/2 | — | — | — | — | — | — | |
| Multiplex PCR Type | Sp. No a | Detection Items | Detection Limit | Detection Method b | Reference or Source |
|---|---|---|---|---|---|
| Septuple | 7 | turkey, goose, pig, sheep, beef, chicken, duck | 0.01–0.05 ng DNA | Gel | This study |
| Multiplex | 4 | ruminant, poultry, pork, and donkey | 0.01–0.1 ng/μL DNA | Gel | [25] |
| Hexaplex | 6 | chicken, cow/buffalo, sheep/goat, horse/donkey, pork, dog | 0.03–0.05 ng DNA | Gel | [31] |
| Multiplex | 5 | sheep/goat, bovine, chicken, duck, pig | 0.5 ng DNA | Gel | [6] |
| Multiplex | 2 | cattle, buffalo | 2.23–2.31 ng/μL DNA | Gel | [34] |
| Quadruple | 4 | fox, mink, or raccoon in beef and mutton | 1% for each species | Gel | [35] |
| Pentaplex | 5 | dog, duck, buffalo, goat, sheep | 0.1–0.32 ng DNA | Gel | [21] |
| Multiplex (two-tube) | 14 | cattle, donkey, Canidae (dog, fox, raccoon-dog), deer and horse, pig, Ovis (sheep, goat), poultry (chicken, duck), cat, mouse | 0.02–0.2 ng DNA | Chip | [11] |
| Quadruplex | 4 | chicken, mutton, beef, pork | 16 pg DNA, 0.01% of each species | Gel | [36] |
| Multiplex (two-tube) | 10 | beef, sheep, pork, chicken, turkey, cat, dog, mouse, rat, human | 30 pg DNA | Gel | [33] |
| Tetraplex | 3 | pig, cattle, fish, eukaryotic18S rRNA | 0.001–0.1 ng DNA | Gel | [37] |
| Hexaplex | 6 | horse, soybean, sheep, poultry, pork, cow | 0.01% for each species | Gel | [38] |
| Octuplex | 8 | dog, chicken, cattle, pig, horse, donkey, fox, and rabbit | 0.05 ng/μL DNA | Gel | [27] |
| Multiplex | 3 | chicken, duck and goose | 0.05 ng DNA, 1% for each species | Gel | [39] |
| Multiplex | 5 | cat, dog, pig, monkey, rat | 0.01–0.02 ng DNA | Chip | [24] |
| Quadruple | 4 | beef, pork, mutton, duck | 0.1 ng DNA | Gel | [40] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cai, Z.; Zhou, S.; Liu, Q.; Ma, H.; Yuan, X.; Gao, J.; Cao, J.; Pan, D. A Simple and Reliable Single Tube Septuple PCR Assay for Simultaneous Identification of Seven Meat Species. Foods 2021, 10, 1083. https://doi.org/10.3390/foods10051083
Cai Z, Zhou S, Liu Q, Ma H, Yuan X, Gao J, Cao J, Pan D. A Simple and Reliable Single Tube Septuple PCR Assay for Simultaneous Identification of Seven Meat Species. Foods. 2021; 10(5):1083. https://doi.org/10.3390/foods10051083
Chicago/Turabian StyleCai, Zhendong, Song Zhou, Qianqian Liu, Hui Ma, Xinyi Yuan, Jiaqi Gao, Jinxuan Cao, and Daodong Pan. 2021. "A Simple and Reliable Single Tube Septuple PCR Assay for Simultaneous Identification of Seven Meat Species" Foods 10, no. 5: 1083. https://doi.org/10.3390/foods10051083
APA StyleCai, Z., Zhou, S., Liu, Q., Ma, H., Yuan, X., Gao, J., Cao, J., & Pan, D. (2021). A Simple and Reliable Single Tube Septuple PCR Assay for Simultaneous Identification of Seven Meat Species. Foods, 10(5), 1083. https://doi.org/10.3390/foods10051083