Products Released from Structurally Different Dextrans by Bacterial and Fungal Dextranases
Abstract
1. Introduction
2. Materials and Methods
2.1. General
2.2. Molecular Cloning and Heterologous Expression
2.3. Optimum Incubation Conditions
2.4. Dextran Production
2.5. Dextranase-Mediated Hydrolysis
2.6. High Performance Anion Exchange Chromatography
3. Results and Discussion
3.1. Dextranase Production and Characterization
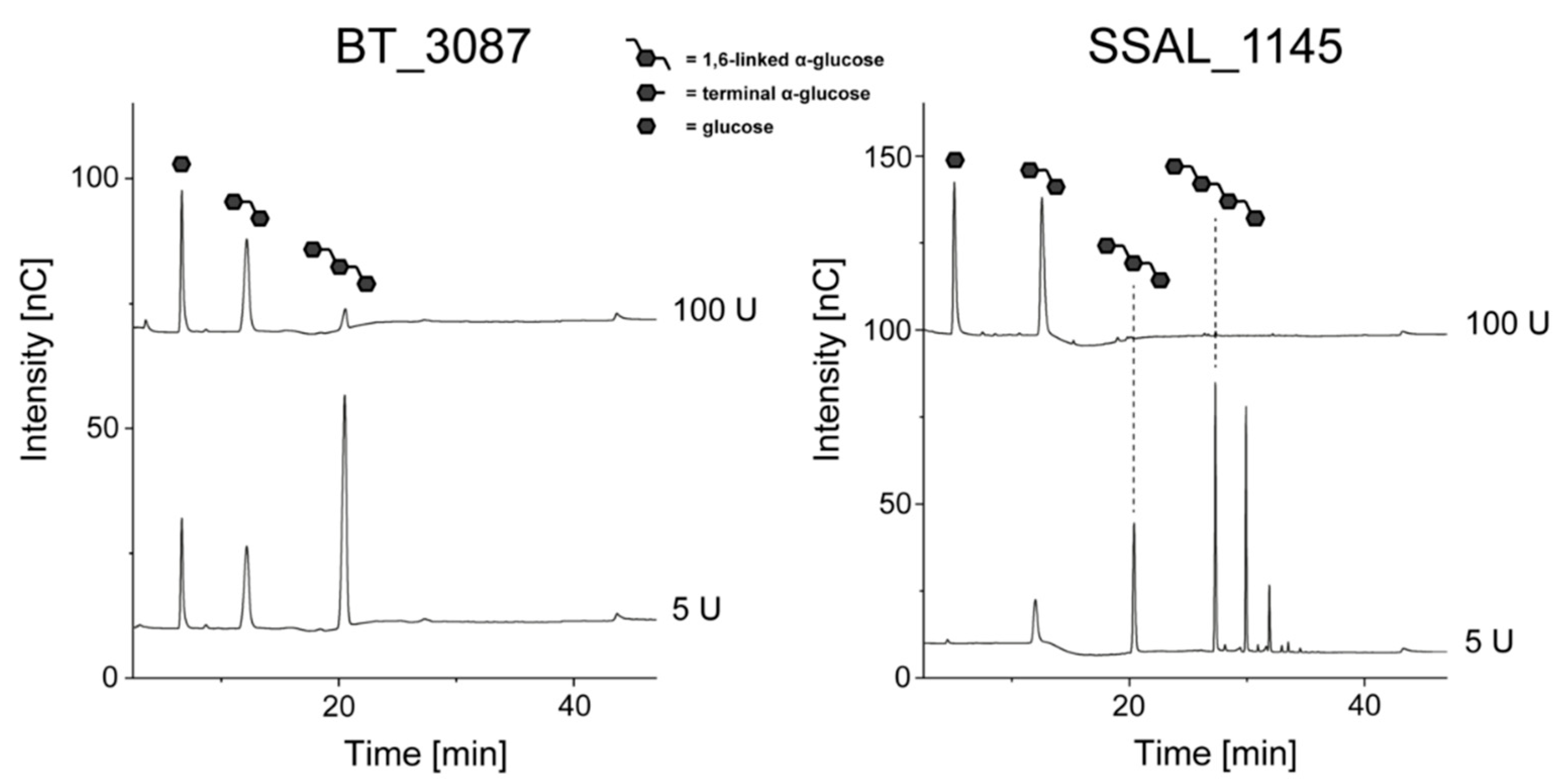
3.2. Hydrolysis of Linear Dextrans
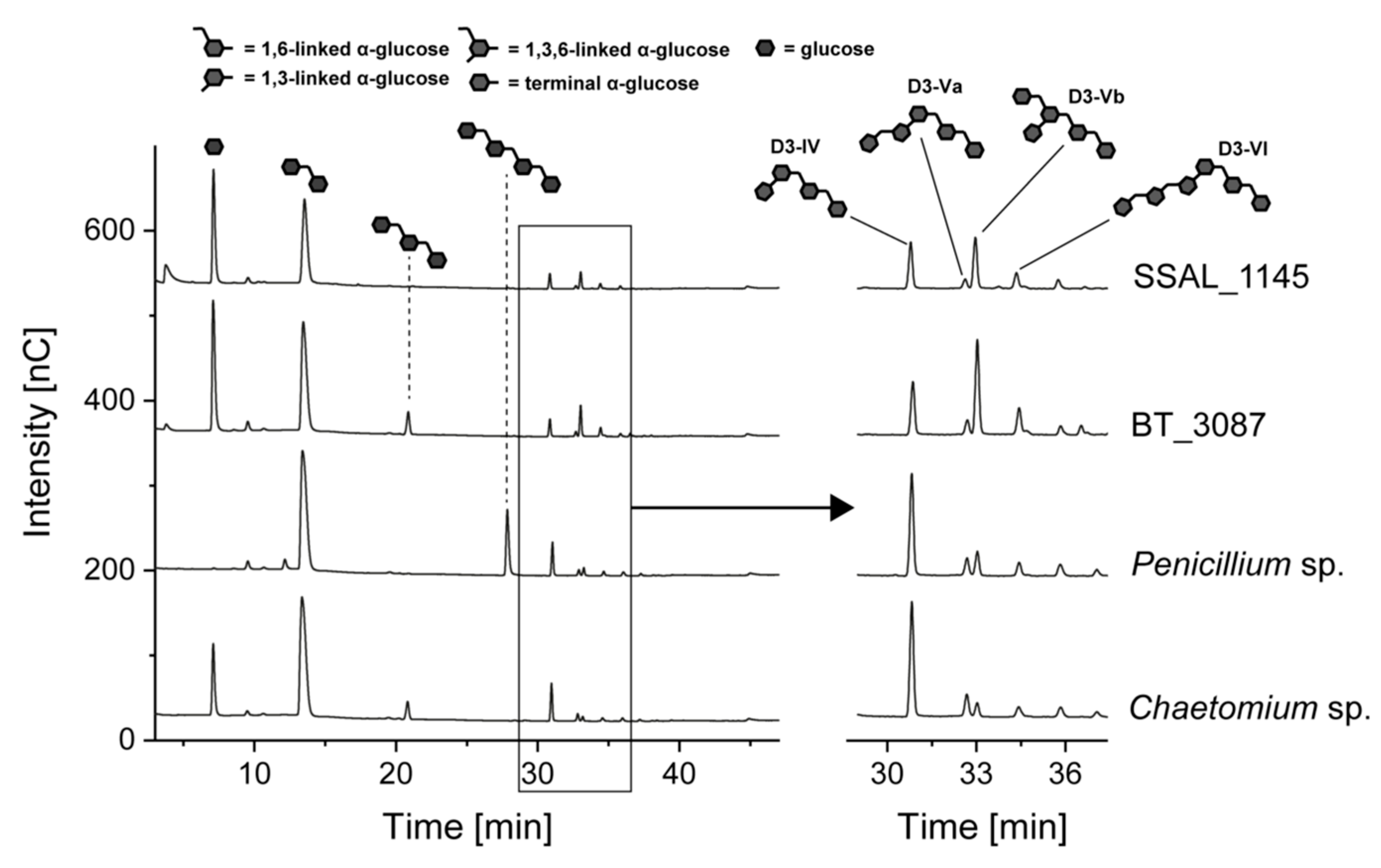
3.3. Hydrolysis of O3- and O4-Branched Dextrans
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Naessens, M.; Cerdobbel, A.; Soetaert, W.; Vandamme, E. Leuconostoc dextransucrase and dextran: Production, properties and applications. J. Chem. Technol. Biotechnol. 2005, 80, 845–860. [Google Scholar] [CrossRef]
- Leemhuis, H.; Pijning, T.; Dobruchowska, J.M.; Van Leeuwen, S.S.; Kralj, S.; Dijkstra, B.W.; Dijkhuizen, L. Glucansucrases: Three-dimensional structures, reactions, mechanism, α-glucan analysis and their implications in biotechnology and food applications. J. Biotechnol. 2013, 163, 250–272. [Google Scholar] [CrossRef]
- Torino, M.I.; Font, G.; Mozzi, F. Biopolymers from lactic acid bacteria. Novel applications in foods and beverages. Front. Microbiol. 2015, 6, 834. [Google Scholar] [CrossRef]
- Zannini, E.; Waters, D.M.; Coffey, A.; Arendt, E.K. Production, properties, and industrial food application of lactic acid bacteria-derived exopolysaccharides. Appl. Microbiol. Biotechnol. 2016, 100, 1121–1135. [Google Scholar] [CrossRef]
- Khalikova, E.; Susi, P.; Korpela, T. Microbial Dextran-Hydrolyzing Enzymes: Fundamentals and Applications. Microbiol. Mol. Biol. Rev. 2005, 69, 306–325. [Google Scholar] [CrossRef]
- Katina, K.; Maina, N.H.; Juvonen, R.; Flander, L.; Johansson, L.; Virkki, L.; Tenkanen, M.; Laitila, A. In situ production and analysis of Weissella confusa dextran in wheat sourdough. Food Microbiol. 2009, 26, 734–743. [Google Scholar] [CrossRef]
- Maina, N.H.; Virkki, L.; Pynnönen, H.; Maaheimo, H.; Tenkanen, M. Structural Analysis of Enzyme-Resistant Isomaltooligosaccharides Reveals the Elongation of α-(1→3)-Linked Branches in Weissella confusa Dextran. Biomacromolecules 2011, 12, 409–418. [Google Scholar] [CrossRef]
- Fels, L.; Jakob, F.; Vogel, R.F.; Wefers, D. Structural characterization of the exopolysaccharides from water kefir. Carbohydr. Polym. 2018, 189, 296–303. [Google Scholar] [CrossRef]
- Xu, D.; Fels, L.; Wefers, D.; Behr, J.; Jakob, F.; Vogel, R.F. Lactobacillus hordei dextrans induce Saccharomyces cerevisiae aggregation and network formation on hydrophilic surfaces. Int. J. Biol. Macromol. 2018, 115, 236–242. [Google Scholar] [CrossRef]
- Bechtner, J.; Wefers, D.; Schmid, J.; Vogel, R.F.; Jakob, F. Identification and comparison of two closely related dextransucrases released by water kefir borne Lactobacillus hordei TMW 1.1822 and Lactobacillus nagelii TMW 1.1827. Microbiology 2019, 165, 956–966. [Google Scholar] [CrossRef]
- Münkel, F.; Wefers, D. Fine structures of different dextrans assessed by isolation and characterization of endo-dextranase liberated isomalto-oligosaccharides. Carbohydr. Polym. 2019, 215, 296–306. [Google Scholar] [CrossRef]
- Münkel, F.; Bechtner, J.; Eckel, V.; Fischer, A.; Herbi, F.; Jakob, F.; Wefers, D. Detailed Structural Characterization of Glucans Produced by Glucansucrases from Leuconostoc citreum TMW 2.1194. J. Agric. Food Chem. 2019, 67, 6856–6866. [Google Scholar] [CrossRef]
- Münkel, F.; Fischer, A.; Wefers, D. Structural characterization of mixed-linkage α-glucans produced by mutants of Lactobacillus reuteri TMW 1.106 dextransucrase. Carbohydr. Polym. 2020, 231, 115697. [Google Scholar] [CrossRef]
- Das, D.K.; Dutta, S.K. Purification, biochemical characterisation and mode of action of an extracellular endo-dextranase from the culture filtrate of Penicillium lilacinum. Int. J. Biochem. Cell Biol. 1996, 28, 107–113. [Google Scholar] [CrossRef]
- Hattori, A.; Ishibashi, K.; Minato, S. The purification and characterization of the dextranase of Chaetomium gracile. Agric. Biol. Chem. 1981, 45, 2409–2416. [Google Scholar] [CrossRef]
- Larsson, A.M.; Andersson, R.; Stahlberg, J.; Kenne, L.; Jones, T.A. Dextranase from Penicillum minioluteum: Reaction course, crystal structure, and product complex. Structure 2003, 11, 1111–1121. [Google Scholar] [CrossRef]
- Sugiura, M.; Ito, A.; Ogiso, T.; Kato, K.; Asano, H. Studies on dextranase—Purification of dextranase from Penicillium funiculosum and its enzymatic properties. Biochim. Biophys. Acta 1973, 309, 357–362. [Google Scholar] [CrossRef]
- Virgen-Ortíz, J.; Ibarra-Junquera, V.; Escalanteminakata, P.; Ornelas-Paz, J.D.J.; Osunacastro, J.A.; González-Potes, A. Kinetics and thermodynamic of the purified dextranase from Chaetomium erraticum. J. Mol. Catal. B Enzym. 2015, 122, 80–86. [Google Scholar] [CrossRef]
- Yang, L.; Zhou, N.; Tian, Y. Purification, characterization, and biocatalytic potential of a novel dextranase from Chaetomium globosum. Biotechnol. Lett. 2018, 40, 1407–1418. [Google Scholar] [CrossRef]
- Yilan, O.; Sun, X.; Du, K.; Ouyang, Y.; Wu, C.; Xu, N.; Linhardt, R.J.; Zhang, Z. UP-HILIC-MS/MS to Determine the Action Pattern of Penicillium sp. Dextranase. J. Am. Soc. Mass Spectrom. 2015, 26, 1174–1185. [Google Scholar] [CrossRef]
- Erhardt, F.A.; Stammen, S.; Jördening, H.-J. Production, characterization and (co-)immobilization of dextranase from Penicillium aculeatum. Biotechnol. Lett. 2008, 30, 1069–1073. [Google Scholar] [CrossRef] [PubMed]
- Walker, G.J.; Dewar, M.D. The action pattern of Penicillium lilacinum dextranase. Carbohydr. Res. 1975, 39, 303–315. [Google Scholar] [CrossRef]
- Hiraoka, N.; Tsuji, H.; Fukumoto, J.; Yamamoto, T.; Tsuru, D. Studies on Mold Dextranases: Some Physicochemical Properties and Substrate Specificity of Dextranases Obtained from Aspergillus carneus and Penicillium luteum. Int. J. Pept. Protein Res. 2009, 5, 161–169. [Google Scholar] [CrossRef] [PubMed]
- Prabhu, M.; Prabhu, K.A. Studies on dextranase from Penicillium aculeatum. Enzyme Microb. Technol. 1984, 6, 217–220. [Google Scholar]
- Kim, Y.-M.; Yamamoto, E.; Kang, M.-S.; Nakai, H.; Saburi, W.; Okuyama, M.; Mori, H.; Funane, K.; Momma, M.; Fujimoto, Z.; et al. Bacteroides thetaiotaomicron VPI-5482 glycoside hydrolase family 66 homolog catalyzes dextranolytic and cyclization reactions. FEBS J. 2012, 279, 3185–3191. [Google Scholar] [CrossRef]
- Igarashi, T.; Morisaki, H.; Goto, N. Molecular Characterization of Dextranase from Streptococcus rattus. Microbiol. Immunol. 2004, 48, 155–162. [Google Scholar] [CrossRef]
- Wanda, S.Y.; Curtiss, R. Purification and characterization of Streptococcus sobrinus dextranase produced in recombinant Escherichia coli and sequence analysis of the dextranase gene. J. Bacteriol. 1994, 176, 3839–3850. [Google Scholar] [CrossRef][Green Version]
- Pulkownik, A.; Walker, G.J. Purification and substrate specificity of an endo-dextranase of Streptococcus mutans K1-R. Carbohydr. Res. 1977, 54, 237–251. [Google Scholar] [CrossRef]
- Van Bueren, A.L.; Saraf, A.; Martens, E.C.; Dijkhuizen, L. Differential Metabolism of Exopolysaccharides from Probiotic Lactobacilli by the Human Gut Symbiont Bacteroides thetaiotaomicron. Appl. Environ. Microbiol. 2015, 81, 3973–3983. [Google Scholar] [CrossRef]
- Rühmkorf, C.; Rübsam, H.; Becker, T.; Bork, C.; Voiges, K.; Mischnick, P.; Brandt, M.J.; Vogel, R.F. Effect of structurally different microbial homoexopolysaccharides on the quality of gluten-free bread. Eur. Food Res. Technol. 2012, 235, 139–146. [Google Scholar] [CrossRef]
- Aslanidis, C.; De Jong, P.J. Ligation-independent cloning of PCR products (LIC-PCR). Nucleic Acids Res. 1990, 18, 6069–6074. [Google Scholar] [CrossRef] [PubMed]
- Burgess-Brown, N.A.; Sharma, S.; Sobott, F.; Loenarz, C.; Oppermann, U.; Gileadi, O. Codon optimization can improve expression of human genes in Escherichia coli: A multi-gene study. Protein Expr. Purif. 2008, 59, 94–102. [Google Scholar] [CrossRef] [PubMed]
- Stols, L.; Gu, M.Y.; Dieckman, L.; Raffen, R.; Collart, F.R.; Donnelly, M.I. A new vector for high-throughput, ligation-independent cloning encoding a tobacco etch virus protease cleavage site. Protein Expr. Purif. 2002, 25, 8–15. [Google Scholar] [CrossRef] [PubMed]
- Rühmkorf, C.; Bork, C.; Mischnick, P.; Rübsam, H.; Becker, T.; Vogel, R.F. Identification of Lactobacillus curvatus TMW 1.624 dextransucrase and comparative characterization with Lactobacillus reuteri TMW 1.106 and Lactobacillus animalis TMW 1.971 dextransucrases. Food Microbiol. 2013, 34, 52–61. [Google Scholar] [CrossRef]
- Sawai, T.; Niwa, Y. Transisomaltosylation activity of a bacterial isomalto-dextranase. Agric. Biol. Chem. 1975, 39, 1077–1083. [Google Scholar]





Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pittrof, S.L.; Kaufhold, L.; Fischer, A.; Wefers, D. Products Released from Structurally Different Dextrans by Bacterial and Fungal Dextranases. Foods 2021, 10, 244. https://doi.org/10.3390/foods10020244
Pittrof SL, Kaufhold L, Fischer A, Wefers D. Products Released from Structurally Different Dextrans by Bacterial and Fungal Dextranases. Foods. 2021; 10(2):244. https://doi.org/10.3390/foods10020244
Chicago/Turabian StylePittrof, Silke L., Larissa Kaufhold, Anja Fischer, and Daniel Wefers. 2021. "Products Released from Structurally Different Dextrans by Bacterial and Fungal Dextranases" Foods 10, no. 2: 244. https://doi.org/10.3390/foods10020244
APA StylePittrof, S. L., Kaufhold, L., Fischer, A., & Wefers, D. (2021). Products Released from Structurally Different Dextrans by Bacterial and Fungal Dextranases. Foods, 10(2), 244. https://doi.org/10.3390/foods10020244





