Combining Protein Content and Grain Yield by Genetic Dissection in Bread Wheat under Low-Input Management
Abstract
1. Introduction
2. Materials and Methods
2.1. Field Conditions
2.2. Experimental Design
2.3. Materials
2.4. Genotyping
2.5. Phenotyping and Statistical Analysis
2.6. Selection of Allelic Combinations with High GY, High PC, and Stability
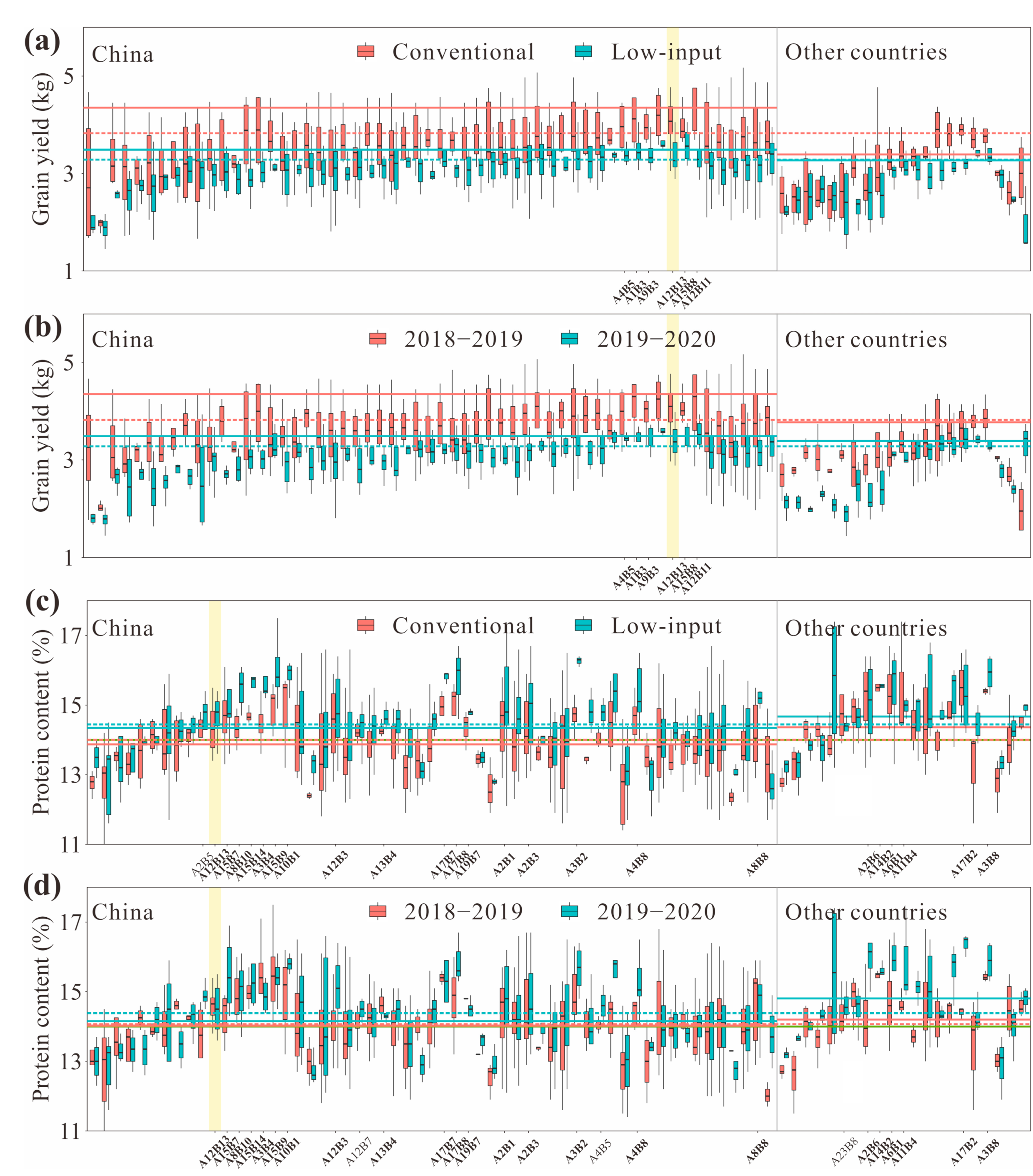
3. Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
References
- FAO. Food and Agriculture Organization of the United Nations. Available online: http://www.fao.org/faostat/en/#data/QC (accessed on 12 July 2020).
- Dixon, J.; Braun, H.J.; Kosina, P.; Crouch, J.H. Wheat Facts and Futures 2009; CIMMYT: Mexico City, Mexico, 2009. [Google Scholar]
- Reynolds, M.P.; Borlaug, N.E. Applying innovations and new technologies for international collaborative wheat improvement. J. Agric. Sci. 2006, 144, 95–110. [Google Scholar] [CrossRef]
- Kahiluoto, H.; Kaseva, J.; Balek, J.; Olesen, J.E.; Ruiz-Ramos, M.; Gobin, A.; Kersebaum, K.C.; Takáč, J.; Ruget, F.; Ferrise, R.; et al. Decline in climate resilience of European wheat. Proc. Natl. Acad. Sci. USA 2019, 116, 123. [Google Scholar] [CrossRef] [PubMed]
- Asif, M.; Tunc, C.E.; Yazici, M.A.; Tutus, Y.; Rehman, R.; Rehman, A.; Ozturk, L. Effect of predicted climate change on growth and yield performance of wheat under varied nitrogen and zinc supply. Plant Soil 2019, 434, 231–244. [Google Scholar] [CrossRef]
- Kumar, A.; Jain, S.; Elias, E.M.; Ibrahim, M.; Sharma, L.K. An Overview of QTL Identification and Marker-Assisted Selection for Grain Protein Content in Wheat. In Eco-Friendly Agro-Biological Techniques for Enhancing Crop Productivity; Springer: Singapore, 2018. [Google Scholar]
- Cohn, A.S.; Vanwey, L.K.; Spera, S.A.; Mustard, J.F. Cropping frequency and area response to climate variability can exceed yield response. Nat. Clim. Chang. 2016, 6, 601–604. [Google Scholar] [CrossRef]
- Koga, S.; Böcker, U.; Moldestad, A.; Tosi, P.; Shewry, P.R.; Mosleth, E.F.; Uhlen, A.K. Influence of temperature on the composition and polymerization of gluten proteins during grain filling in spring wheat (Triticum aestivum L.). J. Cereal Sci. 2015, 65, 1–8. [Google Scholar] [CrossRef]
- Guttieri, M.J.; McLean, R.; Stark, J.C.; Souza, E. Managing irrigation and nitrogen fertility of hard spring wheats for optimum bread and noodle quality. Crop Sci. 2005, 45, 2049–2059. [Google Scholar] [CrossRef]
- Zaveri, E.; Lobell, D.B. The role of irrigation in changing wheat yields and heat sensitivity in India. Nat. Commun. 2019, 10, 4144. [Google Scholar] [CrossRef]
- Carberry, P.S.; Liang, W.L.; Twomlow, S.; Holzworth, D.P.; Dimes, J.P.; McClelland, T.; Huth, N.I.; Chen, F.; Hochman, Z.; Keating, B.A. Scope for improved eco-efficiency varies among diverse cropping systems. Proc. Natl. Acad. Sci. USA 2013, 110, 8381–8386. [Google Scholar] [CrossRef]
- Springmann, M.; Clark, M.; Mason-D’Croz, D.; Wiebe, K.; Bodirsky, B.L.; Lassaletta, L.; de Vries, W.; Vermeulen, S.J.; Herrero, M.; Carlson, K.M.; et al. Options for keeping the food system within environmental limits. Nature 2018, 562, 519–525. [Google Scholar] [CrossRef]
- Reynolds, M.; Braun, H. Benefits to low-input agriculture. Nat. Plants 2019, 5, 652–653. [Google Scholar] [CrossRef]
- Przulj, N.; Momcilovic, V. Genetic variation for dry matter and nitrogen accumulation and translocation in two-rowed spring barley I. Dry matter translocation. Eur. J. Agron. 2001, 15, 241–254. [Google Scholar] [CrossRef]
- Koutroubas, S.D.; Fotiadis, S.; Damalas, C.A. Biomass and nitrogen accumulation and translocation in spelt (Triticum spelta) grown in a Mediterranean area. Field Crop. Res. 2012, 127, 1–8. [Google Scholar] [CrossRef]
- Rozbicki, J.; Ceglińska, A.; Gozdowski, D.; Jakubczak, M.; Cacak-Pietrzak, G.; Mądry, W.; Golba, J.; Piechociński, M.; Sobczynski, G.; Studnicki, M. Influence of the cultivar, environment and management on the grain yield and bread-making quality in winter wheat. J. Cereal Sci. 2015, 61, 126–132. [Google Scholar] [CrossRef]
- Charmet, G.; Robert, N.; Branlard, G.; Linossier, L.; Martre, P.; Triboï, E. Genetic analysis of dry matter and nitrogen accumulation and protein composition in wheat kernels. Theor. Appl. Genet. 2005, 111, 540–550. [Google Scholar] [CrossRef]
- Nicotra, A.B.; Atkin, O.K.; Bonser, S.P.; Davidson, A.M.; Finnegan, E.J.; Mathesius, U.; Poot, P.; Purugganan, M.D.; Richards, C.L.; Valladares, F. Plant phenotypic plasticity in a changing climate. Trends Plant Sci. 2010, 15, 684–692. [Google Scholar] [CrossRef]
- Tack, J.; Barkley, A.; Nalley, L.L. Heterogeneous effects of warming and drought on selected wheat variety yields. Clim. Chang. 2014, 125, 489–500. [Google Scholar] [CrossRef]
- Tahir, I.; Nakata, N. Remobilization of Nitrogen and Carbohydrate from Stems of Bread Wheat in Response to Heat Stress during Grain Filling. J. Agron. Crop Sci. 2005, 191, 106–115. [Google Scholar] [CrossRef]
- Flagella, Z.; Giuliani, M.M.; Giuzio, L.; Volpi, C.; Masci, S. Influence of water deficit on durum wheat storage protein composition and technological quality. Eur. J. Agron. 2010, 33, 197–207. [Google Scholar] [CrossRef]
- Mutava, R.N.; Prasad, P.V.V.; Tuinstra, M.R.; Kofoid, M.D.; Yu, J. Characterization of sorghum genotypes for traits related to drought tolerance. Field Crop. Res. 2011, 123, 10–18. [Google Scholar] [CrossRef]
- Knapp, S.; van der Heijden, M.G.A. A global meta-analysis of yield stability in organic and conservation agriculture. Nat. Commun. 2018, 9, 3632. [Google Scholar] [CrossRef]
- Würschum, T.; Leiser, W.L.; Kazman, E.; Longin, C.F.H. Genetic control of protein content and sedimentation volume in European winter wheat cultivars. Theor. Appl. Genet. 2016, 129, 1685–1696. [Google Scholar] [CrossRef]
- Mosleth, E.F.; Lillehammer, M.; Pellny, T.K.; Wood, A.J.; Richeb, A.B.; Hussain, A.; Griffiths, S.; Hawkesford, M.J.; Shewry, P.R. Genetic variation and heritability of grain protein deviation in European wheat genotypes. Field Crop. Res. 2020, 255, 107896. [Google Scholar] [CrossRef]
- Matus-Cádiz, M.A.; Hucl, P.; Perron, C.E.; Tyler, R.T. Genotype × environment interaction for grain color in hard white spring wheat. Crop Sci. 2003, 43, 219–226. [Google Scholar] [CrossRef]
- Igrejas, G.; Gaborit, T.; Oury, F.-X.; Chiron, H.; Marion, D.; Branlard, G. Genetic and environmental effects on puroindoline-a and puroindoline-b content and their relationship to technological properties in French bread wheats. J. Cereal Sci. 2001, 34, 37–47. [Google Scholar] [CrossRef]
- Williams, R.M.; O’Brien, L.; Eagles, H.A.; Solah, V.A.; Jayasena, V. The influences of genotype, environment, and genotype×environment interaction on wheat quality. Aust. J. Agric. Res. 2008, 59, 95–111. [Google Scholar] [CrossRef]
- Büchi, L.; Charles, R.; Schneider, D.; Sinaj, S.; Maltas, A.; Fossati, D.; Mascher, F. Performance of eleven winter wheat varieties in a long term experiment on mineral nitrogen and organic fertilisation. Field Crop. Res. 2016, 191, 111–122. [Google Scholar] [CrossRef]
- Rempelos, L.; Almuayrifi, M.S.B.; Baranski, M.; Tetard-Jones, C.; Barkla, B.; Cakmak, I.; Ozturk, L.; Cooper, J.; Volakakis, N.; Hall, G.; et al. The effect of agronomic factors on crop health and performance of winter wheat varieties bred for the conventional and the low input farming sector. Field Crop. Res. 2020, 254, 107822. [Google Scholar] [CrossRef]
- Tari, A.F. The effects of different deficit irrigation strategies on yield, quality, and water-use efficiencies of wheat under semi-arid conditions. Agric. Water Manag. 2016, 167, 1–10. [Google Scholar] [CrossRef]
- Lu, D.; Cai, X.; Zhao, J.; Shen, X.; Lu, W. Effects of drought after pollination on grain yield and quality of fresh waxy maize. J. Sci. Food Agric. 2015, 95, 210–215. [Google Scholar] [CrossRef] [PubMed]
- Vollmann, J.; Fritz, C.N.; Wagentristl, H.; Ruckenbauer, P. Environmental and genetic variation of soybean seed protein content under Central European growing conditions. J. Sci. Food Agric. 2000, 80, 1300–1306. [Google Scholar] [CrossRef]
- Oscarsson, M.; Andersson, R.; Åman, P.; Olofsson, S.; Jonsson, A. Effects of cultivar, nitrogen fertilization rate and environment on yield and grain quality of barley. J. Sci. Food Agric. 1998, 78, 359–366. [Google Scholar] [CrossRef]
- Oluwatosin, O.B. Genetic and environmental variation for seed yield, protein, lipid and amino acid composition in cowpea (Vigna unguiculata (L) Walp). J. Sci. Food Agric. 1997, 74, 107–116. [Google Scholar] [CrossRef]
- Brevis, J.C.; Dubcovsky, J. Effects of the chromosome region including the Gpc-B1 locus on wheat grain and protein yield. Crop Sci. 2010, 50, 93–104. [Google Scholar] [CrossRef]
- Mian, M.A.R.; McHale, L.; Li, Z.; Dorrance, A.E. Registration of ‘Highpro1’ Soybean with High Protein and High Yield Developed from a North × South Cross. J. Plant Regist. 2017, 11, 51–54. [Google Scholar] [CrossRef]
- Kumar, J.; Jaiswal, V.; Kumar, A.; Mir, R.; Kumar, S.; Dhariwal, R.; Balyan, H.; Gupta, P. Introgression of a major gene for high grain protein content in some Indian bread wheat cultivars. Field Crop. Res. 2011, 123, 226–233. [Google Scholar] [CrossRef]
- Torrion, J.A.; Walsh, O.S.; Liang, X.; Bicego, B.; Sapkota, A. Managing ‘Egan’ wheat with a gene for high grain protein. Agrosyst. Geosci. Environ. 2019, 2, 190019. [Google Scholar] [CrossRef]
- Michel, S.; Löschenberger, F.; Ametz, C.; Pachler, B.; Sparry, E.; Bürstmayr, H. Combining grain yield, protein content and protein quality by multi-trait genomic selection in bread wheat. Theor. Appl. Genet. 2019, 132, 2767–2780. [Google Scholar] [CrossRef]
- Guillemaut, P.; Maréchal-Drouard, L. Isolation of plant DNA: A fast, inexpensive, and reliable method. Plant Mol. Biol. Rep. 1992, 10, 60–65. [Google Scholar] [CrossRef]
- Khalid, M.; Afzal, F.; Gul, A.; Amir, R.; Subhani, A.; Ahmed, Z.; Mahmood, Z.; Xia, X.; Rasheed, A.; He, Z. Molecular characterization of 87 functional genes in wheat diversity panel and their association with phenotypes under well-watered and water-limited conditions. Front. Plant Sci. 2019, 10, 717. [Google Scholar] [CrossRef] [PubMed]
- Caporaso, N.; Whitworth, M.B.; Fisk, I.D. Near-Infrared spectroscopy and hyperspectral imaging for non-destructive quality assessment of cereal grains. Appl. Spectrosc. Rev. 2018, 53, 667–687. [Google Scholar] [CrossRef]
- Bolker, B.M.; Brooks, M.E.; Clark, C.J.; Geange, S.W.; Poulsen, J.R.; Stevens, M.H.H.; White, J.-S.S. Generalized linear mixed models: A practical guide for ecology and evolution. Trends Ecol. Evol. 2009, 24, 127–135. [Google Scholar] [CrossRef] [PubMed]
- Jia, H.; Wan, H.; Yang, S.; Zhang, Z.; Kong, Z.; Xue, S.; Zhang, L.; Ma, Z. Genetic dissection of yield-related traits in a recombinant inbred line population created using a key breeding parent in China’s wheat breeding. Theor. Appl. Genet. 2013, 126, 2123–2139. [Google Scholar] [CrossRef] [PubMed]
- Döring, T.F.; Annicchiarico, P.; Clarke, S.; Haigh, Z.; Jones, H.E.; Pearce, H.; Snape, J.; Zhan, J.; Wolfe, M.S. Comparative analysis of performance and stability among composite cross populations, variety mixtures and pure lines of winter wheat in organic and conventional cropping systems. Field Crop. Res. 2015, 183, 235–245. [Google Scholar] [CrossRef]
- Rao, M.S.S.; Mullinix, B.G.; Rangappa, M.; Cebert, E.; Bhagsari, A.S.; Sapra, V.T.; Joshi, J.M.; Dadson, R.B. Genotype × environment interactions and yield stability of food-grade soybean genotypes. Agron. J. 2002, 94, 72–80. [Google Scholar] [CrossRef]
- Nishio, Z.; Ito, M.; Tabiki, T.; Nagasawa, K.; Yamauchi, H.; Hirota, T. Influence of higher growing-season temperatures on yield components of winter wheat (Triticum aestivum L.). Crop Sci. 2013, 53, 621–628. [Google Scholar] [CrossRef]
- Zheng, T.C.; Zhang, X.; Yin, G.H.; Wang, L.N.; Han, Y.L.; Chen, L.; Huang, F.; Tang, J.W.; Xia, X.; He, Z. Genetic gains in grain yield, net photosynthesis and stomatal conductance achieved in Henan Province of China between 1981 and 2008. Field Crop. Res. 2011, 122, 225–233. [Google Scholar] [CrossRef]
- Donmez, E.; Sears, R.G.; Shroyer, J.P.; Paulsen, G.M. Genetic gain in yield attributes of winter wheat in the great plains. Crop Sci. 2001, 41, 1412–1419. [Google Scholar] [CrossRef]
- Johansson, E.; Prieto-Linde, M.L.; Gissén, C. Influences of weather, cultivar and fertiliser rate on grain protein polymer accumulation in field-grown winter wheat, and relations to grain water content and falling number. J. Sci. Food Agric. 2008, 88, 2011–2018. [Google Scholar] [CrossRef]
- Weightman, R.M.; Millar, S.; Alava, J.; Foulkes, M.J.; Fish, L.; Snape, J.W. Effects of drought and the presence of the 1BL/1RS translocation on grain vitreosity, hardness and protein content in winter wheat. J. Cereal Sci. 2008, 47, 457–468. [Google Scholar] [CrossRef]
- Galloway, J.N.; Dentener, F.J.; Capone, D.G.; Boyer, E.W.; Howarth, R.W.; Seitzinger, S.P.; Asner, G.P.; Cleveland, C.C.; Green, P.A.; Holland, E.A.; et al. Nitrogen cycles: Past, present, and future. Biogeochemistry 2004, 70, 153–226. [Google Scholar] [CrossRef]
- Kichey, T.; Hirel, B.; Heumez, E.; Dubois, F.; Le Gouis, J. In winter wheat (Triticum aestivum L.), post-anthesis nitrogen uptake and remobilisation to the grain correlates with agronomic traits and nitrogen physiological markers. Field Crop. Res. 2007, 102, 22–32. [Google Scholar] [CrossRef]
- Gaju, O.; Allard, V.; Martre, P.; Gouis, J.L.; Moreau, D.; Bogard, M.; Hubbart, S.; Foulkes, M.J. Nitrogen partitioning and remobilization in relation to leaf senescence, grain yield and grain nitrogen concentration in wheat cultivars. Field Crop. Res. 2014, 155, 213–223. [Google Scholar] [CrossRef]
- Shearman, V.; Sylvester-Bradley, R.; Scott, R.; Foulkes, M. Physiological processes associated with wheat yield progress in the UK. Crop Sci. 2005, 45, 175–185. [Google Scholar] [CrossRef]
- Foulkes, M.J.; Scott, R.K.; Sylvester-Bradley, R. The ability of wheat cultivars to withstand drought in UK conditions: Formation of grain yield. J. Agric. Sci. 2002, 138, 153–169. [Google Scholar] [CrossRef]
- Xu, Z.Z.; Yu, Z.W.; Wang, D.; Zhang, Y.L. Nitrogen accumulation and translocation for winter wheat under different irrigation regimes. J. Agron. Crop Sci. 2005, 191, 439–449. [Google Scholar] [CrossRef]
- Lambers, H.; Simpson, R.J.; Beilharz, V.C.; Dalling, M.J. Growth and translocation of C and N in wheat (Triticum aestivum) grown with a split root system. Physiol. Plant. 1982, 56, 421–429. [Google Scholar] [CrossRef]
- Ozturk, A.; Aydin, F. Effect of water stress at various growth stages on some quality characteristics of winter wheat. J. Agron. Crop Sci. 2004, 190, 93–99. [Google Scholar] [CrossRef]
- Zhao, C.J.; Liu, L.Y.; Wang, J.H.; Huang, W.J.; Song, X.Y.; Li, C.J. Predicting grain protein content of winter wheat using remote sensing data based on nitrogen status and water stress. Int. J. Appl. Earth Obs. Geoinf. 2005, 7, 1–9. [Google Scholar] [CrossRef]
- Wang, X.; Zhao, C.; Müller, C.; Wang, C.; Ciais, P.; Janssens, I.; Peñuelas, J.; Asseng, S.; Li, T.; Elliott, J.; et al. Emergent constraint on crop yield response to warmer temperature from field experiments. Nat. Sustain. 2020, 3, 908–916. [Google Scholar] [CrossRef]
- Savill, G.P.; Michalski, A.; Powers, S.J.; Wan, Y.; Tosi, P.; Buchner, P.; Hawkesford, M.J. Temperature and nitrogen supply interact to determine protein distribution gradients in the wheat grain endosperm. J. Exp. Bot. 2018, 69, 3117–3126. [Google Scholar] [CrossRef]
- Mahmood, N.; Arshad, M.; Kächele, H.; Ma, H.; Ullah, A.; Müller, K. Wheat yield response to input and socioeconomic factors under changing climate: Evidence from rainfed environments of Pakistan. Sci. Total Environ. 2019, 688, 1275–1285. [Google Scholar] [CrossRef] [PubMed]
- Marinaccio, F.; Blandino, M.; Reyneri, A. Effect of nitrogen fertilization on yield and quality of durum wheat cultivated in northern Italy and their interaction with different soils and growing seasons. J. Plant Nutr. 2016, 39, 643–654. [Google Scholar] [CrossRef]
- Kolderup, F. Effects of temperature, photoperiod, and light quantity on protein production in wheat grains. J. Sci. Food Agric. 1975, 26, 583–592. [Google Scholar] [CrossRef]
- Zhang, X.; Shi, Z.; Jiang, D.; Högy, P.; Fangmeier, A. Independent and combined effects of elevated CO2 and post-anthesis heat stress on protein quantity and quality in spring wheat grains. Food Chem. 2019, 277, 524–530. [Google Scholar] [CrossRef] [PubMed]
- Pan, J.; Zhu, Y.; Cao, W.; Dai, T.; Jiang, D. Predicting the protein content of grain in winter wheat with meteorological and genotypic factors. Plant Prod. Sci. 2006, 9, 323–333. [Google Scholar] [CrossRef]
- Li, H.; Zhou, Y.; Xin, W.; Wei, Y.; Zhang, J.; Guo, L. Wheat breeding in northern China: Achievements and technical advances. Crop J. 2019, 7, 718–729. [Google Scholar] [CrossRef]
- Giancaspro, A.; Giove, S.L.; Zacheo, S.A.; Blanco, A.; Gadaleta, A. Genetic variation for protein content and yield-related traits in a durum population derived from an inter-specific cross between hexaploid and tetraploid wheat cultivars. Front. Plant Sci. 2019, 10, 1509. [Google Scholar] [CrossRef]
- He, Z.; Lin, Z.J.; Wang, L.J.; Xiao, Z.M.; Wan, F.S.; Zhuang, Q.S. Classification on Chinese wheat regions based on quality. Sci. Agric. Sin. 2002, 35, 359–364. [Google Scholar]
- Qin, L.; Hao, C.; Hou, J.; Wang, Y.; Li, T.; Wang, L.; Ma, Z.; Zhang, X. Homologous haplotypes, expression, genetic effects and geographic distribution of the wheat yield gene TaGW2. BMC Plant Biol. 2014, 14, 107. [Google Scholar] [CrossRef]
- Hou, J.; Jiang, Q.; Hao, C.; Wang, Y.; Zhang, H.; Zhang, X. Global selection on sucrose synthase haplotypes during a century of wheat breeding. Plant Physiol. 2014, 164, 1918–1929. [Google Scholar] [CrossRef]
- Langner, M.; Krystkowiak, K.; Salmanowicz, B.P.; Adamski, T.; Krajewski, P.; Kaczmarek, Z.; Surma, M. The influence of Glu-1 and Glu-3 loci on dough rheology and bread-making properties in wheat (Triticum aestivum L.) doubled haploid lines. J. Sci. Food Agric. 2017, 97, 5083–5091. [Google Scholar] [CrossRef] [PubMed]
- Oikeh, S.O.; Menkir, A.; Maziya-Dixon, B.; Welch, R.M.; Glahn, R.P.; Gauch, G. Environmental stability of iron and zinc concentrations in grain of elite early-maturing tropical maize genotypes grown under field conditions. J. Agric. Sci. 2004, 142, 543–551. [Google Scholar] [CrossRef]
- Zaim, M.; El Hassouni, K.; Gamba, F.; Filali Maltouf, A.; Belkadi, B.; Sourour, A.; Amri, A.; Nachit, M.; Taghouti, M.; Bassi, F.M. Wide crosses of durum wheat (Triticum durum Desf.) reveal good disease resistance, yield stability, and industrial quality across Mediterranean sites. Field Crop. Res. 2017, 214, 219–227. [Google Scholar] [CrossRef]
- Rharrabti, Y.; del Moral, L.F.G.; Villegas, D.; Royo, C. Durum wheat quality in Mediterranean environments III. Stability and comparative methods in analysing G×E interaction. Field Crop. Res. 2003, 80, 141–146. [Google Scholar] [CrossRef]
- Ruan, Y.; Yu, B.; Knox, R.E.; Singh, A.K.; DePauw, R.; Cuthbert, R.; Zhang, W.; Piche, I.; Gao, P.; Sharpe, A.; et al. High density mapping of quantitative Trait loci conferring gluten strength in canadian durum wheat. Front. Plant Sci. 2020, 11, 170. [Google Scholar] [CrossRef]
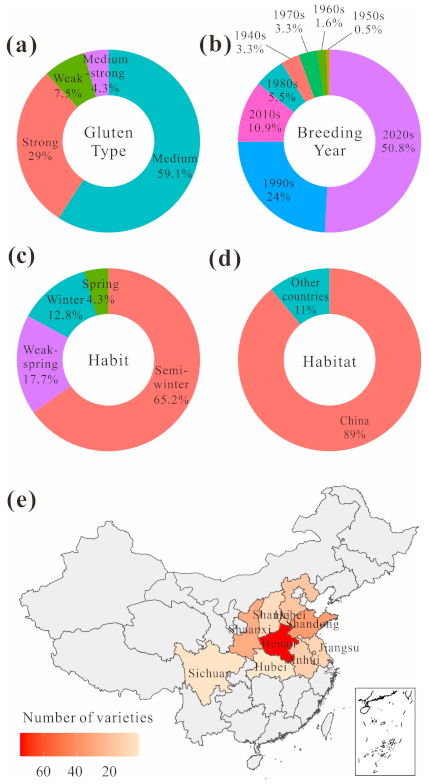
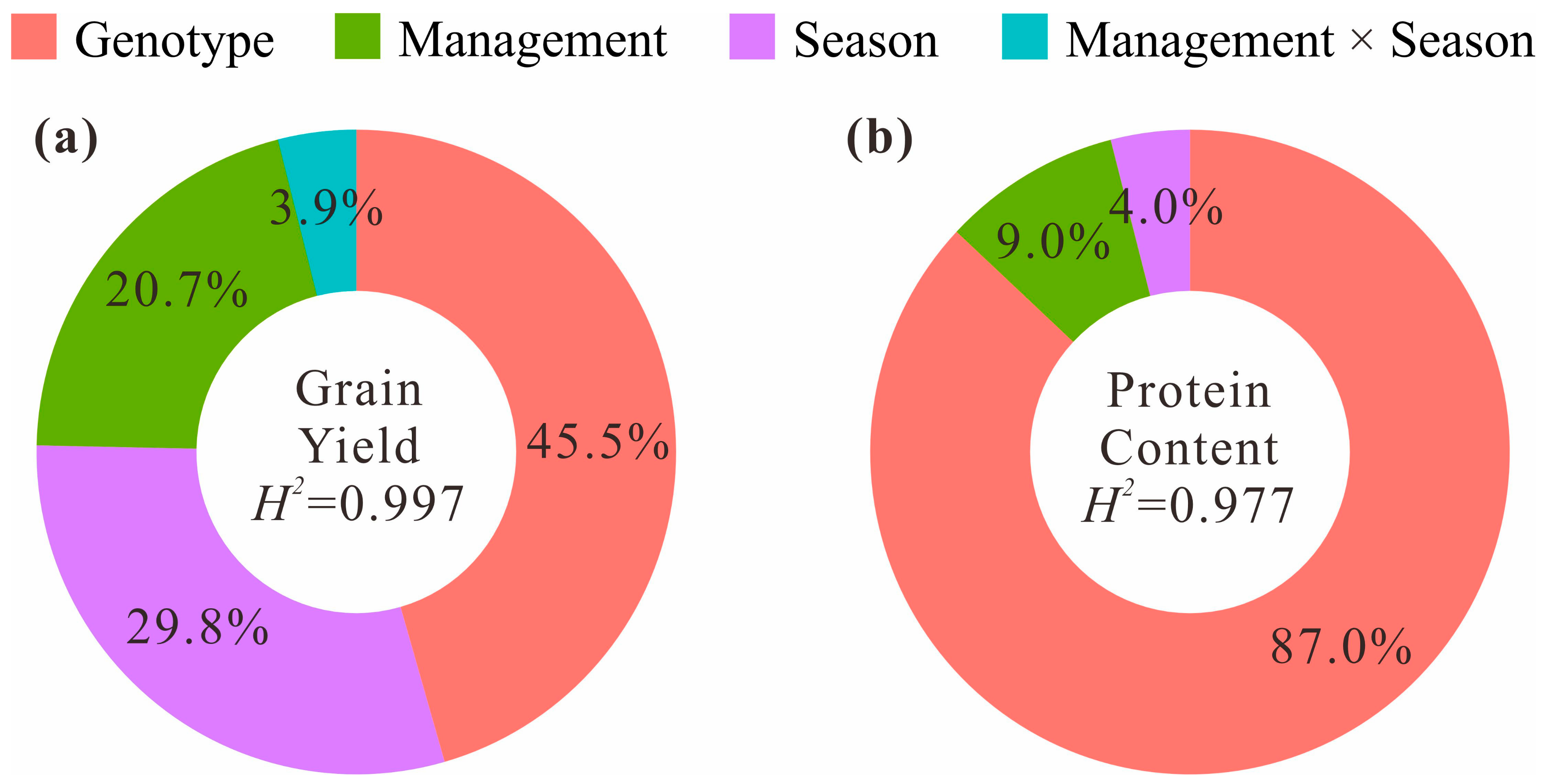
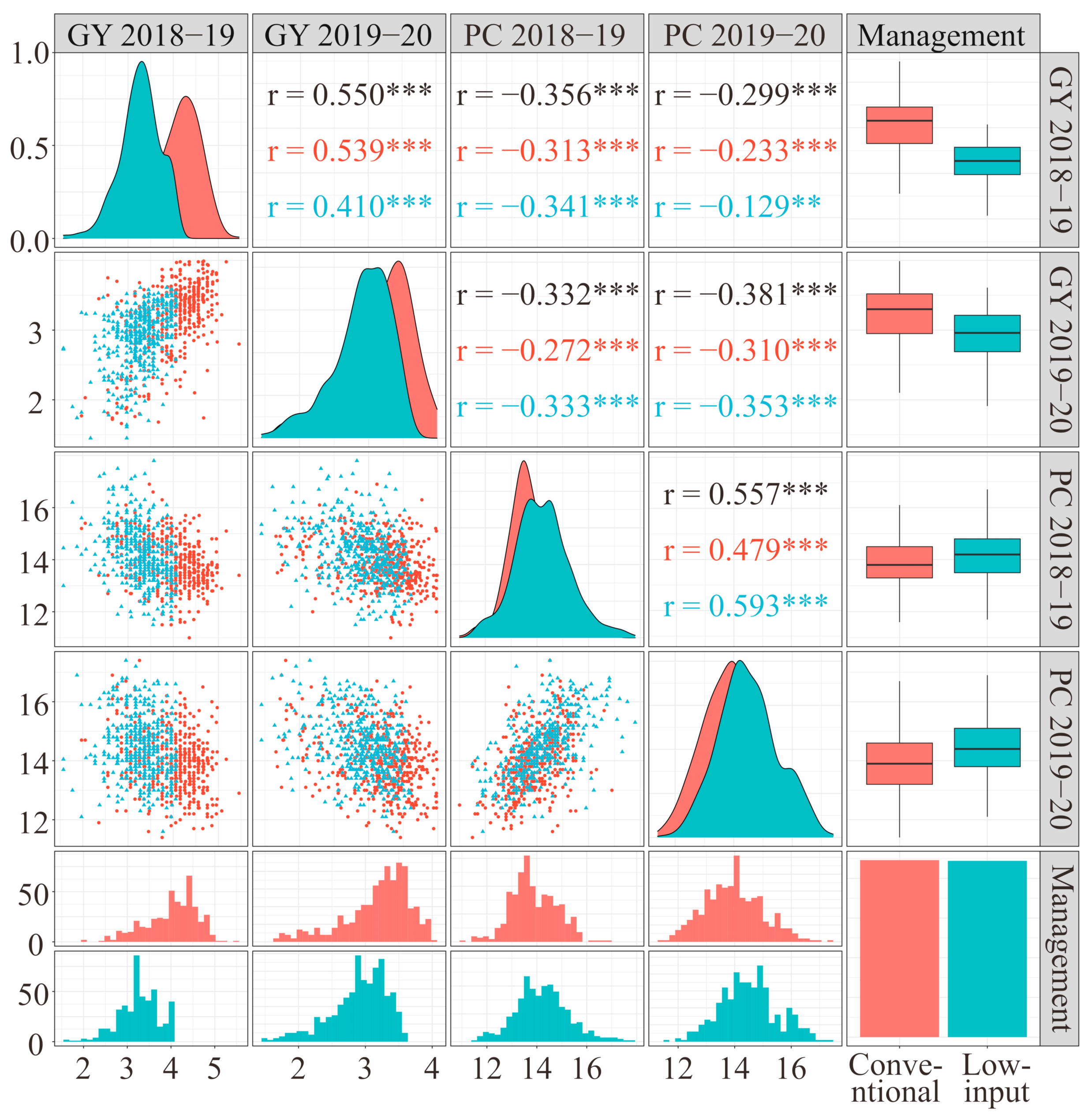
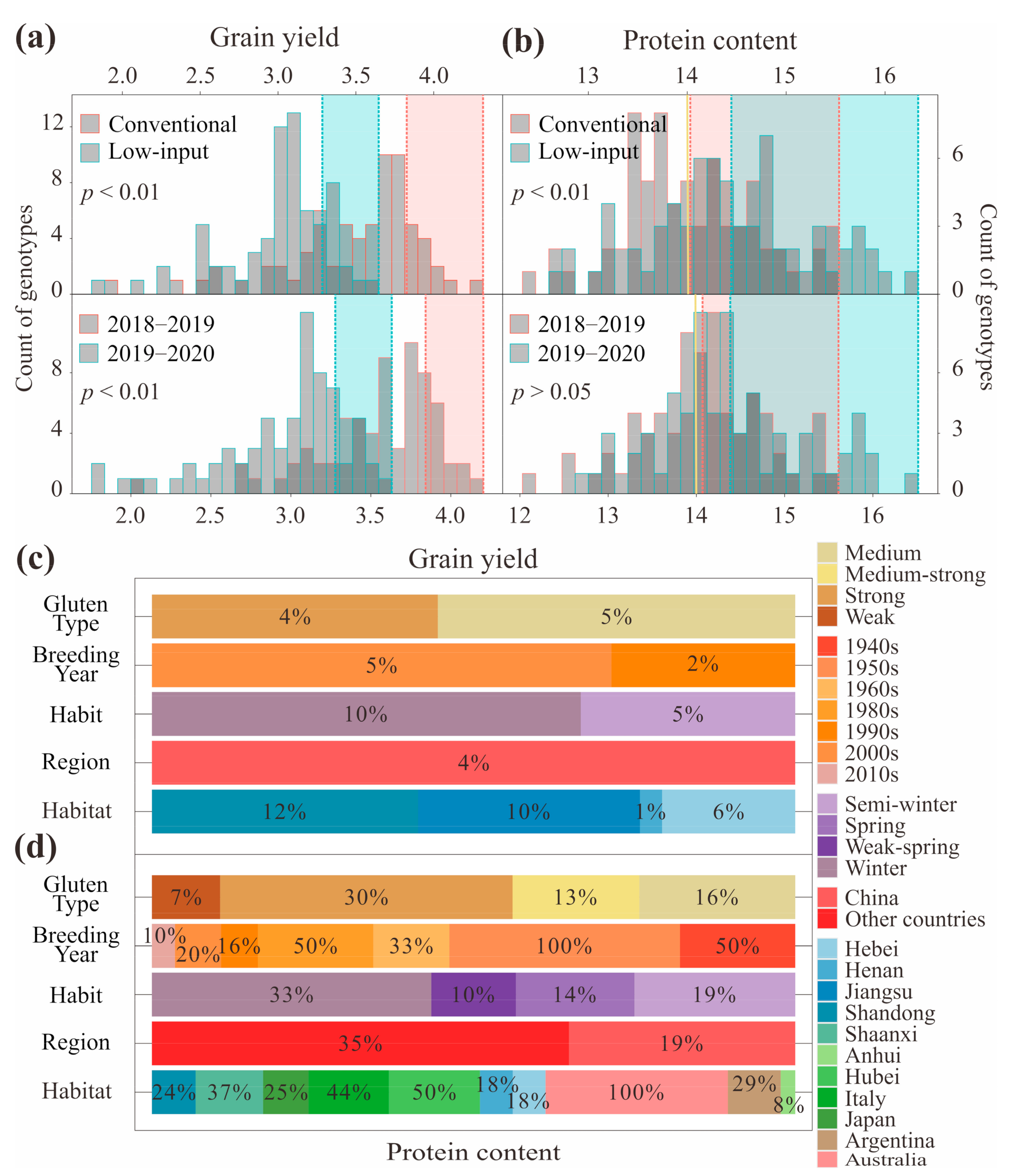
| Trait | Gene | FAM Primer (5′-3′) | HEX Primer (5′-3′) | Common Primer (5′-3′) | Amplification Conditions |
|---|---|---|---|---|---|
| Grain yield | TaCKX-D1 | GAAGGTGACCAAGTTCATGCTCGTCGATAGTCTCATGCATATGC | GAAGGTCGGAGTCAACGGATTATGCATGCATGCATGCGT | AACTTTTCACGGTGAACAG | Primer mixture included 46 μL ddH2O, 30 μL common primer (100 μM) and 12 μL of each tailed primer (100 μM). Assays were tested in 384-well format and set up as 5 μL reaction [2.2 μL DNA (10–20 ng/μL), 2.5 μL of 2XKASP master mixture and 0.056 μL primer mixture]. PCR cycling was performed using following protocol: hot start at 95 °C for 15 min, followed by ten touchdown cycles (95 °C for 20 s; touchdown 65 °C–1 °C per cycle 25 s) further followed by 30 cycles of amplification (95 °C for 10 s; 57 °C for 60 s). Extension step is unnecessary as amplicon is less than 120 bp. Plate was read in BioTek H1 system and data analysis was performed manually using Klustercaller software (version 2.22.0.5; LGC Hoddesdon, United Kingdom). |
| TaGASR7-A1 | GAAGGTGACCAAGTTCATGCTCACGGTAGAGGAGCCGGTTC | GAAGGTCGGAGTCAACGGATTATAACTGCTCACCCCCCACC | ATATGTAGGGCAGGAAGGGC | ||
| TaSus1-7A | GAAGGTGACCAAGTTCATGCTGATTTGATCCATGCCCTCTC | GAAGGTCGGAGTCAACGGATTGATTTGATCCATGCCCTCTT | CTGTCGTTCAACATCATTGTCTG | ||
| TaSus1-7B | GAAGGTGACCAAGTTCATGCTCAATTGCTTATGTTCTGTTGTATGG | GAAGGTCGGAGTCAACGGATTCAATTGCTTATGTTCTGTTGTACAT | ATGGTTATGCTTGAATGGAAGAGC | ||
| TaGS5-A1 | GAAGGTGACCAAGTTCATGCTGTGCAATCTTGGACAAACATCAG | GAAGGTCGGAGTCAACGGATTGTGCAATCTTGGACAAACATCAT | AGTGCTTTGTCAACAACAGATGC | ||
| TaGW2-6A | GAAGGTGACCAAGTTCATGCTTCCCGCTCCAGCTATCTGGTGAAC | GAAGGTCGGAGTCAACGGATTTCCCGCTCCAGCTATCTGGTGAAA | TTCCCAGTCTTTGACATGTTCCGCC | ||
| TaGW2-6B | GAAGGTGACCAAGTTCATGCTTGAGATCCCGTGCAGTAGCTCG | GAAGGTCGGAGTCAACGGATTTGAGATCCCGTGCAGTAGCTCA | TGGCGTGAGCTAGGGTTTGTTG | ||
| Protein content | Glu-A1 | GAAGGTGACCAAGTTCATGCTAAGTGTAACTTCTCCGCAACA | GAAGGTCGGAGTCAACGGATTAAGTGTAACTTCTCCGCAACG | GGCCTGGATAGTATGAAACC | |
| Glu-D1 | GAAGGTGACCAAGTTCATGCTCGCTAATCCTGCGAGCAACAAAT | GAAGGTCGGAGTCAACGGATTGCTAATCCTGCGAGCAACAAAG | AGCCAAGGGCATGTTCTATGTCGAA | ||
| Glu-B3 | GAAGGTGACCAAGTTCATGCTctgttggggttgggaaacG | GAAGGTCGGAGTCAACGGATTctgttggggttgggaaacA | agcagcagcaaccGcaaC |
| Haplotype | TaCKX-D1 | TaGASR7-A1 | TaSus1-7A | TaSus1-7B | TaGS5-A1 | TaGW2-6A | TaGW2-6B | Glu-A1 | Glu-D1 | Glu-B3 | Percent/% |
|---|---|---|---|---|---|---|---|---|---|---|---|
| A10B1 | TaCKX-D1b | H1g | Hap-3/4 | Hap-T | A1b | Hap-6A-A | Hap-1/2 | 1/2* | 5 + 10 | Glu-B3b/d/g/i | 0.48 |
| A11B4 | TaCKX-D1b | H1g | Hap-1/2 | Hap-T | A1b | Hap-6A-G/Hap-6A-A | Hap-1/2 | 1/2* | 2 + 12 | others | 0.48 |
| A12B1 | TaCKX-D1b | H1g | Hap-1/2 | Hap-T | A1a | Hap-6A-A | Hap-1/2 | 1/2* | 5 + 10 | Glu-B3b/d/g/i | 2.42 |
| A12B11 | TaCKX-D1b | H1g | Hap-1/2 | Hap-T | A1a | Hap-6A-A | Hap-1/2 | null | 2 + 12/5 + 10 | others | 0.48 |
| A12B13 | TaCKX-D1b | H1g | Hap-1/2 | Hap-T | A1a | Hap-6A-A | Hap-1/2 | 1/2*/null | 2 + 12 | Glu-B3b/d/g/i | 0.97 |
| A12B15 | TaCKX-D1b | H1g | Hap-1/2 | Hap-T | A1a | Hap-6A-A | Hap-1/2 | 1/2*/null | 5 + 10 | Glu-B3b/d/g/i | 0.48 |
| A12B2 | TaCKX-D1b | H1g | Hap-1/2 | Hap-T | A1a | Hap-6A-A | Hap-1/2 | 1/2* | 5 + 10 | Glu-B3j | 2.42 |
| A12B3 | TaCKX-D1b | H1g | Hap-1/2 | Hap-T | A1a | Hap-6A-A | Hap-1/2 | 1/2* | 2 + 12 | Glu-B3b/d/g/i | 3.38 |
| A12B4 | TaCKX-D1b | H1g | Hap-1/2 | Hap-T | A1a | Hap-6A-A | Hap-1/2 | 1/2* | 2 + 12 | others | 5.31 |
| A12B6 | TaCKX-D1b | H1g | Hap-1/2 | Hap-T | A1a | Hap-6A-A | Hap-1/2 | null | 5 + 10 | others | 0.48 |
| A12B7 | TaCKX-D1b | H1g | Hap-1/2 | Hap-T | A1a | Hap-6A-A | Hap-1/2 | null | 2 + 12 | Glu-B3b/d/g/i | 0.97 |
| A12B8 | TaCKX-D1b | H1g | Hap-1/2 | Hap-T | A1a | Hap-6A-A | Hap-1/2 | null | 2 + 12 | others | 3.38 |
| A12B9 | TaCKX-D1b | H1g | Hap-1/2 | Hap-T | A1a | Hap-6A-A | Hap-1/2 | 1/2* | 2 + 12/5 + 10 | others | 0.97 |
| A13B4 | TaCKX-D1b | H1g | Hap-1/2 | Hap-T | A1a/A1b | Hap-6A-A | Hap-1/2 | 1/2* | 2 + 12 | others | 0.48 |
| A14B2 | TaCKX-D1b | H1g | Hap-1/2 | Hap-C | A1b | Hap-6A-A | Hap-1/2 | 1/2* | 5 + 10 | others | 0.48 |
| A14B4 | TaCKX-D1b | H1g | Hap-1/2 | Hap-C | A1b | Hap-6A-A | Hap-1/2 | 1/2* | 2 + 12 | others | 0.48 |
| A15B14 | TaCKX-D1b | H1g | Hap-1/2 | Hap-T | A1b | Hap-6A-G | Hap-1/2 | 1/2*/null | 2 + 12 | others | 0.48 |
| A15B2 | TaCKX-D1b | H1g | Hap-1/2 | Hap-T | A1b | Hap-6A-G | Hap-1/2 | 1/2* | 5 + 10 | others | 1.47 |
| A15B3 | TaCKX-D1b | H1g | Hap-1/2 | Hap-T | A1b | Hap-6A-G | Hap-1/2 | 1/2* | 2 + 12 | Glu-B3b/d/g/i | 1.47 |
| A15B4 | TaCKX-D1b | H1g | Hap-1/2 | Hap-T | A1b | Hap-6A-G | Hap-1/2 | 1/2* | 2 + 12 | others | 2.42 |
| A15B7 | TaCKX-D1b | H1g | Hap-1/2 | Hap-T | A1b | Hap-6A-G | Hap-1/2 | null | 2 + 12 | Glu-B3b/d/g/i | 0.97 |
| A15B8 | TaCKX-D1b | H1g | Hap-1/2 | Hap-T | A1b | Hap-6A-G | Hap-1/2 | null | 2 + 12 | others | 0.48 |
| A15B9 | TaCKX-D1b | H1g | Hap-1/2 | Hap-T | A1b | Hap-6A-G | Hap-1/2 | 1/2* | 2 + 12/5 + 10 | others | 0.97 |
| A16B2 | TaCKX-D1b | H1g | Hap-3/4 | Hap-T | A1b | Hap-6A-G | Hap-1/2 | 1/2* | 5 + 10 | others | 0.48 |
| A17B2 | TaCKX-D1b | H1g | Hap-1/2 | Hap-C | A1b | Hap-6A-G | Hap-1/2 | 1/2* | 5 + 10 | others | 0.48 |
| A17B4 | TaCKX-D1b | H1g | Hap-1/2 | Hap-T | A1a | Hap-6A-G | Hap-1/2 | 1/2* | 2 + 12 | others | 1.47 |
| A17B7 | TaCKX-D1b | H1g | Hap-1/2 | Hap-T | A1a | Hap-6A-G | Hap-1/2 | null | 2 + 12 | Glu-B3b/d/g/i | 0.48 |
| A17B8 | TaCKX-D1b | H1g | Hap-1/2 | Hap-T | A1a | Hap-6A-G | Hap-1/2 | null | 2 + 12 | others | 0.48 |
| A18B4 | TaCKX-D1b | H1c | Hap-1/2 | Hap-C | A1a | Hap-6A-A | Hap-1/2 | 1/2* | 2 + 12 | others | 0.48 |
| A18B5 | TaCKX-D1b | H1c | Hap-1/2 | Hap-C | A1a | Hap-6A-A | Hap-1/2 | null | 5 + 10 | Glu-B3b/d/g/i | 0.48 |
| A18B8 | TaCKX-D1b | H1c | Hap-1/2 | Hap-C | A1a | Hap-6A-A | Hap-1/2 | null | 2 + 12 | others | 0.48 |
| A19B7 | TaCKX-D1b | H1c | Hap-3/4 | Hap-T | A1a | Hap-6A-A | Hap-1/2 | null | 2 + 12 | Glu-B3b/d/g/i | 0.48 |
| A1B3 | TaCKX-D1a | H1g | Hap-1/2 | Hap-T | A1a | Hap-6A-A | Hap-1/2 | 1/2* | 2 + 12 | Glu-B3b/d/g/i | 0.48 |
| A20B14 | TaCKX-D1b | H1g | Hap-1/2 | Hap-T/Hap-C | A1a | Hap-6A-A | Hap-1/2 | 1/2*/null | 2 + 12 | others | 0.48 |
| A21B4 | TaCKX-D1b | H1g | Hap-1/2 | Hap-T/Hap-C | A1b | Hap-6A-G | Hap-1/2 | 1/2* | 2 + 12 | others | 0.48 |
| A22B9 | TaCKX-D1b | H1g/H1c | Hap-1/2 | Hap-C | A1b | Hap-6A-A | Hap-1/2 | 1/2* | 2 + 12/5 + 10 | others | 0.48 |
| A23B8 | TaCKX-D1a | H1c | Hap-3/4 | Hap-T | A1a | Hap-6A-G | Hap-1/2 | null | 2 + 12 | others | 0.48 |
| A2B1 | TaCKX-D1b | H1c | Hap-1/2 | Hap-T | A1b | Hap-6A-A | Hap-1/2 | 1/2* | 5 + 10 | Glu-B3b/d/g/i | 1.93 |
| A2B2 | TaCKX-D1b | H1c | Hap-1/2 | Hap-T | A1b | Hap-6A-A | Hap-1/2 | 1/2* | 5 + 10 | others | 0.97 |
| A2B3 | TaCKX-D1b | H1c | Hap-1/2 | Hap-T | A1b | Hap-6A-A | Hap-1/2 | 1/2* | 2 + 12 | Glu-B3b/d/g/i | 3.38 |
| A2B4 | TaCKX-D1b | H1c | Hap-1/2 | Hap-T | A1b | Hap-6A-A | Hap-1/2 | 1/2* | 2 + 12 | Glu-B3j | 0.97 |
| A2B5 | TaCKX-D1b | H1c | Hap-1/2 | Hap-T | A1b | Hap-6A-A | Hap-1/2 | null | 5 + 10 | Glu-B3b/d/g/i | 0.48 |
| A2B6 | TaCKX-D1b | H1c | Hap-1/2 | Hap-T | A1b | Hap-6A-A | Hap-1/2 | null | 5 + 10 | others | 0.48 |
| A2B7 | TaCKX-D1b | H1c | Hap-1/2 | Hap-T | A1b | Hap-6A-A | Hap-1/2 | null | 2 + 12 | Glu-B3b/d/g/i | 3.38 |
| A2B8 | TaCKX-D1b | H1c | Hap-1/2 | Hap-T | A1b | Hap-6A-A | Hap-1/2 | null | 2 + 12 | others | 1.93 |
| A3B2 | TaCKX-D1b | H1c | Hap-1/2 | Hap-T | A1b | Hap-6A-G | Hap-1/2 | 1/2* | 5 + 10 | others | 0.48 |
| A3B4 | TaCKX-D1b | H1c | Hap-1/2 | Hap-T | A1b | Hap-6A-G | Hap-1/2 | 1/2* | 2 + 12 | others | 0.48 |
| A3B5 | TaCKX-D1b | H1c | Hap-1/2 | Hap-T | A1b | Hap-6A-G | Hap-1/2 | null | 5 + 10 | Glu-B3b/d/g/i | 0.48 |
| A3B6 | TaCKX-D1b | H1c | Hap-1/2 | Hap-T | A1b | Hap-6A-G | Hap-1/2 | null | 5 + 10 | others | 0.48 |
| A3B8 | TaCKX-D1b | H1c | Hap-1/2 | Hap-T | A1b | Hap-6A-G | Hap-1/2 | null | 2 + 12 | others | 0.48 |
| A4B4 | TaCKX-D1b | H1c | Hap-1/2 | Hap-T | A1a | Hap-6A-A | Hap-1/2 | 1/2* | 2 + 12 | others | 0.48 |
| A4B5 | TaCKX-D1b | H1c | Hap-1/2 | Hap-T | A1a | Hap-6A-A | Hap-1/2 | null | 5 + 10 | Glu-B3b/d/g/i | 0.97 |
| A4B6 | TaCKX-D1b | H1c | Hap-1/2 | Hap-T | A1a | Hap-6A-A | Hap-1/2 | null | 5 + 10 | others | 0.97 |
| A4B7 | TaCKX-D1b | H1c | Hap-1/2 | Hap-T | A1a | Hap-6A-A | Hap-1/2 | null | 2 + 12 | Glu-B3b/d/g/i | 0.97 |
| A4B8 | TaCKX-D1b | H1c | Hap-1/2 | Hap-T | A1a | Hap-6A-A | Hap-1/2 | null | 2 + 12 | others | 0.48 |
| A6B1 | TaCKX-D1b | H1c | Hap-1/2 | Hap-T | A1a | Hap-6A-G | Hap-1/2 | 1/2* | 5 + 10 | Glu-B3b/d/g/i | 0.48 |
| A7B8 | TaCKX-D1b | H1c | Hap-1/2 | Hap-C | A1b | Hap-6A-A | Hap-1/2 | null | 2 + 12 | others | 0.48 |
| A8B1 | TaCKX-D1b | H1g | Hap-1/2 | Hap-T | A1b | Hap-6A-A | Hap-1/2 | 1/2* | 5 + 10 | Glu-B3b/d/g/i | 4.35 |
| A8B10 | TaCKX-D1b | H1g | Hap-1/2 | Hap-T | A1b | Hap-6A-A | Hap-1/2 | 1/2* | 2 + 12/5 + 10 | Glu-B3b/d/g/i | 0.97 |
| A8B12 | TaCKX-D1b | H1g | Hap-1/2 | Hap-T | A1b | Hap-6A-A | Hap-1/2 | null | 2 + 12/5 + 10 | Glu-B3b/d/g/i | 0.97 |
| A8B14 | TaCKX-D1b | H1g | Hap-1/2 | Hap-T | A1b | Hap-6A-A | Hap-1/2 | 1/2*/null | 2 + 12 | others | 0.97 |
| A8B2 | TaCKX-D1b | H1g | Hap-1/2 | Hap-T | A1b | Hap-6A-A | Hap-1/2 | 1/2* | 5 + 10 | others | 2.9 |
| A8B3 | TaCKX-D1b | H1g | Hap-1/2 | Hap-T | A1b | Hap-6A-A | Hap-1/2 | 1/2* | 2 + 12 | Glu-B3b/d/g/i | 9.66 |
| A8B4 | TaCKX-D1b | H1g | Hap-1/2 | Hap-T | A1b | Hap-6A-A | Hap-1/2 | 1/2* | 2 + 12 | others | 13.53 |
| A8B5 | TaCKX-D1b | H1g | Hap-1/2 | Hap-T | A1b | Hap-6A-A | Hap-1/2 | null | 5 + 10 | Glu-B3b/d/g/i | 0.48 |
| A8B6 | TaCKX-D1b | H1g | Hap-1/2 | Hap-T | A1b | Hap-6A-A | Hap-1/2 | null | 5 + 10 | others | 0.97 |
| A8B7 | TaCKX-D1b | H1g | Hap-1/2 | Hap-T | A1b | Hap-6A-A | Hap-1/2 | null | 2 + 12 | Glu-B3b/d/g/i | 2.9 |
| A8B8 | TaCKX-D1b | H1g | Hap-1/2 | Hap-T | A1b | Hap-6A-A | Hap-1/2 | null | 2 + 12 | others | 1.47 |
| A9B3 | TaCKX-D1b | H1g | Hap-1/2 | Hap-T | A1b | Hap-6A-A | Hap-4 | 1/2* | 2 + 12 | Glu-B3b/d/g/i | 0.48 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ma, J.; Xiao, Y.; Hou, L.; He, Y. Combining Protein Content and Grain Yield by Genetic Dissection in Bread Wheat under Low-Input Management. Foods 2021, 10, 1058. https://doi.org/10.3390/foods10051058
Ma J, Xiao Y, Hou L, He Y. Combining Protein Content and Grain Yield by Genetic Dissection in Bread Wheat under Low-Input Management. Foods. 2021; 10(5):1058. https://doi.org/10.3390/foods10051058
Chicago/Turabian StyleMa, Junjie, Yonggui Xiao, Lingling Hou, and Yong He. 2021. "Combining Protein Content and Grain Yield by Genetic Dissection in Bread Wheat under Low-Input Management" Foods 10, no. 5: 1058. https://doi.org/10.3390/foods10051058
APA StyleMa, J., Xiao, Y., Hou, L., & He, Y. (2021). Combining Protein Content and Grain Yield by Genetic Dissection in Bread Wheat under Low-Input Management. Foods, 10(5), 1058. https://doi.org/10.3390/foods10051058






