The Genetic Architecture of Milling Quality in Spring Oat Lines of the Collaborative Oat Research Enterprise
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Materials and Molecular Markers
2.2. Phenotyping
2.3. Statistical Analyses
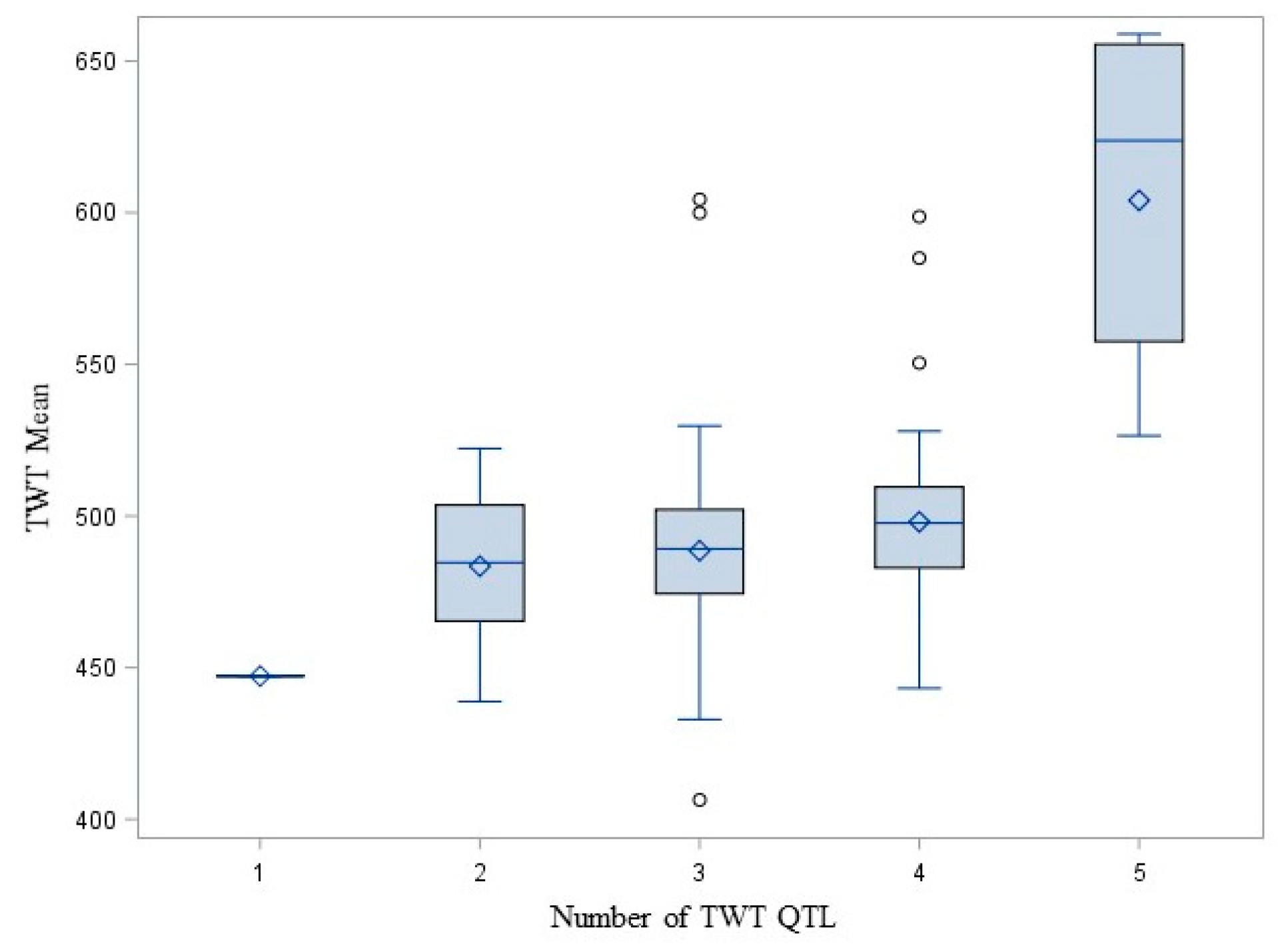
3. Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Thies, F.; Masson, L.F.; Boffetta, P.; Kris-Etherton, P. Oats and CVD risk markers: A systematic literature review. Br. J. Nutr. 2014, 112 (Suppl. 2), S19–S30. [Google Scholar] [CrossRef] [Green Version]
- Roskens, B.; (North American Millers Association, Washington, DC, USA). Personal Communication, 13 May 2019.
- Burnette, D.; Lenz, M.; Sisson, P.F.; Sutherland, S.; Weaver, S.H. Marketing, Processing, and Uses of Oat for Food. In Oat Science and Technology; Marshall, H.G., Sorrels, M.E., Eds.; American Society of Agronomy, Crop Science Society of America: Madison, WI, USA, 1992. [Google Scholar]
- Decker, E.A.; Rose, D.J.; Stewart, D.A. Processing of oats and the impact of processing operations on nutrition and health benefits. Br. J. Nutr. 2014, 112, S58–S64. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Doehlert, D.C.; McMullen, M.S.; Hammond, J.J. Genotype and environmental effects on grain yield and quality of oat grown in North Dakota. Crop Sci. 2001, 41, 1066–1072. [Google Scholar] [CrossRef]
- Doehlert, D.C.; Ohm, J.-B.; McMullen, M.S.; Riveland, N.R. Theoretical and empirical relationships between oat test weight and groat proportion. Cereal Chem. 2009, 86, 239–246. [Google Scholar] [CrossRef] [Green Version]
- Herrmann, M.H.; Yu, J.; Beuch, S.; Weber, W.E. Quantitative trait loci for quality and agronomic traits in two advanced backcross populations in oat (Avena sativa L.). Plant Breed. 2014, 133, 588–601. [Google Scholar] [CrossRef]
- Tanhuanpää, P.; Manninen, O.; Beattie, A.; Eckstein, P.; Scoles, G.; Rossnagel, B.; Kiviharju, E. An updated doubled haploid oat linkage map and QTL mapping of agronomic and grain quality traits from Canadian field trials. Genome 2012, 55, 289–301. [Google Scholar] [CrossRef]
- Pixley, K.V.; Frey, K.J. Inheritance of test weight and its relationship with grain yield of oat. Crop Sci. 1991, 31, 36–40. [Google Scholar] [CrossRef]
- DeKoeyer, D.L.; Tinker, N.A.; Wight, C.P.; Deyl, J.; Burrows, V.D.; O’Donoughue, L.S.; Lybaert, A.; Molnar, S.J.; Armstrong, K.C.; Fedak, G.; et al. A molecular linkage map with associated QTLs from a hulless x covered spring oat population. Theor. Appl. Genet. 2004, 108, 1285–1289. [Google Scholar] [CrossRef]
- Peterson, D.M.; Wesenberg, D.M.; Burrup, D.E.; Erickson, C.A. Relationships among agronomic traits and grain composition in oat genotypes grown in different environments. Crop Sci. 2005, 45, 1249–1255. [Google Scholar] [CrossRef] [Green Version]
- Yan, W.; Mitchell Fetch, J.; Frégeau-Reid, J.; Rossnagel, B.; Ames, N. Genotype X location interaction patterns and testing strategies for oat in the Canadian prairies. Crop Sci. 2011, 51, 1903–1914. [Google Scholar] [CrossRef]
- Doehlert, D.C.; McMullen, M.S.; Riveland, N.R. Sources of variation in oat kernel size. Cereal Chem. 2002, 79, 528–534. [Google Scholar] [CrossRef]
- Yan, W.; Molnar, S.J.; Fregeau-Reid, J.; McElroy, A.; Tinker, N.A. Associations among oat traits and their responses to the environment. J. Crop Improv. 2007, 20, 1–29. [Google Scholar] [CrossRef]
- Doehlert, D.C.; McMullen, D.C. Genotypic and environmental effects on oat milling characteristics and groat hardness. Cereal Chem. 2000, 77, 148–154. [Google Scholar] [CrossRef]
- Siripoonwiwat, W.; O’Donoughue, L.S.; Wesenberg, D.; Hoffman, D.L.; Barbosa-Neto, J.F.; Sorrells, M.E. Chromosomal regions associated with quantitative traits in oat. J. Agric. Genom. 1996, 2, 1–13. [Google Scholar]
- Groh, S.; Kianian, S.F.; Phillips, R.L.; Rines, H.W.; Stuthman, D.D.; Wesenberg, D.M.; Fulcher, R.G. Analysis of factors influencing milling yield and their association to other traits by QTL analysis in two hexaploidy oat populations. Theor. Appl. Genet. 2001, 103, 9–18. [Google Scholar] [CrossRef]
- Blake, V.C.; Woodhouse, M.R.; Lazo, G.R.; Odell, S.G.; Wight, C.P.; Turner, N.A.; Wang, Y.; Gu, Y.Q.; Birkett, C.L.; Jannink, J.-L.; et al. GrainGenes: Centralized small grain resources and digital platform for geneticists and breeders. Database 2019. [Google Scholar] [CrossRef]
- Bekele, W.A.; Wight, C.P.; Chao, S.; Howarth, C.J.; Tinker, N.A. Haplotype-based genotyping-by-sequencing in oat genome research. Plant Biotechnol. J. 2018, 16, 1452–1463. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zimmer, C.M.; McNish, I.G.; Esvelt Klos, K.; Eickholt, D.P.; Arruda, K.M.A.; Pacheco, M.T.; Smith, K.P.; Federizzi, L.C. Genome-wide association mapping for kernel shape and its association with beta-glucan content in oats. Crop Sci. 2021, 1–14. [Google Scholar] [CrossRef]
- Esvelt Klos, K.; Huang, Y.-F.; Bekele, W.A.; Obert, D.E.; Babiker, E.; Beattie, A.D.; Bjornstad, Å.; Bonman, J.M.; Carson, M.L.; Chao, S.; et al. Population genomics related to adaptation in elite oat germplasm. Plant Genome 2016. [Google Scholar] [CrossRef] [Green Version]
- Chaffin, A.S.; Huang, Y.-F.; Smith, S.; Bekele, W.A.; Babiker, E.; Gnanesh, B.N.; Foresman, B.J.; Blanchard, S.G.; Jay, J.J.; Reid, R.W.; et al. A consensus map in cultivated hexaploid oat reveals conserved grass synteny with substantial sub-genome rearrangement. Plant Genome 2016, 9. [Google Scholar] [CrossRef] [Green Version]
- Bradbury, P.J.; Zhang, Z.; Kroon, D.E.; Casstevens, T.M.; Ramdoss, Y.; Buckler, E.S. TASSEL: Software for association mapping of complex traits in diverse samples. Bioinformatics 2007, 23, 2633–2635. [Google Scholar] [CrossRef]
- Korte, A.; Farlow, A. The advantages and limitations of trait analysis with GWAS: A review. Plant Methods 2013, 9, 29. [Google Scholar] [CrossRef] [Green Version]
- Ougham, H.; Latipova, G.; Valentine, J. Morphological and biochemical characterization of spikelet development in naked oats (Avena sativa). New Phytol. 1996, 134, 5–12. [Google Scholar] [CrossRef]
- Esvelt Klos, K.; Yimer, B.A.; Babiker, E.M.; Beattie, A.D.; Bonman, J.M.; Carson, M.L.; Chong, J.; Harrison, S.A.; Ibrahim, A.M.H.; Kolb, F.L.; et al. Genome-wide association mapping of crown rust resistance in oat elite germplasm. Plant Genome 2017. [Google Scholar] [CrossRef]
- Kianian, S.F.; Phillips, R.L.; Rines, H.W.; Fulcher, R.G.; Webster, F.H.; Stuthman, D.D. Identification of quantitative trail loci influencing β-glucan content in oat (Avena sativa, 2n=6x=42). Theor. Appl. Genet. 2000, 101, 1039–1048. [Google Scholar] [CrossRef]
- Fogarty, M.; Smith, S.; Sheridan, J.; Hu, G.; Islamovic, E.; Reid, R.; Jackson, E.; Maughan, J.; Ames, N.; Jellen, R.; et al. Identification of mixed linkage β-glucan quantitative trait loci and evaluation of AsCslF6 homoeologs in hexaploidy oat. Crop Sci. 2020, 60, 914–933. [Google Scholar] [CrossRef]
- Via, S.; Gomulkiewicz, R.; de Jong, G.; Scheiner, S.M.; Schlichting, C.D.; van Tienderen, P.H. Adaptive phenotypic plasticity: Consensus and controversy. Trends Ecol. Evol. 1995, 10, 212–217. [Google Scholar] [CrossRef]
- Kraakman, A.T.W.; Niks, R.E.; Van den Berg, P.M.M.; Stam, P.; Van Eeuwijk, F.A. Linkage disequilibrium mapping of yield and yield stability in modern spring barley cultivars. Genetics 2004, 168, 435–446. [Google Scholar] [CrossRef] [Green Version]
- Lacaze, X.; Hayes, P.M.; Korel, A. Genetics of phenotypic plasticity: QTL analysis in barley, Hordeum vulgare. Heredity 2009, 102, 163–173. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Peltonen-Sainio, P.; Moore, K.; Pehu, E. Phenotypic stability of oats measured with different stability analyses. J. Agric. Sci. 1993, 121, 13–19. [Google Scholar] [CrossRef]
- Wang, Y.; Mette, M.F.; Miedaner, T.; Wilde, P.; Reif, J.C.; Zhao, Y. First insights into the genotype-phenotype map of phenotypic stability in rye. J. Exp. Botany 2015, 66, 3275–3284. [Google Scholar] [CrossRef] [Green Version]

| Br% | GC | Plumps | Thins | TKW | TWT | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Location Year | N | N | N | N | N | N | ||||||
| Ad10 | NA | NA | NA | NA | NA | NA | 22.25 (15.61) | 498 | NA | NA | 533.70 (15.62) | 497 |
| Ad17 | 6.73 (5.19) | 470 | 71.37 (3.42) | 468 | 71.61 (20.27) | 487 | 9.57 (12.10) | 487 | 33.07 (4.30) | 493 | 474 (33.73) | 491 |
| Ay10 | NA | NA | NA | NA | NA | NA | NA | NA | 30.74 (4.49) | 499 | NA | NA |
| Fa10 | NA | NA | NA | NA | NA | NA | 8.25 (10.60) | 499 | NA | NA | 467.37 (32.27) | 499 |
| Fa11 | 4.99 (3.22) | 486 | 73.41 (3.52) | 486 | NA | NA | 14.87 (14.14) | 499 | NA | NA | 447.98 (51.93) | 491 |
| It10 | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | 466.23 (35.52) | 464 |
| La10 | NA | NA | NA | NA | NA | NA | 18.08 (15.67) | 500 | 37.24 (4.18) | 500 | 570.25 (31.95) | 500 |
| La11 | 2.72 (2.71) | 490 | 75.95 (2.54) | 490 | NA | NA | NA | NA | NA | NA | NA | NA |
| Ot10 | NA | NA | NA | NA | NA | NA | 8.85 (8.45) | 398 | 39.15 (6.15019) | 488 | 501.33 (44.76) | 465 |
| Sa10 | NA | NA | 68.34 (3.48) | 483 | 72.12 (21.48) | 496 | 10.64 (13.23) | 496 | 32.97 (5.03) | 496 | 496.38 (44.84) | 496 |
| Sa11 | NA | NA | 72.62 (4.35) | 501 | 63.32 (24.58) | 501 | 10.46 (14.45) | 501 | 32.35 (4.37) | 501 | 513.03 (31.09) | 498 |
| Te10 | 18.48 (10.48) | 387 | 73.15 (2.90) | 387 | 72.14 (15.52) | 395 | 10.27 (10.66) | 395 | NA | NA | 442.87 (23.42) | 394 |
| Te11 | 4.98 (2.64) | 486 | 75.57 (2.66) | 486 | 61.57 (13.81) | 490 | 15.28 (13.81) | 496 | NA | NA | 451.55 (23.30) | 489 |
| Br% Mean | Br% var | GC Mean | GC Var | Plumps Mean | Plumps Var | Thins Mean | Thins Var | TKW Mean | TKW Var | TWT Mean | TWT Var | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Br% mean | −0.10 | 0.07 | 0.08 | 0.08 | −0.03 | −0.03 | 0.12 | 0.10 | 0.12 | −0.08 | −0.05 | |
| Br% var | ns | 0.03 | 0.06 | 0.11 | −0.00 | −0.04 | 0.03 | 0.07 | 0.07 | −0.09 | −0.07 | |
| GC mean | ns | ns | −0.36 | −0.16 | −0.06 | 0.12 | 0.04 | −0.02 | 0.12 | 0.71 | 0.06 | |
| GC var | ns | ns | <0.0001 | −0.19 | 0.11 | 0.18 | −0.10 | −0.17 | 0.07 | −0.40 | 0.08 | |
| Plumps mean | ns | ns | 0.0004 | <0.0001 | −0.35 | −0.93 | 0.40 | 0.76 | −0.21 | 0.01 | −0.10 | |
| Plumps var | ns | ns | ns | ns | <0.0001 | 0.33 | 0.07 | −0.24 | 0.10 | −0.04 | 0.07 | |
| Thins mean | ns | ns | 0.0078 | <0.0001 | <0.0001 | <0.0001 | −0.45 | −0.81 | 0.20 | −0.07 | 0.08 | |
| Thins var | ns | ns | ns | ns | <0.0001 | ns | <0.0001 | 0.35 | −0.04 | 0.11 | −0.00 | |
| TKW mean | ns | ns | ns | 0.0002 | <0.0001 | <0.0001 | <0.0001 | <0.0001 | 0.05 | 0.09 | −0.06 | |
| TKW var | ns | ns | 0.0053 | ns | <0.0001 | ns | <0.0001 | ns | ns | −0.00 | 0.02 | |
| TWT mean | ns | ns | <0.0001 | <0.0001 | ns | ns | ns | ns | ns | ns | −0.12 | |
| TWT var | ns | ns | ns | ns | ns | ns | ns | ns | ns | ns | 0.0060 |
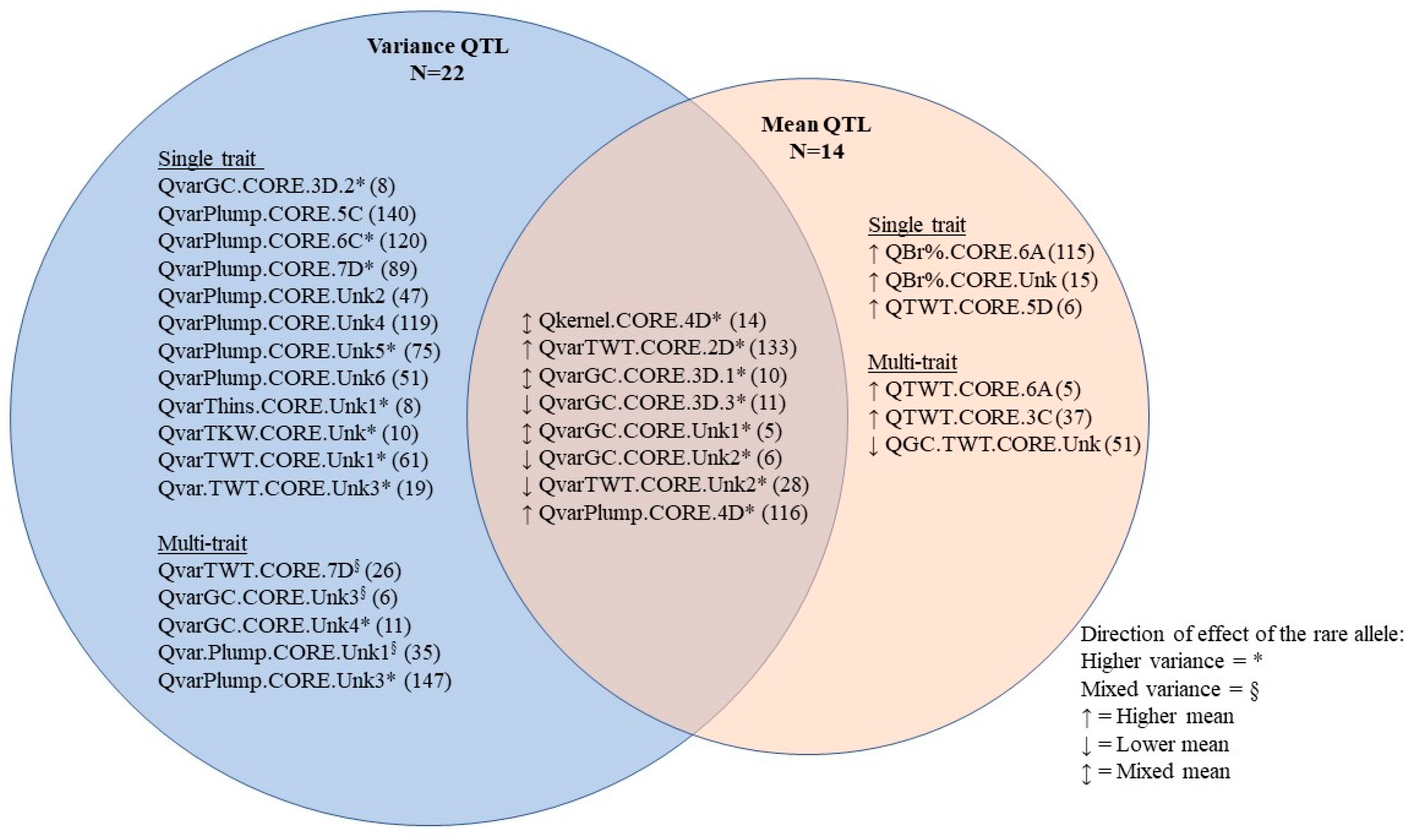
| QTL | Chr | cM Range a | Traits | Representative Marker b | p-Value c |
|---|---|---|---|---|---|
| QTWT.CORE.6A | 6A | 81.7–81.9 | TWT | Avgbs_309316.1.34 | 2.09 × 10−9 |
| QBr%.CORE.6A | 6A | 135.5 | Br% | Avgbs2_58834.1.20 | 1.15 × 10−7 |
| QTWT.CORE.3C | 3C | 70.2 | TWT | Avgbs_cluster_24174.1.59 | 6.06 × 10−8 |
| QvarPlumps.CORE.5C | 5C | 25–28.8 | Plumps variance | Avbgs2_146190.1.36 | 1.13 × 10−7 |
| QvarPlumps.CORE.6C | 6C | 70.5 | Plumps variance | Avgbs_6K_109589.1.51 | 1.73 × 10−10 |
| QvarTWT.CORE.2D | 2D | 151.9 | TWT variance | Avgbs_cluster_27091.1.40 | 3.63 × 10−12 |
| QvarGC.CORE.3D.1 | 3D | 45.4–47 | GC variance | Avgbs_cluster_22727.1.27 | 6.47 × 10−8 |
| QvarGC.CORE.3D.2 | 3D | 45.4 | GC variance | Avgbs_311274.1.63 | 1.24 × 10−7 |
| QvarGC.CORE.3D.3 | 3D | 49.3–49.8 | GC, GC variance | Avgbs_cluster_44297.1.21 | 8.09 × 10−7 |
| QvarPlumps.CORE.4D | 4D | 200.9 | Plumps variance | Avgbs_cluster_35424.1.22 | 6.00 × 10−12 |
| Qkernel.CORE.4D | 4D | 195.7–212.1 | GC, Plump, Thins, TKW, TWT, TWT variance | Avbgs_cluster_7805.1.9 | 1.13 × 10−51 |
| QTWT.CORE.5D | 5D | 26.4 | TWT | Avgbs_405598.1.29 | 3.81 × 10−7 |
| QvarPlumps.CORE.7D | 7D | 85.2 | Plumps variance | Avgbs2_12115.1.19 | 2.85 × 10−8 |
| QvarTWT.CORE.7D | 7D | 85.2–88.7 | TWT variance | Avgbs_cluster_9292.1.49 | 1.77 × 10−7 |
| QTL | # SNPs | # LG a | Trait | Representative Marker b | p-Value c |
|---|---|---|---|---|---|
| QvarTWT.CORE.Unk1 | 56 | 14 | TWT variance | Avgbs2_111600.1.12 | 4.47 × 10−17 |
| QvarTWT.CORE.Unk2 | 66 | 13 | TWT variance | Avgbs2_158635.1.16 | 2.00 × 10−16 |
| QvarPlumps.CORE.Unk1 | 79 | 17 | Plump variance | Avgbs_16668.1.22 | 8.02 × 10−16 |
| QvarPlumps.CORE.Unk2 | 9 | 4 | Plump variance | Avgbs_7838.1.46 | 1.19 × 10−13 |
| QGC.TWT.CORE.Unk | 1 | 0 | GC, TWT | Avgbs2_92005.1.47 | 1.67 × 10−13, 1.94 × 10−8 |
| QvarTWT.CORE.Unk3 | 4 | 4 | TWT variance | Avgbs2_88377.1.31 | 8.65 × 10−13 |
| QvarGC.CORE.Unk1 | 54 | 8 | GC variance, TWT | Avgbs2_134662.1.14 | 7.60 × 10−8, 1.60 × 10−8 |
| QvarThins.CORE.Unk1 | 15 | 5 | Thins variance | Avgbs2_147274.1.11 | 2.54 × 10−11 |
| QvarPlumps.CORE.Unk3 | 2 | 2 | Plumps variance | Avgbs_cluster_20790.1.15 | 2.76 × 10−11 |
| QvarPlumps.CORE.Unk4 | 13 | 6 | Plumps variance | Avgbs2_31344.1.24 | 6.37 × 10−11 |
| QvarPlumps.CORE.Unk5 | 19 | 9 | Plumps variance | Avgbs_cluster_18028.1.19 | 2.11 × 10−10 |
| QvarGC.CORE.Unk2 | 7 | 0 | GC variance | Avgbs2_198262.1.30 | 2.04 × 10−9 |
| QvarTKW.CORE.Unk | 2 | 0 | TKW variance | Avgbs2_33384.1.6 | 7.84 × 10−9 |
| QvarGC.CORE.Unk3 | 12 | 7 | GC variance | Avgbs2_198688.1.12 | 1.83 × 10−8 |
| QvarGC.CORE.Unk4 | 2 | 0 | GC variance | Avgbs_45323.1.23 | 2.37 × 10−8 |
| QvarPlumps.CORE.Unk6 | 4 | 3 | Plumps variance | Avgbs2_23356.1.24 | 2.22 × 10−7 |
| QBr%.CORE.Unk | 1 | 0 | Br% | Avgbs2_60654.1.6 | 2.92 × 10−7 |
| QTL | # SNPs | # LG a | Chr (cM) | Trait (Location Year) | Representative Marker b | p-Value c |
|---|---|---|---|---|---|---|
| QTWT.Saska11.Unk1 | 3 | 3 | NA | TWT (Sa11) | Avgbs_38660.1.16 | 9.89 × 10−17 |
| QTWT.Farge11.Unk1 | 9 | 3 | NA | TWT (Fa11) | Avgbs_581244 | 4.48 × 10−10 |
| QTWT.Ottaw10.Unk1 | 1 | 0 | NA | TWT (Ot10) | Avgbs_413385.1.50 | 7.56 × 10−11 |
| QTWT.Fargo11.2D.1 | 3 | 1 | 2D (142.3) | TWT (Fa11) | Avgbs_cluster_39595.1.45 | 1.63 × 10−9 |
| QTWT.Fargo11.7D | 1 | 1 | 7D (87.3) | TWT (Fa11) | Avgbs_cluster_1682.1.6 | 2.67 × 10−9 |
| QGC.Lacom11.Unk1 | 241 | 16 | NA | GC (La11) | Avgbs_cluster_17564.1.10 | 1.05 × 10−8 |
| QTWT.Fargo11.Unk2 | 1 | 0 | NA | TWT (Fa11) | Avgbs_288442 | 1.83 × 10−8 |
| QTWT.Teton11.7A | 1 | 1 | 7A (36.3) | TWT (Te11) | Avgbs_99656.1.17 | 2.24 × 10−8 |
| QGC.Lacom11.Unk2 | 1 | 1 | NA | GC (La11) | Avgbs_392008.1.17 | 3.05 × 10−8 |
| QTWT.Saska11.Unk2 | 1 | 0 | NA | TWT (Sa11) | Avgbs2_104381.1.22 | 3.33 × 10−8 |
| QPlumps.Aberd17.2A | 1 | 1 | 2A (59.3) | Plumps (Ad17) | Avgbs2_190926.2.35 | 4.11 × 10−8 |
| QGC.Lacom11.3C | 5 | 1 | 3C (35.1) | GC (La11) | Avgbs2_40053.2.56 | 4.93 × 10−8 |
| QGC.Teton11.Unk | 2 | 2 | NA | GC (Te11) | Avgbs2_191851 | 7.16 × 10−8 |
| QTWT.Fargo11.Unk3 | 1 | 0 | NA | TWT (Fa11) | Avgbs_cluster_15112.1.10 | 8.56 × 10−8 |
| QPlumps.Saska10.7A | 1 | 1 | 7A (61.4) | Plumps (Sa10) | Avgbs2_181490.2.45 | 1.16 × 10−7 |
| QTWT.Teton10.Unk | 15 | 8 | NA | TWT (Te10) | Avgbs2_130697.1.9 | 1.20 × 10−7 |
| QTWT.Ottaw10.Unk2 | 4 | 2 | NA | TWT (Ot10) | Avgbs_34428 | 2.60 × 10−7 |
| QTKW.Abery10.2A | 1 | 1 | 2A (34) | TKW (Ay10) | Avgbs_cluster_7015.1.27 | 2.97 × 10−7 |
| QPlumps.Saska11.Unk | 3 | 2 | NA | Plumps (Sa11) | Avgbs_4871 | 3.07 × 10−7 |
| QThins.Saska10.Unk | 1 | 0 | NA | Thins (Sa10) | Avgbs_cluster_20577.1.16 | 6.25 × 10−7 |
| QBr%.Lacom11.4C | 2 | 1 | 4C (52.8) | Br% (La11) | Avgbs_cluster_14607.1.44 | 6.73 × 10−7 |
| QTWT.Ithac11.Unk | 1 | 0 | NA | TWT (It10) | Avgbs_75885.1.16 | 6.97 × 10−7 |
| QTWT.Saska11.Unk3 | 1 | 0 | NA | TWT (Sa11) | Avgbs_32105.1.60 | 7.20 × 10−7 |
| QTWT.Teton10.1D | 1 | 1 | 1D (118.9) | TWT (Te10) | Avgbs2_80976.1.6 | 7.48 × 10−7 |
| QPlumps.Teton10.3A | 2 | 1 | 3A (104.9) | Plumps (Te10) | Avgbs_cluster_3791.1.28 | 7.33 × 10−7 |
| QTWT.Fargo11.2D.2 | 1 | 1 | 2D (142) | TWT (Fa11) | Avgbs2_115576.1.14 | 9.08 × 10−7 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Esvelt Klos, K.; Yimer, B.A.; Howarth, C.J.; McMullen, M.S.; Sorrells, M.E.; Tinker, N.A.; Yan, W.; Beattie, A.D. The Genetic Architecture of Milling Quality in Spring Oat Lines of the Collaborative Oat Research Enterprise. Foods 2021, 10, 2479. https://doi.org/10.3390/foods10102479
Esvelt Klos K, Yimer BA, Howarth CJ, McMullen MS, Sorrells ME, Tinker NA, Yan W, Beattie AD. The Genetic Architecture of Milling Quality in Spring Oat Lines of the Collaborative Oat Research Enterprise. Foods. 2021; 10(10):2479. https://doi.org/10.3390/foods10102479
Chicago/Turabian StyleEsvelt Klos, Kathy, Belayneh A. Yimer, Catherine J. Howarth, Michael S. McMullen, Mark E. Sorrells, Nicholas A. Tinker, Weikai Yan, and Aaron D. Beattie. 2021. "The Genetic Architecture of Milling Quality in Spring Oat Lines of the Collaborative Oat Research Enterprise" Foods 10, no. 10: 2479. https://doi.org/10.3390/foods10102479
APA StyleEsvelt Klos, K., Yimer, B. A., Howarth, C. J., McMullen, M. S., Sorrells, M. E., Tinker, N. A., Yan, W., & Beattie, A. D. (2021). The Genetic Architecture of Milling Quality in Spring Oat Lines of the Collaborative Oat Research Enterprise. Foods, 10(10), 2479. https://doi.org/10.3390/foods10102479






