Comprehensive Metabolite Profiling and Microbial Communities of Doenjang (Fermented Soy Paste) and Ganjang (Fermented Soy Sauce): A Comparative Study
Abstract
1. Introduction
2. Materials and Methods
2.1. Chemicals and Reagents
2.2. Materials
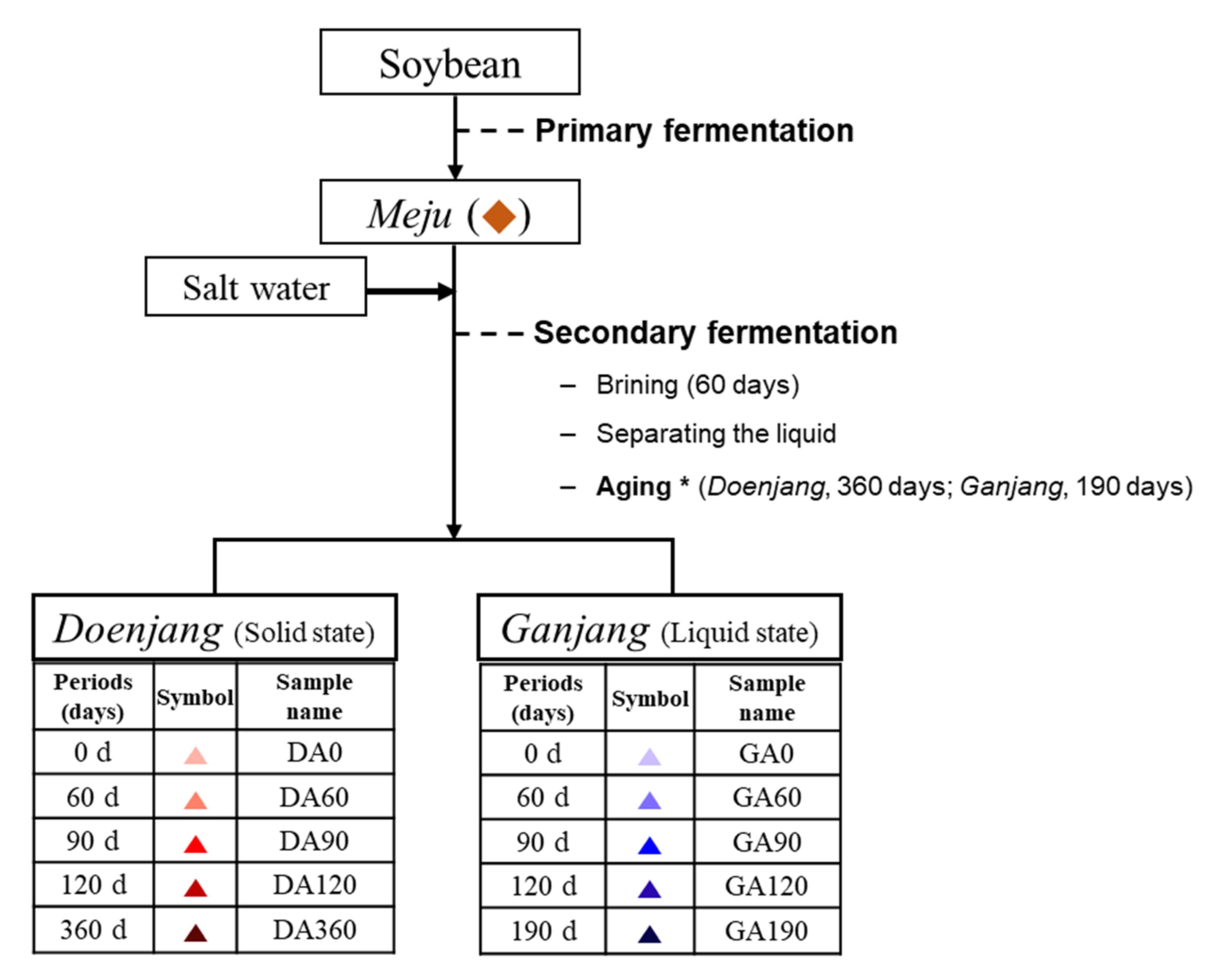
2.3. Preparation of Doenjang and Ganjang
2.4. Bacterial and Fungal Community Analysis
2.5. Gas Chromatography Time-of-Flight Mass Spectrometry (GC-TOF-MS) Analysis
2.6. Ultrahigh Performance Liquid Chromatography-Orbitrap-Mass Spectrometry/Mass Spectrometry (UHPLC-orbitrap-MS/MS) Analysis
2.7. Data Processing and Multivariate Statistical Analysis
2.8. Bioactivity Assay Analysis
3. Results and Discussion
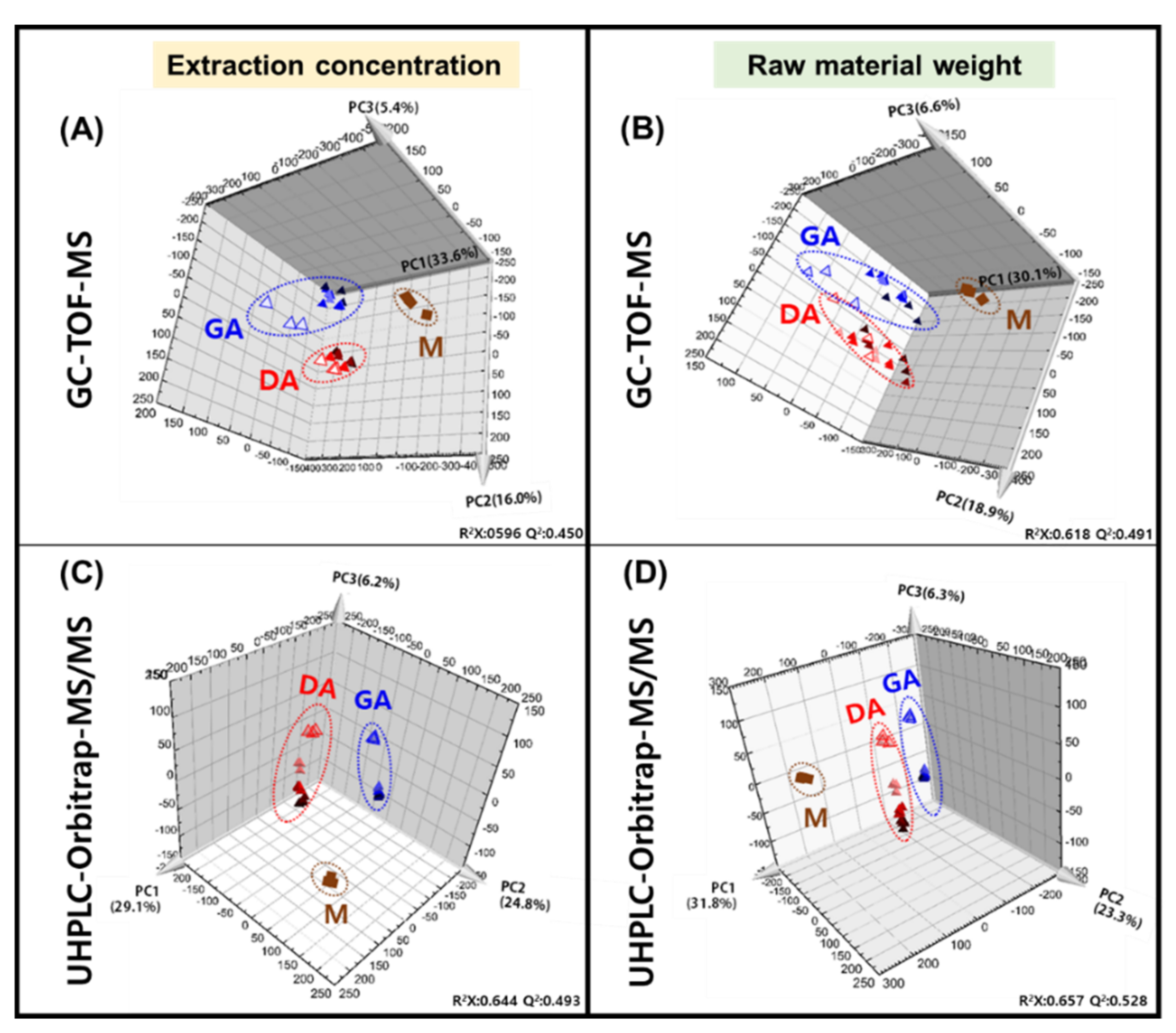
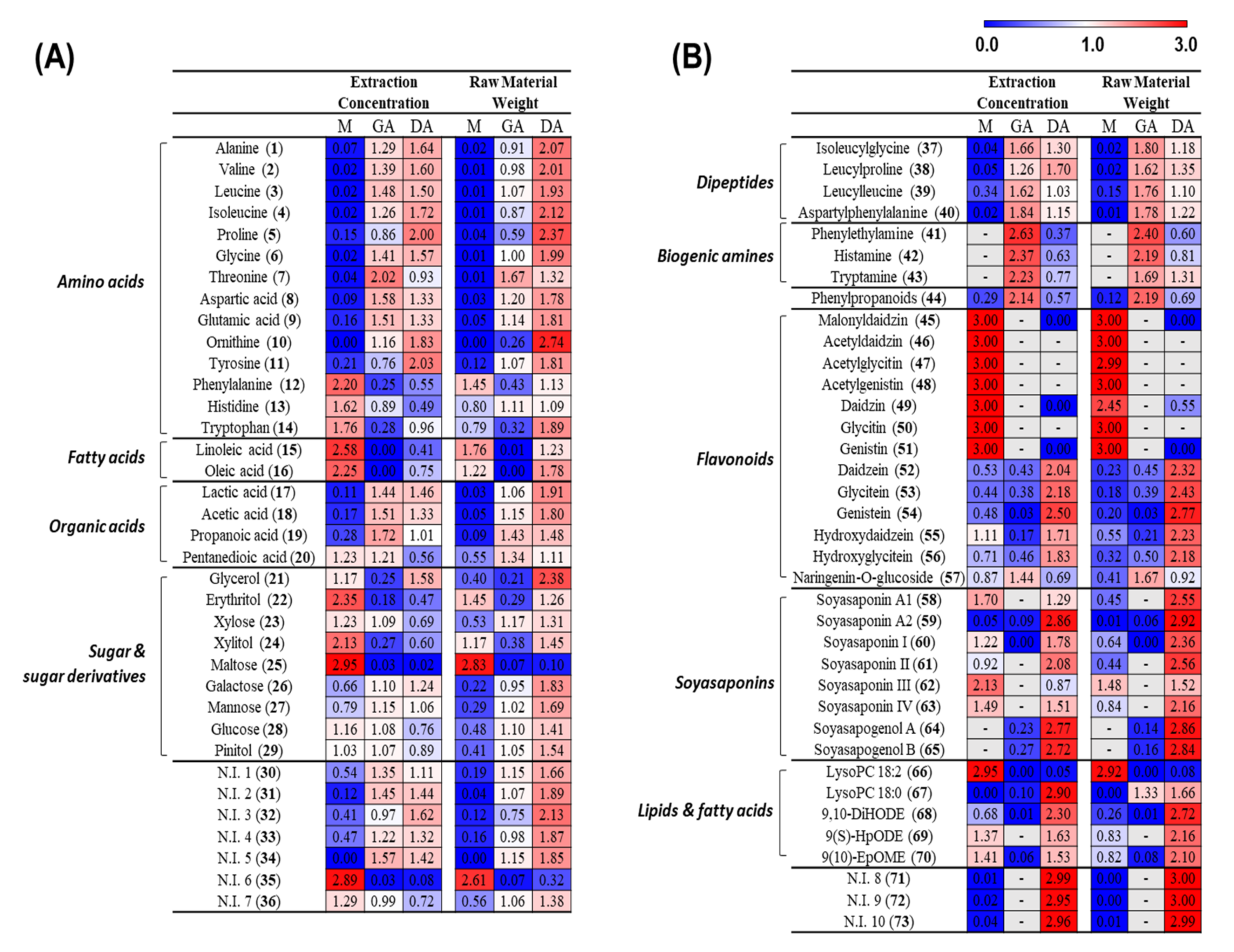
3.1. Metabolite Profiling of Doenjang (Solid) and Ganjang (Liquid)
3.2. Comparison of Metabolite and Microbial Changes during Doenjang and Ganjang Fermentation
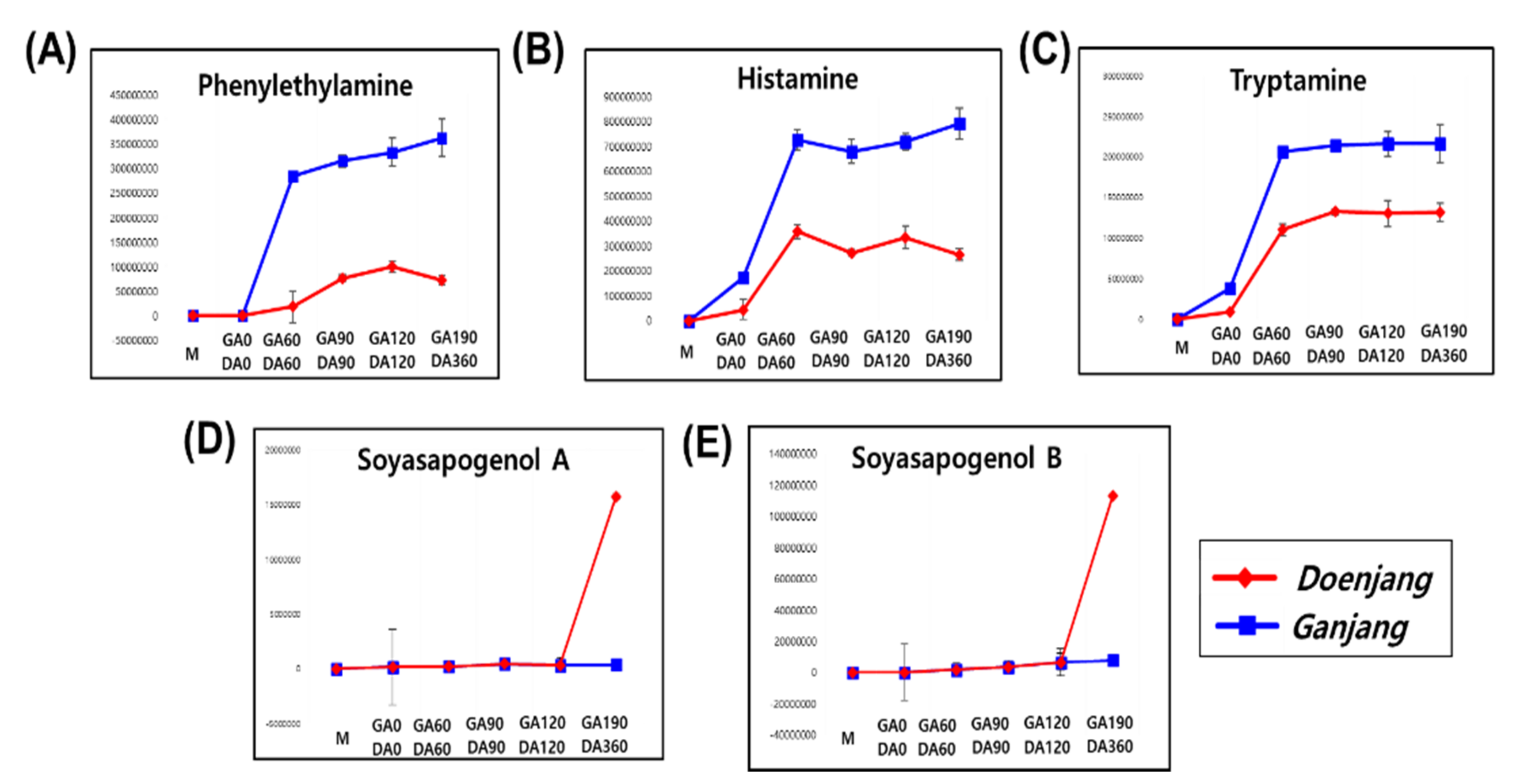
3.2.1. Metabolite Change
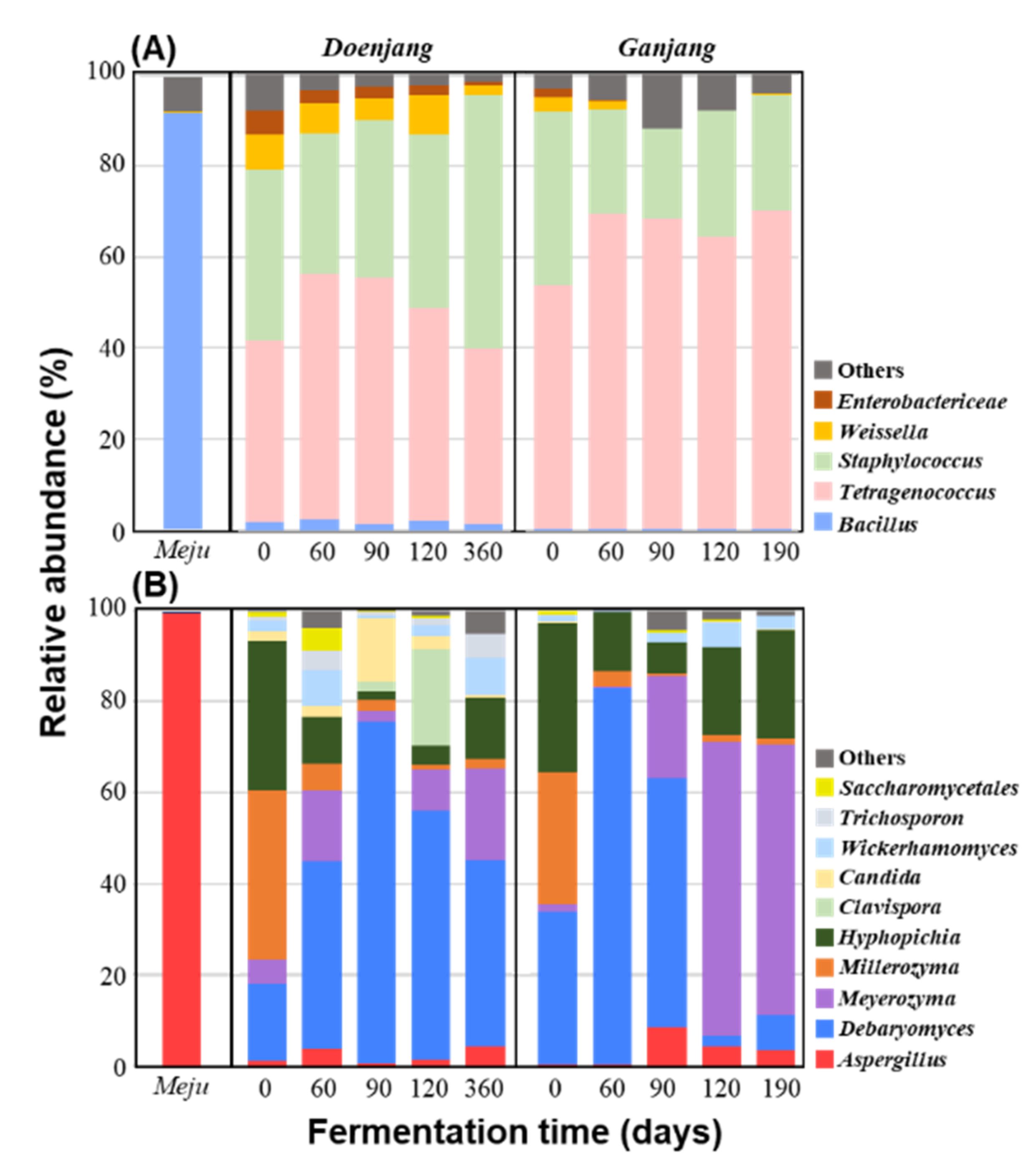
3.2.2. Microbial Community Changes
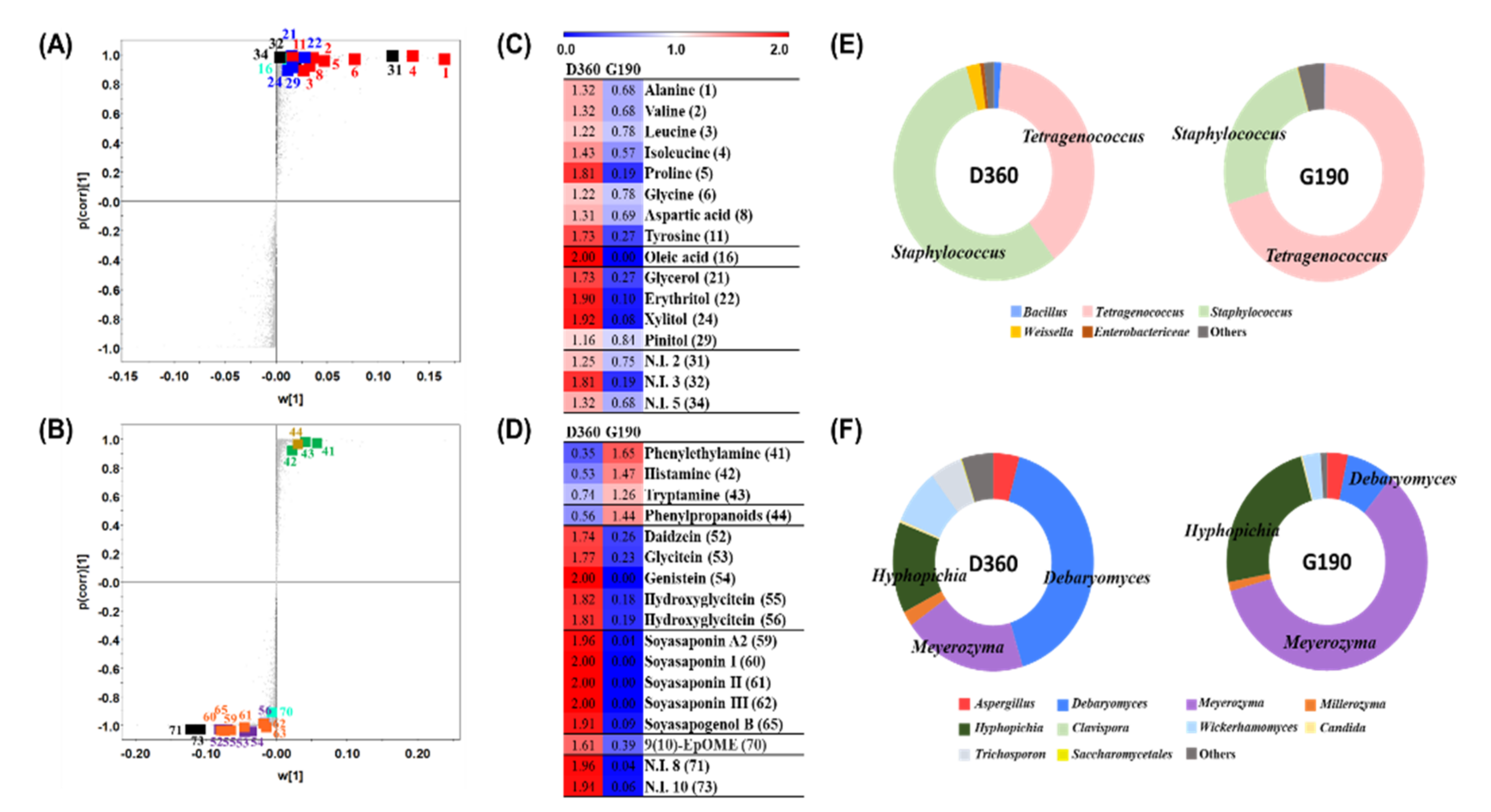
3.3. Comparative Analysis of the Doenjang and Ganjang End Products
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Yokota, T.; Hattori, T.; Ohishi, H.; Hasegawa, K.; Watanabe, K. The effect of antioxidant-containing fraction from fermented soybean food on atherosclerosis development in cholesterol-fed rabbits. LWT-Food Sci. Technol. 1996, 29, 751–755. [Google Scholar] [CrossRef]
- Lee, S.; Lee, S.; Singh, D.; Oh, J.Y.; Jeon, E.J.; Ryu, H.S.; Lee, D.W.; Kim, B.S.; Lee, C.H. Comparative evaluation of microbial diversity and metabolite profiles in doenjang, a fermented soybean paste, during the two different industrial manufacturing processes. Food Chem. 2017, 221, 1578–1586. [Google Scholar] [CrossRef]
- Jung, J.Y.; Lee, S.H.; Jeon, C.O. Microbial community dynamics during fermentation of doenjang-meju, traditional Korean fermented soybean. Int. J. Food Microbiol. 2014, 185, 112–120. [Google Scholar] [CrossRef]
- Cho, K.-M.; Seo, W.-T. Bacterial diversity in a Korean traditional soybean fermented food (doenjang and ganjang) by 16S rRNA gene sequence analysis. Food Sci. Biotechnol. 2007, 16, 320–324. [Google Scholar]
- John, K.M.; Jung, E.S.; Lee, S.; Kim, J.-S.; Lee, C.H. Primary and secondary metabolites variation of soybean contaminated with Aspergillus sojae. Food Res. Int. 2013, 54, 487–494. [Google Scholar] [CrossRef]
- Baek, J.G.; Shim, S.-M.; Kwon, D.Y.; Choi, H.-K.; Lee, C.H.; Kim, Y.-S. Metabolite profiling of Cheonggukjang, a fermented soybean paste, inoculated with various Bacillus strains during fermentation. Biosci. Biotechnol. Biochem. 2010, 74, 1860–1868. [Google Scholar] [CrossRef]
- Gibbons, H.; O’Gorman, A.; Brennan, L. Metabolomics as a tool in nutritional research. Curr. Opin. Lipidol. 2015, 26, 30–34. [Google Scholar] [CrossRef]
- Kim, M.J.; Rhee, H.S. Studies on the change of taste compounds during soy paste fermentation. Korean J. Soc. Food Sci. 1990, 6, 1–8. [Google Scholar]
- Kim, M.K.; Seo, W.T.; Lee, Y.B.; Cho, K.M. Analyses of archaeal communities in Doenjang and Ganjang using a culture-independent manner based on 16S rRNA sequences. Food Sci. Biotechnol. 2013, 22, 449–454. [Google Scholar] [CrossRef]
- Mun, E.; Park, J.E.; Cha, Y. Effects of Doenjang, a traditional Korean soybean paste, with high-salt diet on blood pressure in Sprague–Dawley rats. Nutrients 2019, 11, 2745. [Google Scholar] [CrossRef]
- Kim, K.H.; Lee, S.H.; Chun, B.H.; Jeong, S.E.; Jeon, C.O. Tetragenococcus halophilus MJ4 as a starter culture for repressing biogenic amine (cadaverine) formation during saeu-jeot (salted shrimp) fermentation. Food Microbiol. 2019, 82, 465–473. [Google Scholar] [CrossRef]
- Chun, B.H.; Kim, K.H.; Jeong, S.E.; Jeon, C.O. The effect of salt concentrations on the fermentation of doenjang, a traditional Korean fermented soybean paste. Food Microbiol. 2020, 86, 103329. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.; Oh, D.-G.; Lee, S.; Kim, G.R.; Lee, J.S.; Son, Y.K.; Bae, C.-H.; Yeo, J.; Lee, C.H. Chemotaxonomic metabolite profiling of 62 indigenous plant species and its correlation with bioactivities. Molecules 2015, 20, 19719–19734. [Google Scholar] [CrossRef]
- Son, S.Y.; Kim, N.K.; Lee, S.; Singh, D.; Kim, G.R.; Lee, J.S.; Yang, H.-S.; Yeo, J.; Lee, S.; Lee, C.H. Metabolite fingerprinting, pathway analyses, and bioactivity correlations for plant species belonging to the Cornaceae, Fabaceae, and Rosaceae families. Plant Cell Rep. 2016, 35, 1917–1931. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.Y.; Lee, S.; Lee, S.; Oh, J.Y.; Jeon, E.J.; Ryu, H.S.; Lee, C.H. Primary and secondary metabolite profiling of doenjang, a fermented soybean paste during industrial processing. Food Chem. 2014, 165, 157–166. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.Y.; Kim, H.Y.; Lee, S.; Lee, J.M.; Muthaiya, M.J.; Kim, B.S.; Oh, J.Y.; Song, C.K.; Jeon, E.J.; Ryu, H.S. Mass spectrometry-based metabolite profiling and bacterial diversity characterization of Korean traditional meju during fermentation. J. Microbiol. Biotechnol. 2012, 22, 1523–1531. [Google Scholar] [CrossRef]
- Namgung, H.J.; Park, H.J.; Cho, I.H.; Choi, H.K.; Kwon, D.Y.; Shim, S.M.; Kim, Y.S. Metabolite profiling of doenjang, fermented soybean paste, during fermentation. J. Sci. Food Agric. 2010, 90, 1926–1935. [Google Scholar] [CrossRef]
- Nelson, G.; Chandrashekar, J.; Hoon, M.A.; Feng, L.; Zhao, G.; Ryba, N.J.; Zuker, C.S. An amino-acid taste receptor. Nature 2002, 416, 199–202. [Google Scholar] [CrossRef] [PubMed]
- Jeong, S.W.; Kwon, D.J.; Gu, M.S.; Kim, Y.S. Quality characteristics and preference of Doenjang using rice. Agric. Chem. Biotechnol. 1994, 37, 266–271. [Google Scholar]
- Munro, I.C.; Harwood, M.; Hlywka, J.J.; Stephen, A.M.; Doull, J.; Flamm, W.G.; Adlercreutz, H. Soy isoflavones: A safety review. Nutr. Rev. 2003, 61, 1–33. [Google Scholar] [CrossRef]
- Rostagno, M.A.; Villares, A.; Guillamón, E.; García-Lafuente, A.; Martínez, J. Sample preparation for the analysis of isoflavones from soybeans and soy foods. J. Chromatogr. A 2009, 1216, 2–29. [Google Scholar] [CrossRef]
- Lee, S.; Seo, M.-H.; Oh, D.-K.; Lee, C.H. Targeted metabolomics for Aspergillus oryzae-mediated biotransformation of soybean isoflavones, showing variations in primary metabolites. Biosci. Biotechnol. Biochem. 2014, 78, 167–174. [Google Scholar] [CrossRef] [PubMed]
- Rupasinghe, H.V.; Jackson, C.-J.C.; Poysa, V.; Di Berardo, C.; Bewley, J.D.; Jenkinson, J. Soyasapogenol A and B distribution in soybean (Glycine max L. Merr.) in relation to seed physiology, genetic variability, and growing location. J. Agric. Food Chem. 2003, 51, 5888–5894. [Google Scholar] [CrossRef] [PubMed]
- Jung, J.Y.; Chun, B.H.; Jeon, C.O. Chromohalobacter is a causing agent for the production of organic acids and putrescine during fermentation of ganjang, a Korean traditional soy sauce. J. Food Sci. 2015, 80, M2853–M2859. [Google Scholar] [CrossRef] [PubMed]
- Shukla, S.; Park, H.-K.; Kim, J.-K.; Kim, M. Determination of biogenic amines in Korean traditional fermented soybean paste (Doenjang). Food Chem. Toxicol. 2010, 48, 1191–1195. [Google Scholar] [CrossRef] [PubMed]
- Chang, M.; Chang, H.C. Development of a screening method for biogenic amine producing Bacillus spp. Int. J. Food Microbiol. 2012, 153, 269–274. [Google Scholar] [CrossRef]
- Han, D.M.; Chun, B.H.; Feng, T.; Kim, H.M.; Jeon, C.O. Dynamics of microbial communities and metabolites in ganjang, a traditional Korean fermented soy sauce, during fermentation. Food Microbiol. 2020, 92, 103591. [Google Scholar] [CrossRef] [PubMed]
- Pannala, A.S.; Chan, T.S.; O’Brien, P.J.; Rice-Evans, C.A. Flavonoid Bring chemistry and antioxidant activity: Fast reaction kinetics. Biochem. Biophys. Res. Commun. 2001, 282, 1161–1168. [Google Scholar] [CrossRef]
- Takaaki, Y.; Michikatsu, S. Isolation and properties of β-glucosidase produced by Debaryomyces hansenii and its application in winemaking. Am. J. Enol. Vitic. 1999, 50, 231–235. [Google Scholar]
- Pyo, Y.-H.; Lee, T.-C.; Lee, Y.-C. Enrichment of bioactive isoflavones in soymilk fermented with b-glucosidase-producing lactic acid bacteria. Food Res. Int. 2005, 38, 551–559. [Google Scholar] [CrossRef]
- Jeong, D.-W.; Kim, H.-R.; Jung, G.; Han, S.; Kim, C.-T.; Lee, J.-H. Bacterial community migration in the ripening of doenjang, a traditional Korean fermented soybean food. J. Microbiol. Biotechnol. 2014, 24, 648–660. [Google Scholar] [CrossRef] [PubMed]
- Coda, R.; Rizzello, C.G.; Di Cagno, R.; Trani, A.; Cardinali, G.; Gobbetti, M. Antifungal activity of Meyerozyma guilliermondii: Identification of active compounds synthesized during dough fermentation and their effect on long-term storage of wheat bread. Food Microbiol. 2013, 33, 243–251. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.; Wu, B.; Zhao, W.; Lao, F.; Chen, F.; Liao, X.; Wu, J. Shifts in autochthonous microbial diversity and volatile metabolites during the fermentation of chili pepper (Capsicum frutescens L.). Food Chem. 2020, 335, 127512. [Google Scholar] [CrossRef] [PubMed]
- Shim, J.M.; Lee, K.W.; Yao, Z.; Kim, H.-J.; Kim, J.H. Properties of doenjang (soybean paste) prepared with different types of salts. J. Microbiol. Biotechnol. 2016, 26, 1533–1541. [Google Scholar] [CrossRef]
- Kim, T.-W.; Lee, J.-H.; Kim, S.-E.; Park, M.-H.; Chang, H.C.; Kim, H.-Y. Analysis of microbial communities in doenjang, a Korean fermented soybean paste, using nested PCR-denaturing gradient gel electrophoresis. Int. J. Food Microbiol. 2009, 131, 265–271. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Song, D.H.; Chun, B.H.; Lee, S.; Son, S.Y.; Reddy, C.K.; Mun, H.I.; Jeon, C.O.; Lee, C.H. Comprehensive Metabolite Profiling and Microbial Communities of Doenjang (Fermented Soy Paste) and Ganjang (Fermented Soy Sauce): A Comparative Study. Foods 2021, 10, 641. https://doi.org/10.3390/foods10030641
Song DH, Chun BH, Lee S, Son SY, Reddy CK, Mun HI, Jeon CO, Lee CH. Comprehensive Metabolite Profiling and Microbial Communities of Doenjang (Fermented Soy Paste) and Ganjang (Fermented Soy Sauce): A Comparative Study. Foods. 2021; 10(3):641. https://doi.org/10.3390/foods10030641
Chicago/Turabian StyleSong, Da Hye, Byung Hee Chun, Sunmin Lee, Su Young Son, Chagam Koteswara Reddy, Ha In Mun, Che Ok Jeon, and Choong Hwan Lee. 2021. "Comprehensive Metabolite Profiling and Microbial Communities of Doenjang (Fermented Soy Paste) and Ganjang (Fermented Soy Sauce): A Comparative Study" Foods 10, no. 3: 641. https://doi.org/10.3390/foods10030641
APA StyleSong, D. H., Chun, B. H., Lee, S., Son, S. Y., Reddy, C. K., Mun, H. I., Jeon, C. O., & Lee, C. H. (2021). Comprehensive Metabolite Profiling and Microbial Communities of Doenjang (Fermented Soy Paste) and Ganjang (Fermented Soy Sauce): A Comparative Study. Foods, 10(3), 641. https://doi.org/10.3390/foods10030641







