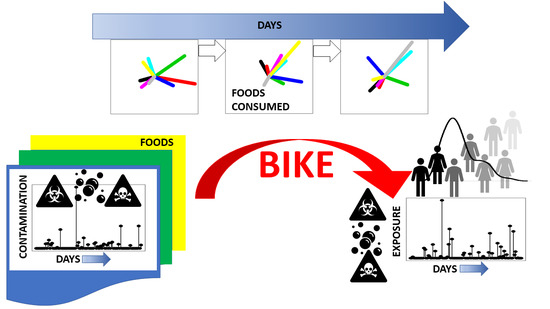
BIKE: Dietary Exposure Model for Foodborne Microbiological and Chemical Hazards
Abstract
:1. Introduction
2. Materials and Methods
Quantifying Uncertainty with Probability: Theory in Short
3. Bayesian Inference from Chemical and Microbiological Occurrence Data
3.1. Option 1: Distribution of Positive Concentrations and Contamination Prevalence Estimated Separately
3.2. Option 2: Distribution of Positive Concentrations and Contamination Prevalence Estimated Jointly
4. Bayesian Inference from Consumption Data
- between-foods correlation in expected (long-term average) consumption amounts,
- between-foods correlation in one-day consumption amounts,
- between-foods correlation in consumption frequency, and
- either dependency or independency of consumption decisions between days.
4.1. Model for Consumption Amounts on Actual Consumption Days
4.2. Option 1: Consumption Frequencies Assuming Days Are Independent of Each Other
4.3. Option 2: Consumption Frequencies Assuming the Consumption on a Day Depends on the Previous Day
4.4. Posterior Distribution for Consumption Model Parameters
5. Exposure Assessment Implied by Bayesian Inference
5.1. Univariate (Marginal) Acute Positive Exposure for Single Food Type, Single Microbiological Hazard
5.2. Univariate (Marginal) Chronic Positive Exposure for Single Food Type, Single Chemical Hazard
5.3. Multiple Exposure from a Subset of Selected Foods among All Food Types, Single Hazard
5.4. Posterior Predictive Distributions
5.5. Microbiological Acute Exposures
5.5.1. Exposure to a Hazard-Food Pair
5.5.2. Exposure to Several Food Types Summed up
5.6. Chemical Chronic Exposures
5.6.1. Exposure to a Hazard-Food Pair
5.6.2. Exposure to Several Food Types
5.7. Posterior Predictive Distributions for Acute and Chronic Exposure
5.7.1. Is Separation of Uncertainty and Variability Always Worth It?
5.7.2. Posterior Predictive Outputs in BIKE
- (1) uncertain population parameters ,
- (2a) consumers’ variable parameters , given the population parameters,
- (2b) consumers’ variable chronic exposures , given the population parameters (if assessment of chronic exposure),
- (3) consumers’ variable acute exposures , given the parameters specific for consumer and population (if assessment of acute exposure),
6. Results: Driving BIKE
6.1. From Inputs to Bayesian Computations and Results: Shiny App
6.1.1. Input Data Format
6.1.2. Automatic Model Construction and Simulation
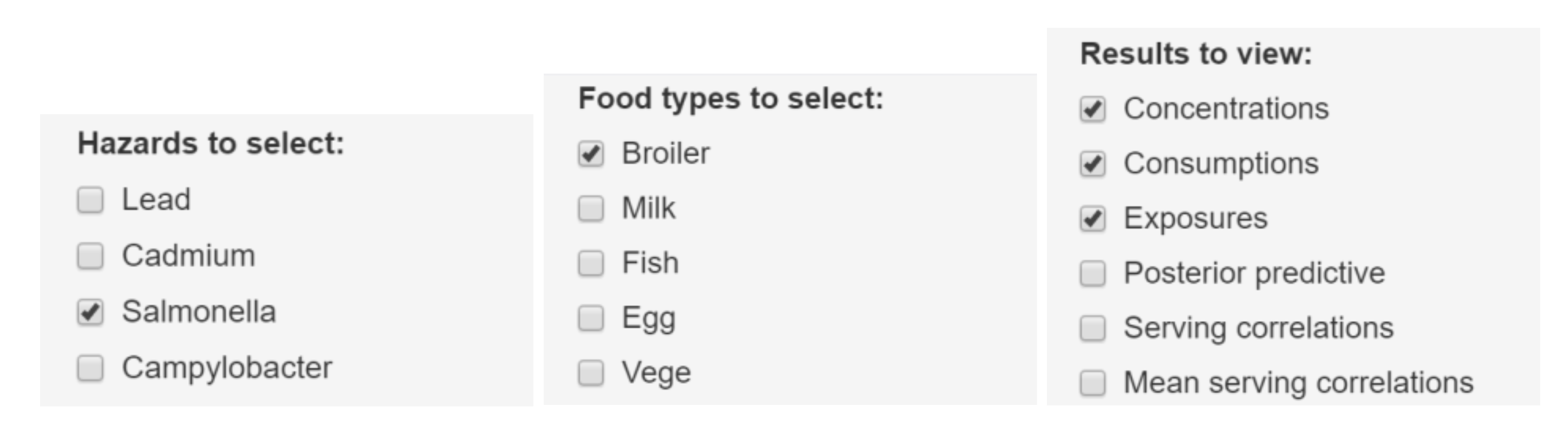
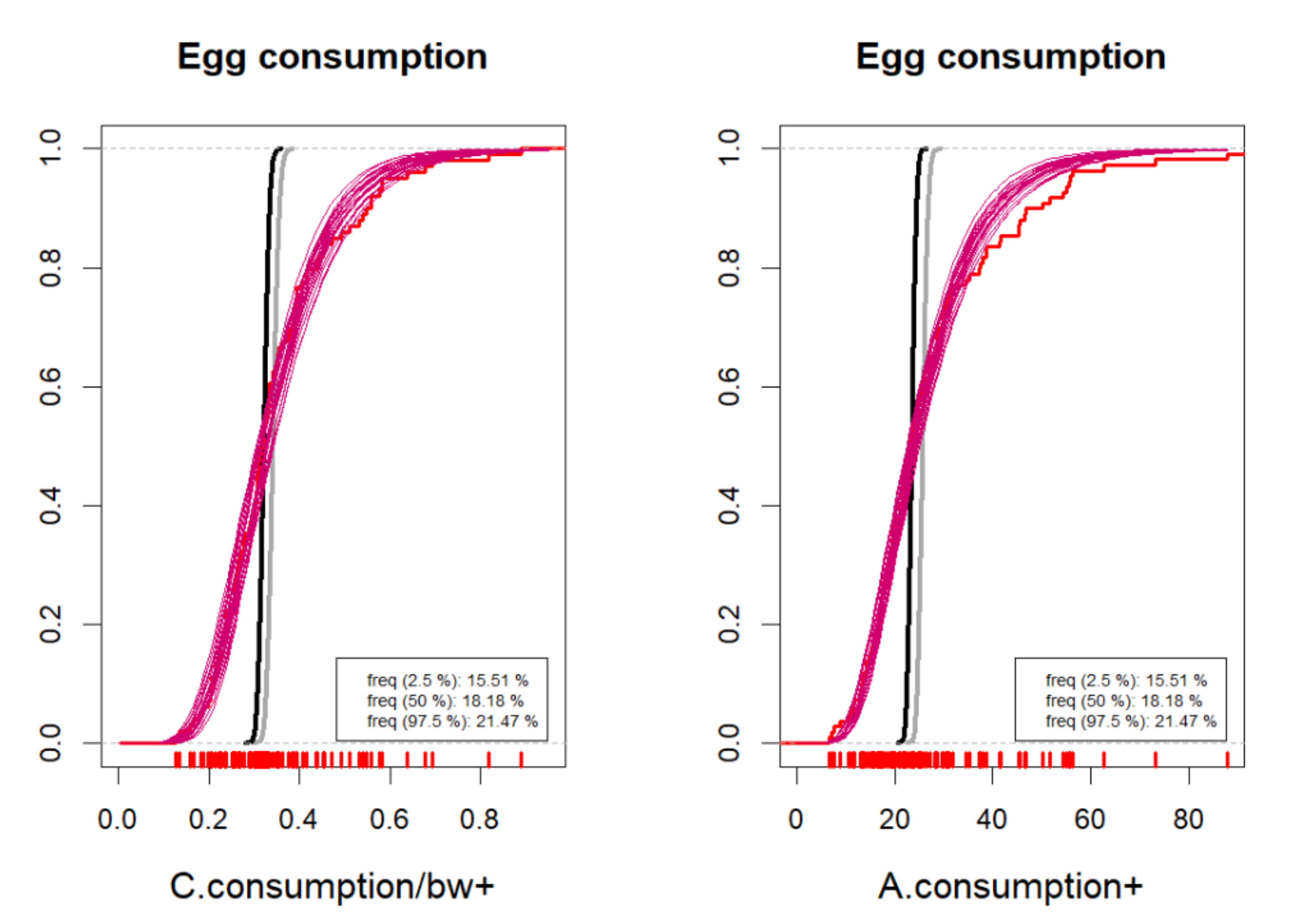
6.1.3. Selection of Plots for Results
6.1.4. Validation against Data
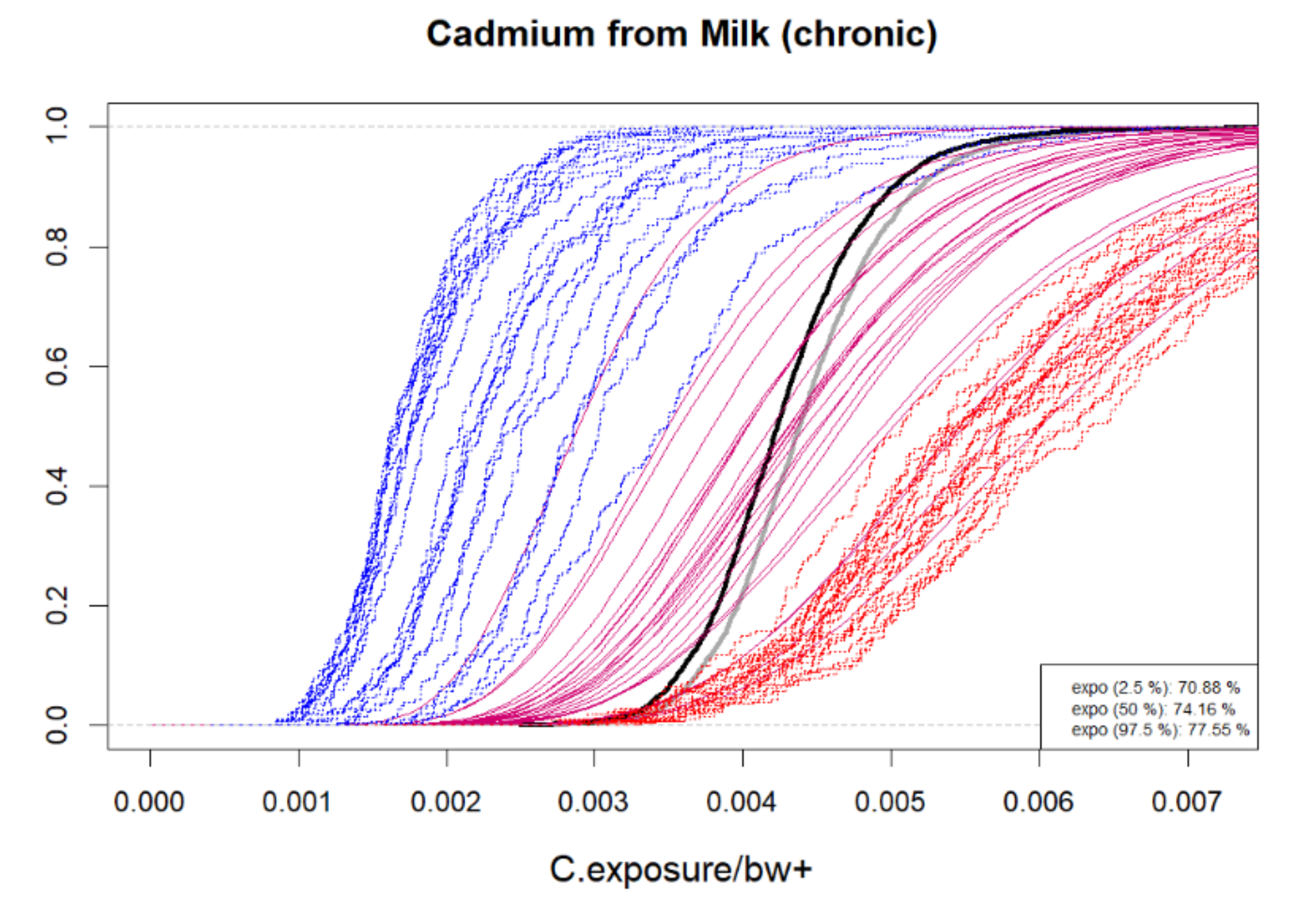
6.1.5. Inspecting Uncertainty and Variability
6.1.6. Adjustment Factors
7. Discussion
7.1. The Realm of Modeling in Dietary Exposure Assessment
7.2. Approaches to Uncertainty
7.3. Parametric or Non-Parametric?
7.4. Advantages in Multivariate Multiparameter Exposure Assessments
7.5. Further Issues
7.6. Obtaining BIKE
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| bw | body weight. |
| E(·) | expected value of ·. |
| f | probability density function. |
| F | cumulative probability function. |
| L | likelihood function, i.e., conditional probability of data. |
| LOD | Limit of detection. |
| LOQ | Limit of quantification. |
| LN | Log-normal distribution. |
| MCMC | Markov chain Monte Carlo simulation method. |
| MN | Multinormal distribution. |
Appendix A. Synthetic Data
References
- Dodd, K.W.; Guenther, P.M.; Freedman, L.S.; Subar, A.F.; Kipnis, V.; Midthune, D.; Tooze, J.A.; Krebs-Smith, S.M. Statistical Methods for Estimating Usual Intake of Nutrients and Foods: A Review of the Theory. J. Am. Diet. Assoc. 2006, 106, 1640–1650. [Google Scholar] [CrossRef]
- Hoffmann, K.; Boeing, H.; Dufour, A.; Volatier, J.L.; Telman, J.; Virtanen, M.; Becker, W.; De Henauw, S. Estimating the distribution of usual dietary intake by short-term measurements. Eur. J. Clin. Nutr. 2002, 56 (Suppl. 2), S53–S62. [Google Scholar] [CrossRef] [Green Version]
- Tooze, J.A.; Midthune, D.; Dodd, K.W.; Freedman, L.S.; Krebs-Smith, S.M.; Subar, A.F.; Guenther, P.M.; Carroll, R.J.; Kipnis, V. A New Statistical Method for Estimating the Usual Intake of Episodically Consumed Foods with Application to Their Distribution. J. Am. Diet. Assoc. 2006, 106, 1575–1587. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- van der Voet, H.; de Boer, W.J.; Kruisselbrink, J.W.; Goedhart, P.W.; van der Heijden, G.W.A.M.; Kennedy, M.C.; Boon, P.E.; van Klaveren, J.D. The MCRA model for probabilistic single-compound and cumulative risk assessment of pesticides. Food Chem. Toxicol. 2015, 79, 5–12. [Google Scholar] [CrossRef] [PubMed]
- Dekkers, A.L.M.; Verkaik-Kloosterman, J.; Rossum, C.T.M.V.; Ocké, M.C. SPADE, a New Statistical Program to Estimate Habitual Dietary Intake from Multiple Food Sources and Dietary Supplements. J. Nutr. 2014, 144, 2083–2091. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cowles, K.; Carlin, B.P. Markov Chain Monte Carlo Convergence Diagnostics: A Comparative Review. J. Am. Stat. Assoc. 1996, 91, 883–904. [Google Scholar] [CrossRef]
- European Food Safety Authority. Management of left-censored data in dietary exposure assessment of chemical substances. EFSA J. 2010, 8, 1557. [Google Scholar] [CrossRef] [Green Version]
- Kennedy, M.C.; van der Voet, H.; Roelofs, V.J.; Roelofs, W.; Glass, C.R.; de Boer, W.J.; Kruisselbrink, J.W.; Hart, A.D.M. New approaches to uncertainty analysis for use in aggregate and cumulative risk assessment of pesticides. Food Chem. Toxicol. 2015, 79, 54–64. [Google Scholar] [CrossRef]
- EFSA Panel on Plant Protection Products and their Residues (PPR). Guidance on the Use of Probabilistic Methodology for Modelling Dietary Exposure to Pesticide Residues. EFSA J. 2012, 10, 2839. [Google Scholar] [CrossRef]
- Lindqvist, R.; Langerholc, T.; Ranta, J.; Hirvonen, T.; Sand, S. A common approach for ranking of microbiological and chemical hazards in foods based on risk assessment–Useful but is it possible? Crit. Rev. Food Sci. Nutr. 2020, 60, 3461–3474. [Google Scholar] [CrossRef]
- Kennedy, M. Bayesian modelling of long-term dietary intakes from multiple sources. Food Chem. Toxicol. 2010, 48, 250–263. [Google Scholar] [CrossRef]
- Paulo, M.J.; Voet, H.; Jansen, M.J.W.; Braak, C.J.F.; Klaveren, J.D. Risk assessment of dietary exposure to pesticides using a Bayesian method. Pest Manag. Sci. 2005, 61, 759–766. [Google Scholar] [CrossRef]
- Chatterjee, A.; Horgan, G.; Theobald, C. Exposure Assessment for Pesticide Intake from Multiple Food Products: A Bayesian Latent-Variable Approach. Risk Anal. 2008, 28, 1727–1736. [Google Scholar] [CrossRef] [Green Version]
- Theobald, C.; Chatterjee, A.; Horgan, G. A hierarchical Bayesian mixture model for repeated dietary records. Food Chem. Toxicol. 2012, 50, 320–327. [Google Scholar] [CrossRef]
- Tressou, J.; Ben Abdallah, N.; Planche, C.; Dervilly-Pinel, G.; Sans, P.; Engel, E.; Albert, I. Exposure assessment for dioxin-like PCBs intake from organic and conventional meat integrating cooking and digestion effects. Food Chem. Toxicol. 2017, 110, 251–261. [Google Scholar] [CrossRef]
- Lunn, D.; Jackson, C.; Best, N.; Thomas, A.; Spiegelhalter, D. The BUGS Book. A Practical Introduction to Bayesian Analysis; Chapman & Hall/CRC: London, UK, 2013. [Google Scholar]
- R Development Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2008; ISBN 3-900051-07-0. [Google Scholar]
- Kruschke, J. Doing Bayesian Data Analysis: A Tutorial with R, JAGS, and Stan, 2nd ed.; Academic Press: London, UK, 2015. [Google Scholar]
- Gelman, A.; Carlin, J.B.; Stern, H.S.; Dunson, D.B.; Vehtari, A.; Rubin, D.B. Bayesian Data Analysis, 3rd ed.; Chapman & Hall/CRC: London, UK, 2013. [Google Scholar]
- Lunn, D.; Spiegelhalter, D.; Thomas, A.; Best, N. The BUGS project: Evolution, critique and future directions. Stat. Med. 2009, 28, 3049–3067. [Google Scholar] [CrossRef]
- Plummer, M. JAGS: A Program for Analysis of Bayesian Graphical Models Using Gibbs Sampling. In Proceedings of the 3rd International Workshop on Distributed Statistical Computing (DSC 2003), Vienna, Austria, 20–22 March 2003. ISSN 1609-395X. [Google Scholar]
- Carpenter, B.; Gelman, A.; Hoffman, M.D.; Lee, D.; Goodrich, B.; Betancourt, M.; Brubaker, M.; Guo, J.; Li, P.; Riddell, A. Stan: A Probabilistic Programming Language. J. Stat. Softw. 2017, 76, 1–32. [Google Scholar] [CrossRef] [Green Version]
- Cox, D.R.; Oakes, D. Analysis of Survival Data; Chapman & Hall: London, UK, 1984. [Google Scholar]
- Armbruster, D.A.; Pry, T. Limit of Blank, Limit of Detection and Limit of Quantification. Clin. Biochem. Rev. 2008, 29 (Suppl. 1), S49–S52. [Google Scholar] [PubMed]
- Belter, M.; Sajnóg, A.; Barałkiewicz, D. Over a century of detection and quantification capabilities in analytical chemistry—Historical overview and trends. Talanta 2014, 129, 606–616. [Google Scholar] [CrossRef] [PubMed]
- Wenzl, T.; Haedrich, J.; Schaechtele, A.; Robouch, P.; Stroka, J. Guidance Document on the Estimation of LOD and LOQ for Measurements in the Field of Contaminants in Feed and Food; EUR 28099; Publications Office of the European Union: Luxembourg, 2016; ISBN 978-92-79-61768-3. [Google Scholar] [CrossRef]
- Lorimer, M.F.; Kiermeier, A. Analysing microbiological data: Tobit or not Tobit? Int. J. Food Microbiol. 2007, 116, 313–318. [Google Scholar] [CrossRef] [PubMed]
- Busschaert, P.; Geeraerd, A.H.; Uyttendaele, M.; Van Impe, J.F. Estimating distributions out of qualitative and (semi)quantitative microbiological contamination data for use in risk assessment. Int. J. Food Microbiol. 2010, 138, 260–269. [Google Scholar] [CrossRef]
- Duarte, A.S.R.; Stockmarr, A.; Nauta, M.J. Fitting a distribution to microbial counts: Making sense of zeroes. Int. J. Food Microbiol. 2015, 196, 40–50. [Google Scholar] [CrossRef] [PubMed]
- Duarte, A.S.R.; Nauta, M.J. Impact of microbial count distributions on human health risk estimates. Int. J. Food Microbiol. 2015, 195, 48–57. [Google Scholar] [CrossRef] [PubMed]
- Chik, A.H.S.; Schmidt, P.J.; Emelko, M.B. Learning Something from Nothing: The Critical Importance of Rethinking Microbial Non-detects. Front. Microbiol. 2018, 9, 2304. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pouillot, R.; Hoelzer, K.; Chen, Y.; Dennis, S. Estimating probability distributions of bacterial concentrations in food based on data generated using the most probable number (MPN) method for use in risk assessment. Food Control 2013, 29, 350–357. [Google Scholar] [CrossRef]
- Helsel, D.R. Fabricating data: How substituting values for nondetects can ruin results, and what can be done about it. Chemosphere 2006, 65, 2434–2439. [Google Scholar] [CrossRef]
- LaFleur, B.; Lee, W.; Billhiemer, D.; Lockhart, C.; Liu, J.; Merchant, N. Statistical methods for assays with limits of detection: Serum bile acid as a differentiator between patients with normal colons, adenomas, and colorectal cancer. J. Carcinog. 2011, 10, 12. [Google Scholar] [CrossRef]
- Ranta, J. Estimating concentration distributions: The effect of measurement limits with small data. In Chapter 6, Risk Assessment Methods for Biological and Chemical Hazards in Food, 1st ed.; CRC Press, Taylor & Francis Group: Boca Raton, FL, USA, 2020; ISBN 9781498762021. [Google Scholar]
- Office of Pesticide Programs. Assigning Values to Nondetected/Non-Quantified Pesticide Residues in Human Health Food Exposure Assessments; U.S. Environmental Protection Agency: Washington, DC, USA, 2000.
- European Food Safety Authority. Guidance on the EU Menu methodology. EFSA J. 2014, 12, 3944. [Google Scholar] [CrossRef] [Green Version]
- Pasonen, P.; Ranta, J.; Tapanainen, H.; Valsta, L.; Tuominen, P. Listeria monocytogenes risk assessment on cold smoked and salt-cured fishery products in Finland—A repeated exposure model. Int. J. Food Microbiol. 2019, 304, 97–105. [Google Scholar] [CrossRef]
- Pouillot, R.; Delignette-Muller, M.L. Evaluating variability and uncertainty separately in microbial quantitative risk assessment using two R packages. Int. J. Food Microbiol. 2010, 142, 330–340. [Google Scholar] [CrossRef]
- Scholz, R. European database of processing factors for pesticides. Efsa Support. Publ. 2018, 15. [Google Scholar] [CrossRef]
- Ashby, D. Bayesian statistics in medicine: A 25 year review. Stat. Med. 2006, 25, 3589–3631. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, P.J.; Emelko, M.B.; Thompson, M.E. Recognizing Structural Nonidentifiability: When Experiments Do Not Provide Information About Important Parameters and Misleading Models Can Still Have Great Fit. Risk Anal. 2020, 40, 352–369. [Google Scholar] [CrossRef] [PubMed]
- Busschaert, P.; Geeraerd, A.H.; Uyttendaele, M.; Van Impe, J.F. Hierarchical Bayesian analysis of censored microbiological contamination data for use in risk assessment and mitigation. Food Microbiol. 2011, 28, 712–719. [Google Scholar] [CrossRef] [PubMed]
- Xie, M.; Singh, K. Confidence Distribution, the Frequentist Distribution Estimator of a Parameter: A Review. Int. Stat. Rev. 2013, 81, 3–39. [Google Scholar] [CrossRef]
- Seidenfeld, T. RA Fisher’s Fiducial Argument and Bayes’ Theorem. Stat. Sci. 1992, 7, 358–368. [Google Scholar] [CrossRef]
- Ranta, J.; Lindqvist, H.I.; Tuominen, P.; Nauta, M. A Bayesian approach to the evaluation of risk-based microbiological criteria for Campylobacter in broiler meat. Ann. Appl. Stat. 2015, 9, 1415–1432. [Google Scholar] [CrossRef]
- Kansallinen FINRISKI 2012-Terveystutkimus. Osa2: Tutkimuksen Taulukkoliite. Raportti 22/2013. THL. Available online: https://www.julkari.fi/handle/10024/114942 (accessed on 22 September 2021).
- Straver, J.M.; Janssen, A.F.W.; Linnemann, A.R.; Van Boekel, M.A.J.S.; Beumer, R.R.; Zwietering, M.H. Number of Salmonella on Chicken Breast Filet at Retail Level and Its Implications for Public Health Risk. J. Food Prot. 2007, 70, 2045–2055. [Google Scholar] [CrossRef]
- Busani, L.; Cigliano, A.; Taioli, E.; Caligiuri, V.; Chiavacci, L.; Di Bella, C.; Battisti, A.; Duranti, A.; Gianfranceschi, M.; Nardella, M.C.; et al. Prevalence of Salmonella enterica and Listeria monocytogenes Contamination in Foods of Animal Origin in Italy. J. Food Prot. 2005, 68, 1729–1733. [Google Scholar] [CrossRef] [Green Version]
- Risk Assessment Microbiology Section. Food Standards Australia New Zealand. Microbiological Risk Assessment of Raw Cow Milk. 2009. Available online: https://www.foodstandards.gov.au/Pages/default.aspx (accessed on 22 September 2021).
- Seo, K.H.; Valentin-Bon, I.E.; Brackett, R.E. Detection and Enumeration of Salmonella Enteritidis in Homemade Ice Cream Associated with an Outbreak: Comparison of Conventional and Real-Time PCR Methods. J. Food Prot. 2006, 69, 639–643. [Google Scholar] [CrossRef]
- EFSA and ECDC (European Food Safety Authority and European Centre for Disease Prevention and Control). The European Union summary report on trends and sources of zoonoses, zoonotic agents and food-borne outbreaks in 2017. EFSA J. 2018, 16, e5500. [Google Scholar]
- Humphrey, T.J.; Whitehead, A.; Gawler, A.H.L.; Henley, A.; Rowe, B. Numbers of Salmonella enteritidis in the contents of naturally contaminated hens’ eggs. Epidemiol. Infect 1991, 106, 489–496. [Google Scholar] [CrossRef] [Green Version]
- Asai, Y.; Kaneko, M.; Ohtsuka, K.; Morita, Y.; Kaneko, S.; Noda, H.; Furukawa, I.; Takatori, K.; Hara-Kudo, Y. Salmonella Prevalence in Seafood Imported into Japan. J. Food Prot. 2008, 71, 1460–1464. [Google Scholar] [CrossRef]
- Pielaat, A.; Wijnands, L.M.; Fitz-James, I.; van Leusden, F.M. Survey Analysis of Microbial Contamination of Fresh Produce and Ready-to-Eat Salads, and the Associated Risk to Consumers in The Netherlands. 2008. Available online: https://www.rivm.nl/bibliotheek/rapporten/330371002.html (accessed on 22 September 2021).
- Mikkelä, A.; Ranta, J.; González, M.; Hakkinen, M.; Tuominen, P. Campylobacter QMRA: A Bayesian Estimation of Prevalence and Concentration in Retail Foods Under Clustering and Heavy Censoring. Risk Anal. 2016, 36, 2065–2080. [Google Scholar] [CrossRef] [PubMed]
- Humphrey, T.J.; Beckett, P. Campylobacterjejuni in dairy cows and raw milk. Epidemiol. Infect. 1987, 98, 263–269. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Messelhäusser, U.; Thärigen, D.; Elmer-Englhard, D.; Bauer, H.; Schreiner, H.; Höller, C. Occurrence of Thermotolerant Campylobacter spp. on Eggshells: A Missing Link for Food-Borne Infections. Appl. Environ. Microbiol. 2011, 77, 3896–3897. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sato, M.; Sashihara, N. Occurrence of Campylobacter in Commercially Broken Liquid Egg in Japan. J. Food Prot. 2010, 73, 412–417. [Google Scholar] [CrossRef]
- Novotny, L.; Dvorska, L.; Lorencova, A.; Beran, V.; Pavlik, I. Fish: A potential source of bacterial pathogens for human beings. Vet. Med. Czech 2004, 49, 343–358. [Google Scholar] [CrossRef]
- Reinhard, R.G.; McAdam, T.J.; Flick, G.J.; Croonenberghs, R.E.; Wittman, R.F.; Diallo, A.A.; Fernandes, C. Analysis of Campylobacter jejuni, Campylobacter coli, Salmonella, Klebsiella pneumaniae, and Escherichia coli 0157:H7 in Fresh Hand-Picked Blue Crab (Callinectes sapidus) Meat. J. Food Prot. 1996, 59, 803–807. [Google Scholar] [CrossRef]
- European Food Safety Authority. Lead dietary exposure in the European population. EFSA J. 2012, 10, 2831. [Google Scholar] [CrossRef]
- European Food Safety Authority. Cadmium dietary exposure in the European population. EFSA J. 2012, 10, 2551. [Google Scholar] [CrossRef]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ranta, J.; Mikkelä, A.; Suomi, J.; Tuominen, P. BIKE: Dietary Exposure Model for Foodborne Microbiological and Chemical Hazards. Foods 2021, 10, 2520. https://doi.org/10.3390/foods10112520
Ranta J, Mikkelä A, Suomi J, Tuominen P. BIKE: Dietary Exposure Model for Foodborne Microbiological and Chemical Hazards. Foods. 2021; 10(11):2520. https://doi.org/10.3390/foods10112520
Chicago/Turabian StyleRanta, Jukka, Antti Mikkelä, Johanna Suomi, and Pirkko Tuominen. 2021. "BIKE: Dietary Exposure Model for Foodborne Microbiological and Chemical Hazards" Foods 10, no. 11: 2520. https://doi.org/10.3390/foods10112520
APA StyleRanta, J., Mikkelä, A., Suomi, J., & Tuominen, P. (2021). BIKE: Dietary Exposure Model for Foodborne Microbiological and Chemical Hazards. Foods, 10(11), 2520. https://doi.org/10.3390/foods10112520