Component Identification of Phenolic Acids in Cell Suspension Cultures of Saussureainvolucrata and Its Mechanism of Anti-Hepatoma Revealed by TMT Quantitative Proteomics
Abstract
:1. Introduction
2. Materials and Methods
2.1. Cell Suspension Culture of S. involucrata
2.2. Extraction of the Phenolic Acids in Cell Suspension Cultures of S. involucrata (EPSI)
2.3. Component Identification by Liquid Chromatography-Mass Spectrometry (LC-MS)
2.4. Effect of EPSI on the Multiplication of HepG2 Cells In Vitro
2.5. Acute Toxic Test
2.6. Establishment of the Animal Model
2.7. Hematoxylin and Eosin (H&E) Staining
2.8. Enzyme Linked Immunosorbent Assay (ELISA) analysis
2.9. Tandem Mass Tag (TMT) Quantitative Proteomics
2.9.1. Total Protein Extraction
2.9.2. Protein Quality Test
2.9.3. TMT Labeling of Peptides
2.9.4. Separation of Fractions
2.9.5. LC-MS/MS Analysis
2.9.6. Data Analysis
Identification and Quantitation of Protein
Functional Analysis of Protein and DEP
2.10. Statistical Analysis
3. Results and Discussion
3.1. Composition of the Extract of the Phenolic Acids in Cell Suspension Cultures from S. involucrata (EPSI)
3.2. Growth Inhibition of EPSI in HepG2 Cells
3.3. Acute Toxicity Test
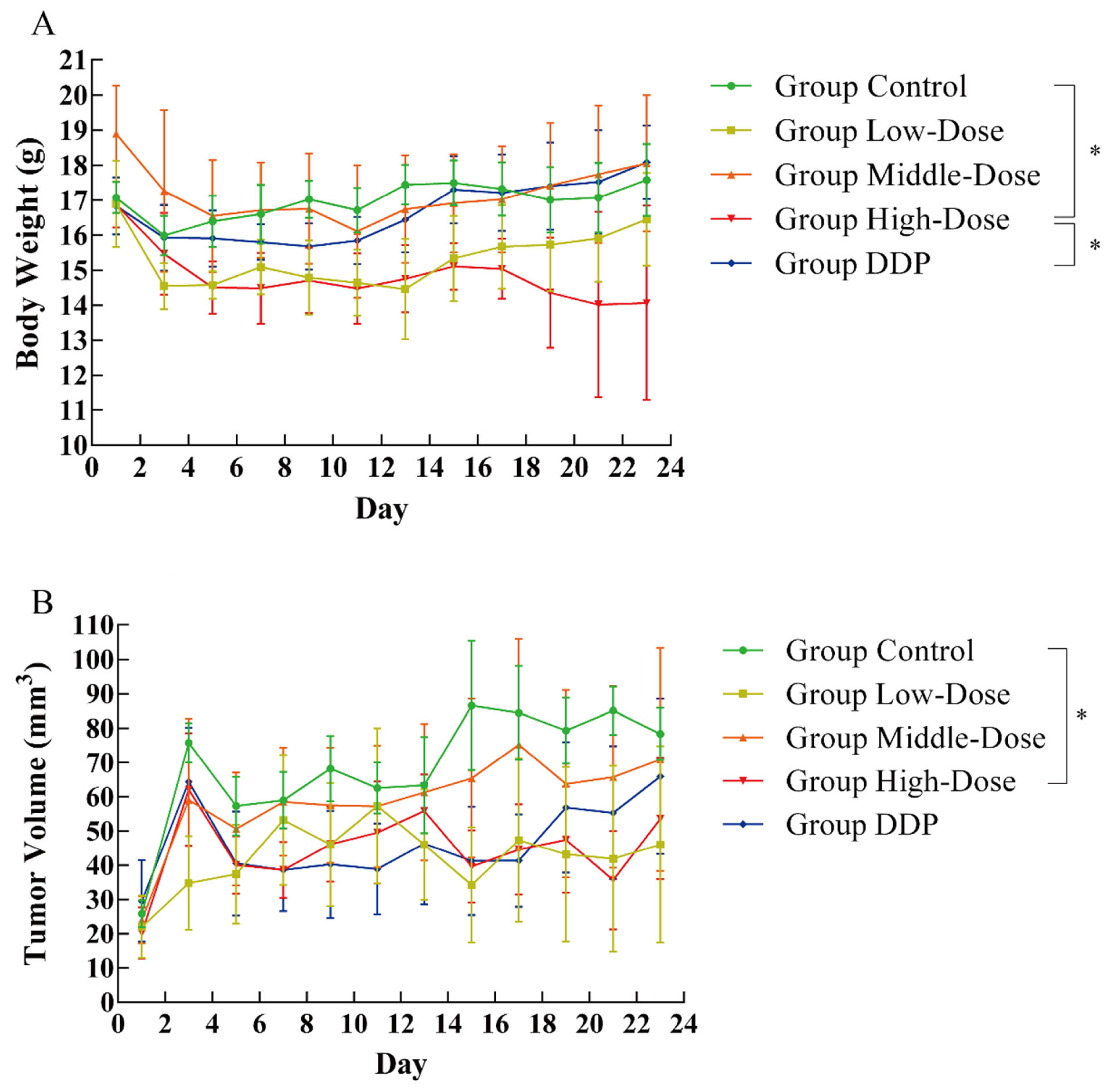
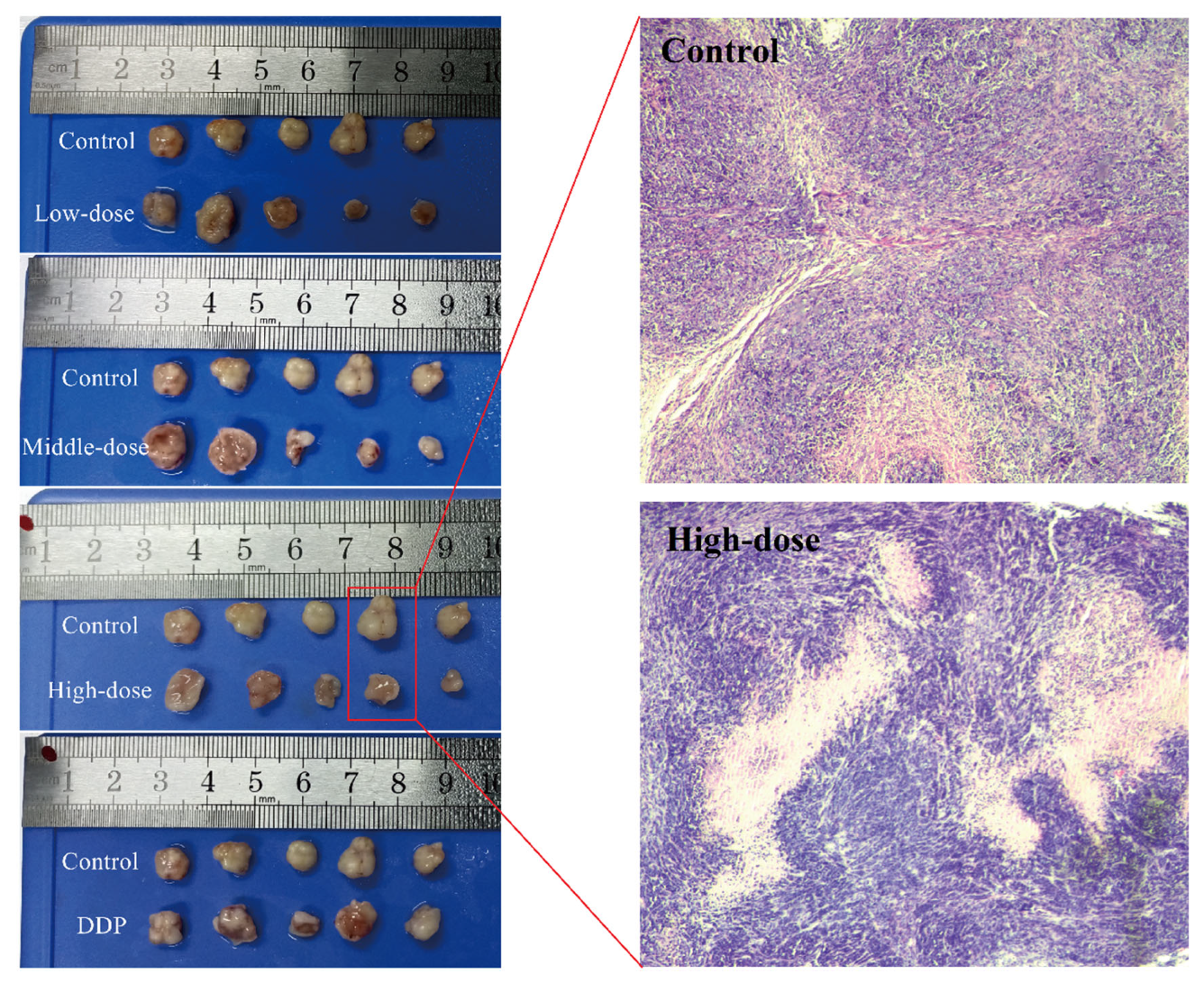
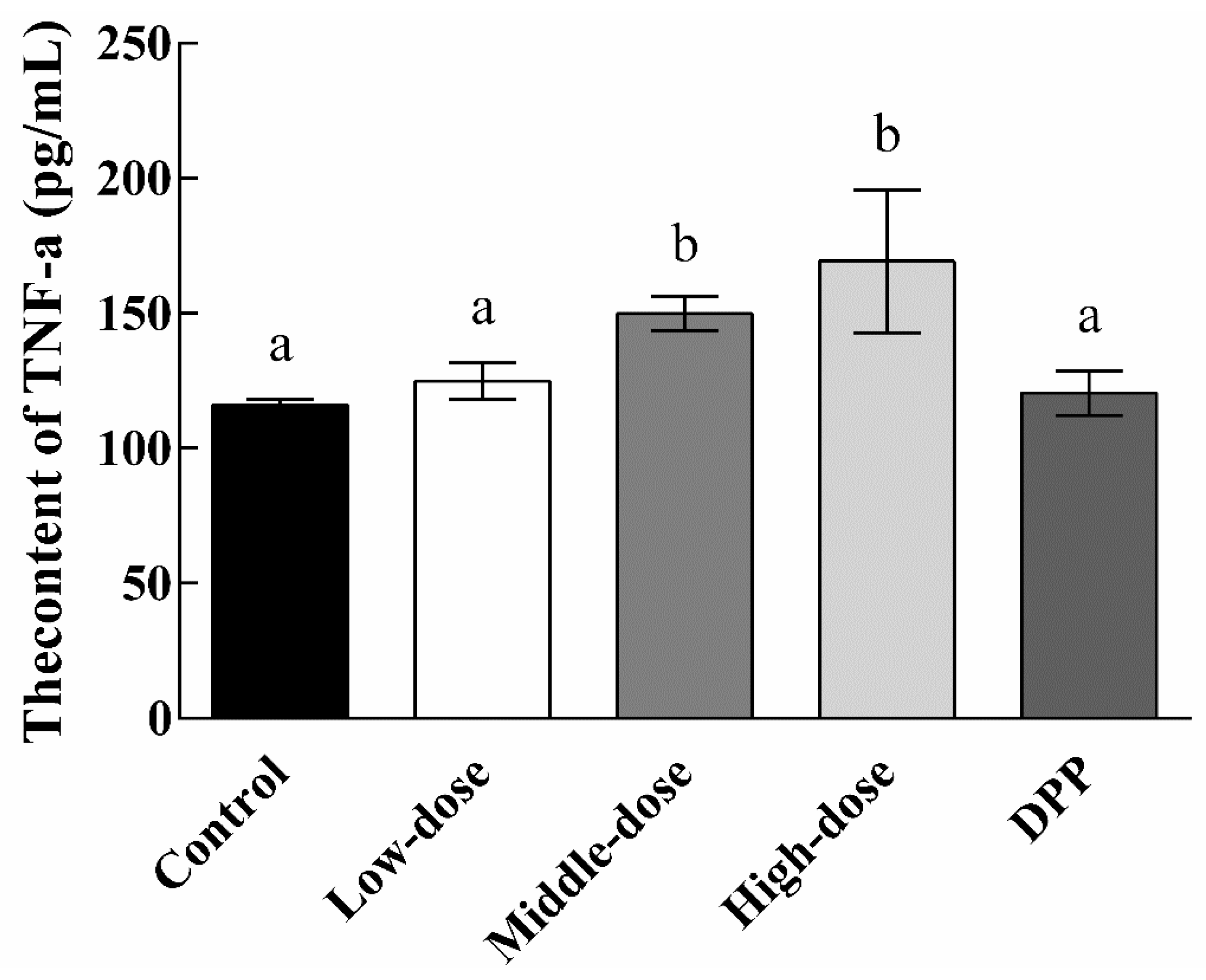
3.4. Effect of EPSI on Tumors in BALB/c Nude Mice
3.5. Protein Expression Differences Induced by EPSI
3.6. GO Enrichment of Differentially Quantified Proteins
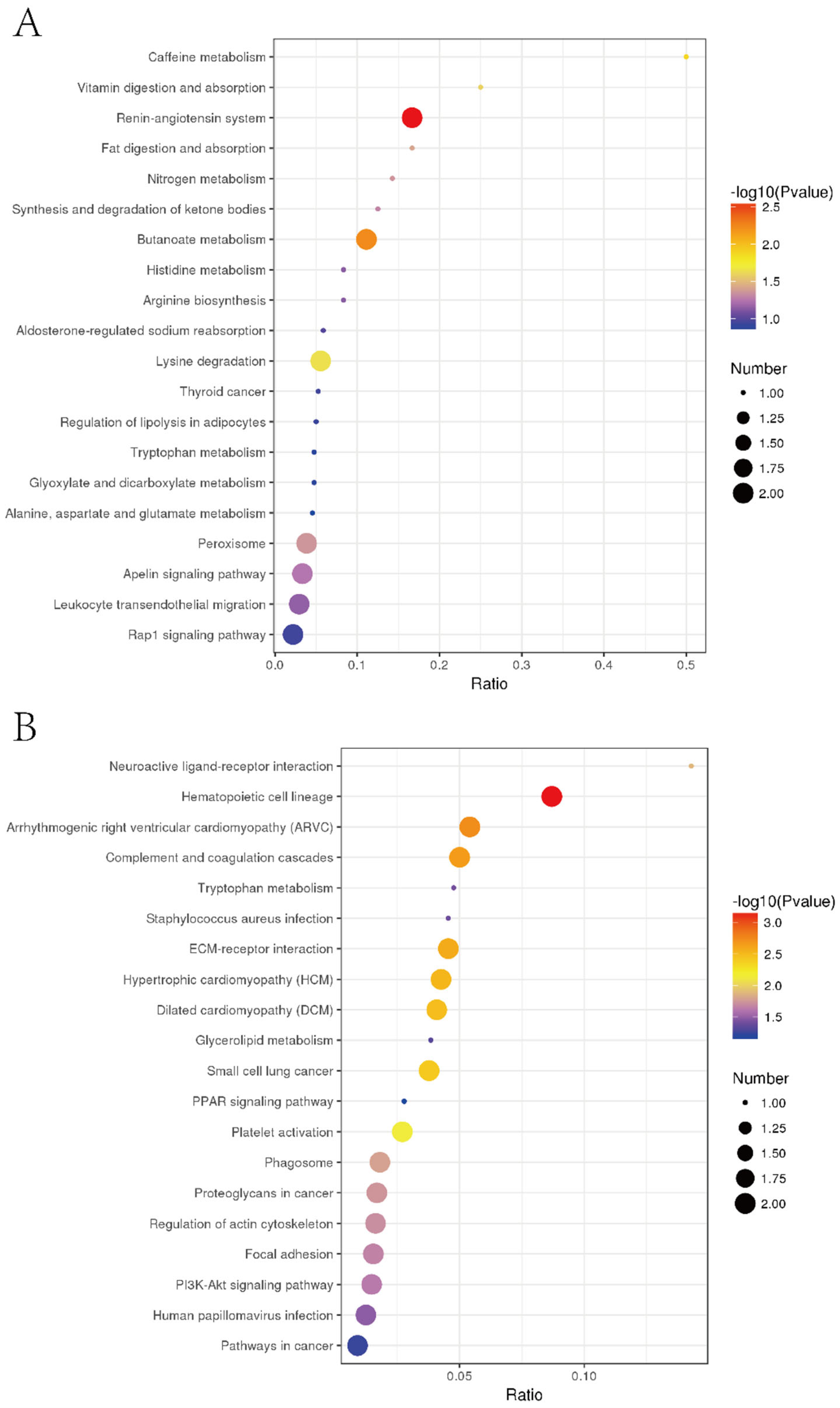
3.7. Specific Regulation Pathways for Inhibiting Liver Tumor Proliferation by EPSI
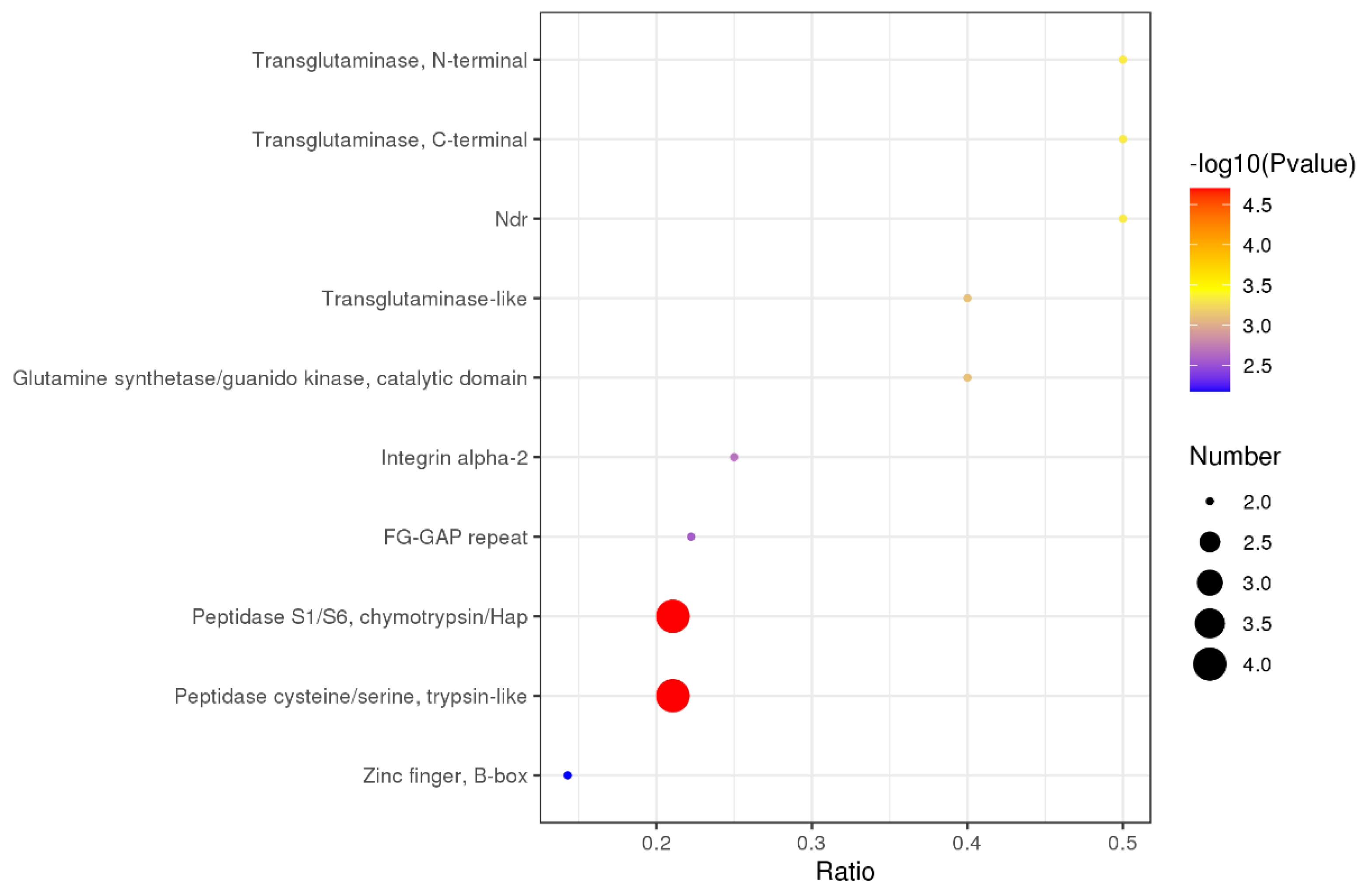
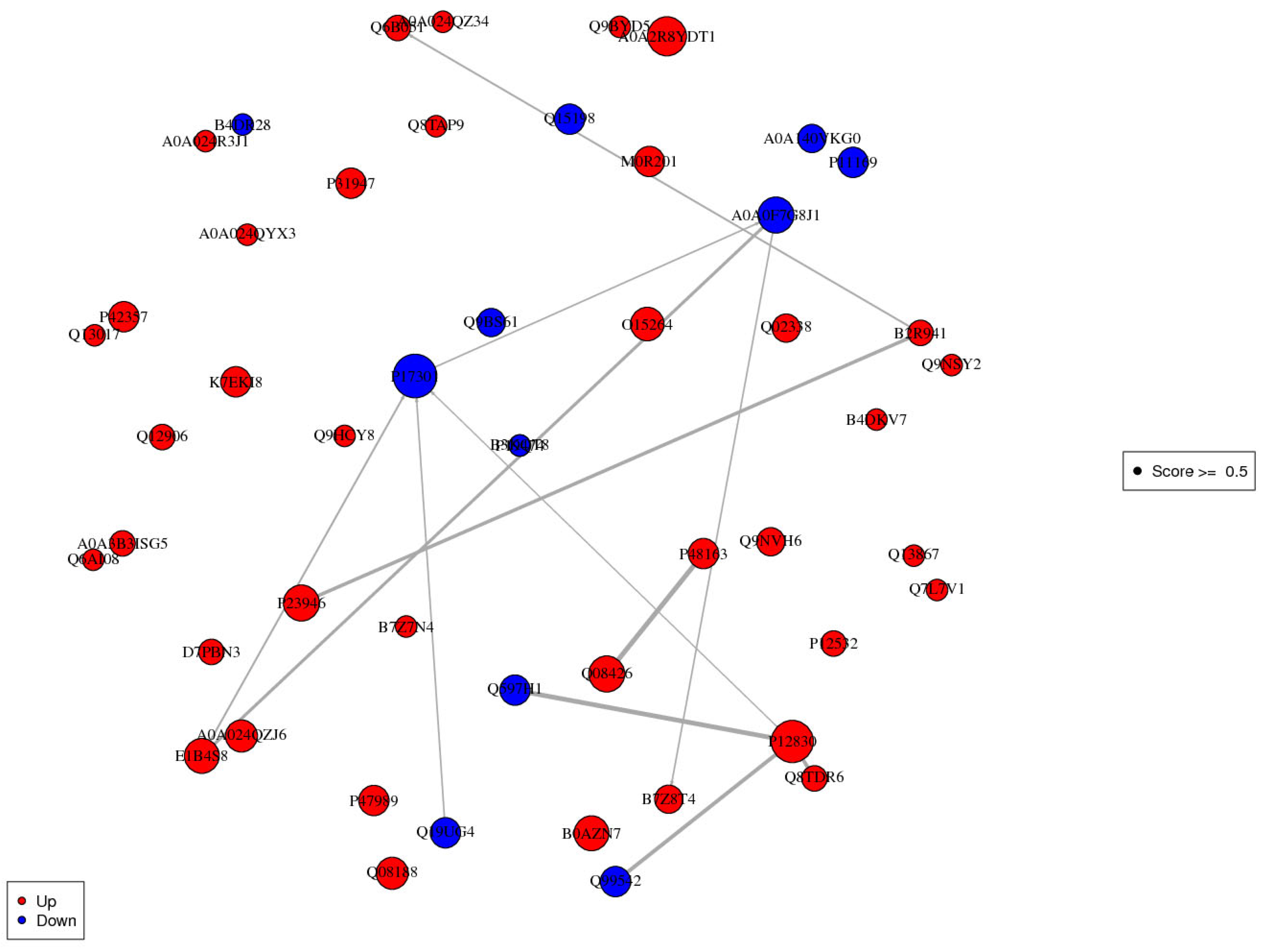
3.8. Interaction Analysis of Differentially Expressed Proteins
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Li, G.; Liu, F.; Zhao, R. Studies on pharmacological actions of Saussurea involucrata Kar et Kir ex Maxim (author’s transl). Yao Xue Xue Bao Acta Pharm. Sin. 1980, 15, 368–370. [Google Scholar]
- Chik, W.-I.; Zhu, L.; Fan, L.-L.; Yi, T.; Zhu, G.-Y.; Gou, X.-J.; Tang, Y.-N.; Xu, J.; Yeung, W.-P.; Zhao, Z.-Z.; et al. Saussurea involucrata: A review of the botany, phytochemistry and ethnopharmacology of a rare traditional herbal medicine. J. Ethnopharmacol. 2015, 172, 44–60. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, Y.; Wang, C.; Guo, S.; Yang, J.; Xiao, P. Three guaianolides from Saussurea involucrata and their contents determination by HPLC. J. Pharm. Biomed. Anal. 2007, 44, 288–292. [Google Scholar] [CrossRef] [PubMed]
- Chaudhary, N.; Aparoy, P. Deciphering the mechanism behind the varied binding activities of COXIBs through Molecular Dynamic Simulations, MM-PBSA binding energy calculations and per-residue energy decomposition studies. J. Biomol. Struct. Dyn. 2017, 35, 868–882. [Google Scholar] [CrossRef] [PubMed]
- Yu, L.; Zhong-Jian, J. Guaianolides from Saussurea involucrata. Phytochemistry 1989, 28, 3395–3397. [Google Scholar] [CrossRef]
- Seilgazy, M.; Li, J.; Aisa, H.A. Isolation of Steroidal Esters from Seeds of Saussurea involucrata. Chem. Nat. Compd. 2017, 53, 1196–1198. [Google Scholar] [CrossRef]
- Ma, H.-P.; Fan, P.-C.; Jing, L.-L.; Yao, J.; He, X.-R.; Yang, Y.; Chen, K.-M.; Jia, Z.-P. Anti-hypoxic activity at simulated high altitude was isolated in petroleum ether extract of Saussurea involucrata. J. Ethnopharmacol. 2011, 137, 1510–1515. [Google Scholar] [CrossRef]
- Zhai, K.-F.; Duan, H.; Xing, J.-G.; Huang, H. Study on the anti-inflammatory and analgesic effects of various parts from Saussurea involucrate. Chin. J. Hosp. Pharm. 2010, 5, 374–377. [Google Scholar]
- Way, T.-D.; Lee, J.-C.; Kuo, D.-H.; Fan, L.-L.; Huang, C.-H.; Lin, H.-Y.; Shieh, P.-C.; Kuo, P.-T.; Liao, C.-F.; Liu, H.; et al. Inhibition of Epidermal Growth Factor Receptor Signaling by Saussurea involucrata, a Rare Traditional Chinese Medicinal Herb, in Human Hormone-Resistant Prostate Cancer PC-3 Cells. J. Agric. Food Chem. 2010, 58, 3356–3365. [Google Scholar] [CrossRef] [PubMed]
- Gong, G.; Xie, F.; Zheng, Y.; Hu, W.; Qi, B.; He, H.; Dong, T.T.; Tsim, K.W. The effect of methanol extract from Saussurea involucrata in the lipopolysaccharide-stimulated inflammation in cultured RAW 264.7 cells. J. Ethnopharmacol. 2020, 251, 112532. [Google Scholar] [CrossRef] [PubMed]
- Paek, K.-Y.; Murthy, H.N.; Zhong, J.-J. Production of Biomass and Bioactive Compounds Using Bioreactor Technology; Springer: Berlin/Heidelberg, Germany, 2014. [Google Scholar]
- Wang, J.; Li, J.-L.; Li, J.; Li, J.-X.; Liu, S.-J.; Huang, L.-Q.; Gao, W.-Y. Production of active compounds in medicinal plants: From plant tissue culture to biosynthesis. Chin. Herb. Med. 2017, 9, 115–125. [Google Scholar] [CrossRef]
- Rhiouani, H.; EL Hilaly, J.; Israili, Z.H.; Lyoussi, B. Acute and sub-chronic toxicity of an aqueous extract of the leaves of Herniaria glabra in rodents. J. Ethnopharmacol. 2008, 118, 378–386. [Google Scholar] [CrossRef]
- Stalikas, C.D. Extraction, separation, and detection methods for phenolic acids and flavonoids. J. Sep. Sci. 2007, 30, 3268–3295. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.; Gong, L.; Guo, Z.; Wang, W.; Zhang, H.; Liu, X.; Yu, S.; Xiong, L.; Luo, J. A Novel Integrated Method for Large-Scale Detection, Identification, and Quantification of Widely Targeted Metabolites: Application in the Study of Rice Metabolomics. Mol. Plant 2013, 6, 1769–1780. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Garcia, A.; Barbas, C. Gas Chromatography-Mass Spectrometry (GC-MS)-Based Metabolomics. In Metabolic Profiling; Springer: Berlin/Heidelberg, Germany, 2011; pp. 191–204. [Google Scholar]
- Chen, Y.; Zhang, R.; Song, Y.; He, J.; Sun, J.; Bai, J.; An, Z.; Dong, L.; Zhan, Q.; Abliz, Z. RRLC-MS/MS-based metabonomics combined with in-depth analysis of metabolic correlation network: Finding potential biomarkers for breast cancer. Analyst 2009, 134, 2003–2011. [Google Scholar] [CrossRef]
- Van den Heuvel, M.; Clark, D.; Fielder, R.; Koundakjian, P.; Oliver, G.; Pelling, D.; Tomlinson, N.; Walker, A. The international validation of a fixed-dose procedure as an alternative to the classical LD50 test. Food Chem. Toxicol. 1990, 28, 469–482. [Google Scholar] [CrossRef]
- Stallard, N.; Whitehead, A. Reducing animal numbers in the fixed-dose procedure. Hum. Exp. Toxicol. 1995, 14, 315–323. [Google Scholar] [CrossRef] [PubMed]
- Wen, Y.; Jin, L.; Zhang, D.; Zhang, L.; Xie, C.; Guo, D.; Wang, Y.; Wang, L.; Zhu, M.; Tong, J.; et al. Quantitative proteomic analysis of scleras in guinea pig exposed to wavelength defocus. J. Proteom. 2021, 243, 104248. [Google Scholar] [CrossRef]
- Kachuk, C.; Stephen, K.; Doucette, A. Comparison of sodium dodecyl sulfate depletion techniques for proteome analysis by mass spectrometry. J. Chromatogr. A 2015, 1418, 158–166. [Google Scholar] [CrossRef]
- Wiśniewski, J.R.; Zougman, A.; Nagaraj, N.; Mann, M. Universal sample preparation method for proteome analysis. Nat. Methods 2009, 6, 359–362. [Google Scholar] [CrossRef] [PubMed]
- Gillette, M.A.; Satpathy, S.; Cao, S.; Dhanasekaran, S.M.; Vasaikar, S.V.; Krug, K.; Petralia, F.; Li, Y.; Liang, W.-W.; Reva, B.; et al. Proteogenomic Characterization Reveals Therapeutic Vulnerabilities in Lung Adenocarcinoma. Cell 2020, 182, 200–225. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Liu, T.; Zhang, Z.; Payne, S.H.; Zhang, B.; McDermott, J.E.; Zhou, J.-Y.; Petyuk, V.A.; Chen, L.; Ray, D.; et al. Integrated Proteogenomic Characterization of Human High-Grade Serous Ovarian Cancer. Cell 2016, 166, 755–765. [Google Scholar] [CrossRef] [Green Version]
- Yin, X.-L.; Xu, B.-Q.; Zhang, Y.-Q. Gynura divaricata rich in 3, 5−/4, 5-dicaffeoylquinic acid and chlorogenic acid reduces islet cell apoptosis and improves pancreatic function in type 2 diabetic mice. Nutr. Metab. 2018, 15, 73. [Google Scholar] [CrossRef] [PubMed]
- Lodise, O.; Patil, K.; Karshenboym, I.; Prombo, S.; Chukwueke, C.; Pai, S.B. Inhibition of Prostate Cancer Cells by 4,5-Dicaffeoylquinic Acid through Cell Cycle Arrest. Prostate Cancer 2019, 2019, 4520645. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Byambaragchaa, M.; de la Cruz, J.; Yang, S.H.; Hwang, S.-G. Anti-metastatic potential of ethanol extract of Saussurea involucrata against hepatic cancer in vitro. Asian Pac. J. Cancer Prev. 2013, 14, 5397–5402. [Google Scholar] [CrossRef] [PubMed]
- Crout, R.; Gilbertson, J.R.; Gilbertson, J.D.; Platt, D.; Langkamp, H.; Connamacher, R. Effect of linolenyl alcohol on the in-vitro growth of the oral bacterium Streptococcus mutans. Arch. Oral Biol. 1982, 27, 1033–1037. [Google Scholar] [CrossRef]
- Sola, A.; Vilas, P.; Rodriguez, S.; Gancedo, A.G.; Gil-Fernández, C. Inactivation and inhibition of African swine fever virus by monoolein, monolinolein, and γ-linolenyl alcohol. Arch. Virol. 1986, 88, 285–292. [Google Scholar] [CrossRef]
- Marshall, M.E.; Kervin, K.; Benefield, C.; Umerani, A.; Albainy-Jenei, S.; Zhao, Q.; Khazaeli, M.B. Growth-inhibitory effects of coumarin (1,2-benzopyrone) and 7-hydroxycoumarin on human malignant cell lines in vitro. J. Cancer Res. Clin. Oncol. 1994, 120, S3–S10. [Google Scholar] [CrossRef] [PubMed]
- Timonen, J.M.; Nieminen, R.M.; Sareila, O.; Goulas, A.; Moilanen, L.J.; Haukka, M.; Vainiotalo, P.; Moilanen, E.; Aulaskari, P.H. Synthesis and anti-inflammatory effects of a series of novel 7-hydroxycoumarin derivatives. Eur. J. Med. Chem. 2011, 46, 3845–3850. [Google Scholar] [CrossRef]
- Kostova, I.P.; Manolov, I.I.; Nicolova, I.N.; Danchev, N.D. New metal complexes of 4-methyl-7-hydroxycoumarin sodium salt and their pharmacological activity. Il Farm. 2001, 56, 707–713. [Google Scholar] [CrossRef]
- Sato, Y.; Itagaki, S.; Kurokawa, T.; Ogura, J.; Kobayashi, M.; Hirano, T.; Sugawara, M.; Iseki, K. In vitro and in vivo antioxidant properties of chlorogenic acid and caffeic acid. Int. J. Pharm. 2011, 403, 136–138. [Google Scholar] [CrossRef]
- Lou, Z.; Wang, H.; Zhu, S.; Ma, C.; Wang, Z. Antibacterial Activity and Mechanism of Action of Chlorogenic Acid. J. Food Sci. 2011, 76, M398–M403. [Google Scholar] [CrossRef]
- Kim, Y.I.; Hirai, S.; Takahashi, H.; Goto, T.; Ohyane, C.; Tsugane, T.; Konishi, C.; Fujii, T.; Inai, S.; Iijima, Y.; et al. 9-oxo-10(E),12(E)-octadecadienoic acid derived from tomato is a potent PPAR α agonist to decrease triglyceride accumulation in mouse primary hepatocytes. Mol. Nutr. Food Res. 2011, 55, 585–593. [Google Scholar] [CrossRef] [PubMed]
- Zhao, B.; Tomoda, Y.; Mizukami, H.; Makino, T. 9-Oxo-(10E, 12E)-octadecadienoic acid, a cytotoxic fatty acid ketodiene isolated from eggplant calyx, induces apoptosis in human ovarian cancer (HRA) cells. J. Nat. Med. 2015, 69, 296–302. [Google Scholar] [CrossRef] [PubMed]
- Xargay-Torrent, S.; López-Guerra, M.; Saborit-Villarroya, I.; Rosich, L.; Navarro, A.; Villamor, N.; Aymerich, M.; Perez-Galan, P.; Roue, G.; Campo, E.; et al. The Multi-Kinase Inhibitor Sorafenib Blocks Migration, BCR Survival Signals, Protein Translation and Stroma-Mediated Bortezomib Resistance in Mantle Cell Lymphoma. Blood 2012, 120, 1647. [Google Scholar] [CrossRef]
- Li, H.-T.; Zhu, X. Quinoline-based compounds with potential activity against drugresistant cancers. Curr. Top. Med. Chem. 2021, 21, 426–437. [Google Scholar] [CrossRef]
- Mirmalek, S.A.; Jangholi, E.; Jafari, M.; Yadollah-Damavandi, S.; Javidi, M.A.; Parsa, Y.; Parsa, T.; Salimi-Tabatabaee, S.A.; Kolagar, H.G.; Jalil, S.K.; et al. Comparison of in Vitro Cytotoxicity and Apoptogenic Activity of Magnesium Chloride and Cisplatin as Conventional Chemotherapeutic Agents in the MCF-7 Cell Line. Asian Pac. J. Cancer Prev. 2016, 17, 131–134. [Google Scholar] [CrossRef] [Green Version]
- Spindler, S.R. Rapid and reversible induction of the longevity, anticancer and genomic effects of caloric restriction. Mech. Ageing Dev. 2005, 126, 960–966. [Google Scholar] [CrossRef] [PubMed]
- Rephaeli, A.; Waks-Yona, S.; Nudelman, A.; Tarasenko, I.; Tarasenko, N.; Phillips, D.; Cutts, S.; Kessler-Icekson, G. Anticancer prodrugs of butyric acid and formaldehyde protect against doxorubicin-induced cardiotoxicity. Br. J. Cancer 2007, 96, 1667–1674. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gionfra, F.; De Vito, P.; Pallottini, V.; Lin, H.-Y.; Davis, P.J.; Pedersen, J.Z.; Incerpi, S. The Role of Thyroid Hormones in Hepatocyte Proliferation and Liver Cancer. Front. Endocrinol. 2019, 10, 532. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kowalik, M.A.; Columbano, A.; Perra, A. Thyroid Hormones, Thyromimetics and Their Metabolites in the Treatment of Liver Disease. Front. Endocrinol. 2018, 9, 382. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Baker, S.D.; Grochow, L.B.; Donehower, R.C. Should Anticancer Drug Doses Be Adjusted in the Obese Patient? J. Natl. Cancer Inst. 1995, 87, 333–334. [Google Scholar] [CrossRef]
- Felici, A.; Verweij, J.; Sparreboom, A. Dosing strategies for anticancer drugs: The good, the bad and body-surface area. Eur. J. Cancer 2002, 38, 1677–1684. [Google Scholar] [CrossRef]
- Ueno, N.; Rizzo, J.; Demirer, T.; Cheng, Y.; Hegenbart, U.; Zhang, M.; Bregni, M.; Carella, A.; Blaise, D.; Bashey, A. Allogeneic hematopoietic cell transplantation for metastatic breast cancer. Bone Marrow Transplant. 2008, 41, 537–545. [Google Scholar] [CrossRef]
- Osada, J.; Rusak, M.; Kamocki, Z.; Dabrowska, M.I.; Kedra, B. Platelet activation in patients with advanced gastric cancer. Neoplasma 2010, 57, 145–150. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bao, Y.; Wang, L.; Shi, L.; Yun, F.; Liu, X.; Chen, Y.; Chen, C.; Ren, Y.; Jia, Y. Transcriptome profiling revealed multiple genes and ECM-receptor interaction pathways that may be associated with breast cancer. Cell. Mol. Biol. Lett. 2019, 24, 38. [Google Scholar] [CrossRef]
- Van Meerbeeck, J.P.; Fennell, D.A.; De Ruysscher, D.K. Small-cell lung cancer. Lancet 2011, 378, 1741–1755. [Google Scholar] [CrossRef]
- Iozzo, R.V.; Sanderson, R.D. Proteoglycans in cancer biology, tumour microenvironment and angiogenesis. J. Cell. Mol. Med. 2011, 15, 1013–1031. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Guan, J.-L. Signal transduction by focal adhesion kinase in cancer. Cancer Metastasis Rev. 2009, 28, 35–49. [Google Scholar] [CrossRef]
- Chen, W.Y.; Wu, F.; You, Z.Y.; Zhang, Z.M.; Guo, Y.L.; Zhong, L.X. Analyzing the differentially expressed genes and pathway cross-talk in aggressive breast cancer. J. Obstet. Gynaecol. Res. 2015, 41, 132–140. [Google Scholar] [CrossRef]
- Imai-Sumida, M.; Chiyomaru, T.; Majid, S.; Saini, S.; Nip, H.; Dahiya, R.; Tanaka, Y.; Yamamura, S. Silibinin suppresses bladder cancer through down-regulation of actin cytoskeleton and PI3K/Akt signaling pathways. Oncotarget 2017, 8, 92032–92042. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Grm, H.S.; Bergant, M.; Banks, L. Human papillomavirus infection, cancer & therapy. Indian J. Med. Res. 2009, 130, 277. [Google Scholar] [PubMed]
- Ding, X.; Zhang, W.; Li, S.; Yang, H. The role of cholesterol metabolism in cancer. Am. J. Cancer Res. 2019, 9, 219–227. [Google Scholar] [PubMed]
- Kälin, R.E.; Kretz, M.P.; Meyer, A.M.; Kispert, A.; Heppner, F.L.; Brändli, A.W. Paracrine and autocrine mechanisms of apelin signaling govern embryonic and tumor angiogenesis. Dev. Biol. 2007, 305, 599–614. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bailey, C.L.; Kelly, P.; Casey, P.J. Activation of Rap1 Promotes Prostate Cancer Metastasis. Cancer Res. 2009, 69, 4962–4968. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jain, S.; Jagtap, V.; Pise, N. Computer Aided Melanoma Skin Cancer Detection Using Image Processing. Procedia Comput. Sci. 2015, 48, 735–740. [Google Scholar] [CrossRef] [Green Version]
- Makrilia, N.; Kollias, A.; Manolopoulos, L.; Syrigos, K. Cell adhesion molecules: Role and clinical significance in cancer. Cancer Investig. 2009, 27, 1023–1037. [Google Scholar] [CrossRef] [PubMed]
- Veziant, J.; Gagnière, J.; Jouberton, E.; Bonnin, V.; Sauvanet, P.; Pezet, D.; Barnich, N.; Miot-Noirault, E.; Bonnet, M. Association of colorectal cancer with pathogenic Escherichia coli: Focus on mechanisms using optical imaging. World J. Clin. Oncol. 2016, 7, 293. [Google Scholar] [CrossRef]
- Bajenova, O.; Chaika, N.; Tolkunova, E.; Davydov-Sinitsyn, A.; Gapon, S.; Thomas, P.; O’Brien, S. Carcinoembryonic antigen promotes colorectal cancer progression by targeting adherens junction complexes. Exp. Cell Res. 2014, 324, 115–123. [Google Scholar] [CrossRef]
- Pan, D. The Hippo Signaling Pathway in Development and Cancer. Dev. Cell 2010, 19, 491–505. [Google Scholar] [CrossRef] [Green Version]
- Liu, Y.; Xiong, S.; Liu, S.; Chen, J.; Yang, H.; Liu, G.; Li, G. Analysis of gene expression in bladder cancer: Possible involvement of mitosis and complement and coagulation cascades signaling pathway. J. Comput. Biol. 2020, 27, 987–998. [Google Scholar] [CrossRef] [PubMed]
- Huan, J.; Wang, L.; Xing, L.; Qin, X.; Feng, L.; Pan, X.; Zhu, L. Insights into significant pathways and gene interaction networks underlying breast cancer cell line MCF-7 treated with 17β-estradiol (E2). Gene 2014, 533, 346–355. [Google Scholar] [CrossRef]
- Raad, I.; Narro, J.; Khan, A.; Tarrand, J.; Vartivarian, S.; Bodey, G. Serious complications of vascular catheter-related Staphylococcus aureus bacteremia in cancer patients. Eur. J. Clin. Microbiol. Infect. Dis. 1992, 11, 675–682. [Google Scholar] [CrossRef]
- Pinter, M.; Jain, R.K. Targeting the renin-angiotensin system to improve cancer treatment: Implications for immunotherapy. Sci. Transl. Med. 2017, 9, eaan5616. [Google Scholar] [CrossRef] [Green Version]
- Reber, H.A.; Johnson, F.E.; Montgomery, C.; Carl, W.R. Pancreatic secretion in hamsters with pancreatic cancer. Surgery 1977, 82, 34–41. [Google Scholar]
- Platten, M.; Nollen, E.A.A.; Röhrig, U.F.; Fallarino, F.; Opitz, C.A. Tryptophan metabolism as a common therapeutic target in cancer, neurodegeneration and beyond. Nat. Rev. Drug Discov. 2019, 18, 379–401. [Google Scholar] [CrossRef]
- Li, X.; Zhang, C.; Zhao, T.; Su, Z.; Li, M.; Hu, J.; Wen, J.; Shen, J.; Wang, C.; Pan, J.; et al. Lysine-222 succinylation reduces lysosomal degradation of lactate dehydrogenase a and is increased in gastric cancer. J. Exp. Clin. Cancer Res. 2020, 39, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Budczies, J.; Brockmöller, S.F.; Müller, B.M.; Barupal, D.K.; Richter-Ehrenstein, C.; Kleine-Tebbe, A.; Griffin, J.L.; Orešič, M.; Dietel, M.; Denkert, C.; et al. Comparative metabolomics of estrogen receptor positive and estrogen receptor negative breast cancer: Alterations in glutamine and beta-alanine metabolism. J. Proteom. 2013, 94, 279–288. [Google Scholar] [CrossRef]
- Newman, A.C.; Maddocks, O.D.K. One-carbon metabolism in cancer. Br. J. Cancer 2017, 116, 1499–1504. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mikalayeva, V.; Ceslevičienė, I.; Sarapinienė, I.; Žvikas, V.; Skeberdis, V.A.; Jakštas, V.; Bordel, S. Fatty acid synthesis and degradation interplay to regulate the oxidative stress in cancer cells. Int. J. Mol. Sci. 2019, 20, 1348. [Google Scholar] [CrossRef] [Green Version]
- Low, E.N.D.; Mokhtar, N.M.; Wong, Z.; Ali, R.A.R. Colonic Mucosal Transcriptomic Changes in Patients with Long-Duration Ulcerative Colitis Revealed Colitis-Associated Cancer Pathways. J. Crohn’s Coliti 2019, 13, 755–763. [Google Scholar] [CrossRef] [PubMed]
- Conde, V.R.; Oliveira, P.F.; Nunes, A.R.; Rocha, C.S.; Ramalhosa, E.; Pereira, J.A.; Alves, M.G.; Silva, B.M. The progression from a lower to a higher invasive stage of bladder cancer is associated with severe alterations in glucose and pyruvate metabolism. Exp. Cell Res. 2015, 335, 91–98. [Google Scholar] [CrossRef] [PubMed]
- Malehmir, M.; Pfister, D.; Gallage, S.; Szydlowska, M.; Inverso, D.; Kotsiliti, E.; Leone, V.; Peiseler, M.; Surewaard, B.G.J.; Rath, D.; et al. Platelet GPIbα is a mediator and potential interventional target for NASH and subsequent liver cancer. Nat. Med. 2019, 25, 641–655. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zheng, L.; Gong, W.; Liang, P.; Huang, X.; You, N.; Han, K.Q.; Li, Y.M.; Li, J. Effects of AFP-activated PI3K/Akt signaling pathway on cell proliferation of liver cancer. Tumor Biol. 2014, 35, 4095–4099. [Google Scholar] [CrossRef]
- Ferrara-Romeo, I.; Martínez, P.; Blasco, M.A. Mice lacking RAP1 show early onset and higher rates of DEN-induced hepatocellular carcinomas in female mice. PLoS ONE 2018, 13, e0204909. [Google Scholar] [CrossRef] [PubMed] [Green Version]
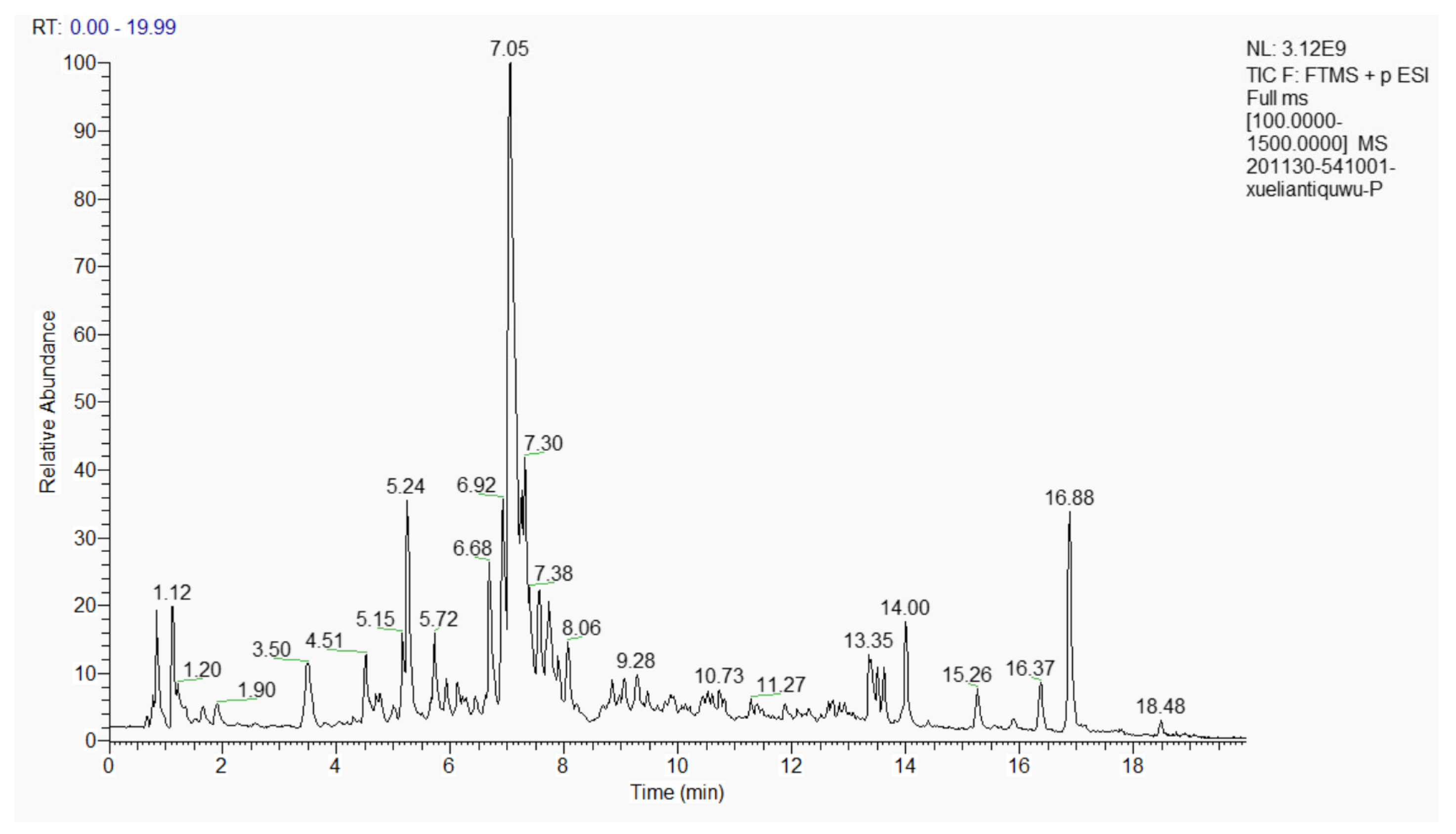



| Time (min) | Current Speed (μL/mL) | Gradient | B (%) |
|---|---|---|---|
| 0–2 | 300 | - | 5 |
| 2–6 | 300 | Linear | 30 |
| 6–7 | 300 | - | 30 |
| 7–12 | 300 | Linear | 78 |
| 12–14 | 300 | - | 78 |
| 14–17 | 300 | Linear | 95 |
| 17–20 | 300 | - | 95 |
| 20–21 | 300 | Linear | 5 |
| 21–25 | 300 | - | 5 |
| Time (min) | Flow Rate (mL/min) | Mobile Phase A (%) | Mobile Phase B (%) |
|---|---|---|---|
| 0 | 1 | 97 | 3 |
| 10 | 1 | 95 | 5 |
| 30 | 1 | 80 | 20 |
| 48 | 1 | 60 | 40 |
| 50 | 1 | 50 | 50 |
| 53 | 1 | 30 | 70 |
| 54 | 1 | 0 | 100 |
| Time (min) | Flow Rate (nL/min) | Mobile Phase A (%) | Mobile Phase B (%) |
|---|---|---|---|
| 0 | 600 | 94 | 6 |
| 2 | 600 | 85 | 15 |
| 48 | 600 | 60 | 40 |
| 50 | 600 | 50 | 50 |
| 51 | 600 | 45 | 55 |
| 60 | 600 | 0 | 100 |
| Name | Formula | CAS | Content (%) |
|---|---|---|---|
| 4,5-Dicaffeoylquinic acid | C25H24O12 | 14534-61-3 | 29.000 |
| Linolenyl alcohol | C18H32O | 506-44-5 | 8.453 |
| 7-Hydroxycoumarine | C9H6O3 | 93-35-6 | 7.691 |
| Chlorogenic acid | C16H18O9 | 327-97-9 | 6.134 |
| Metronidazole | C6H9N3O3 | 443-48-1 | 3.523 |
| Apigenin 7- (6”-crotonylglucoside) | C25H24O11 | NA | 2.814 |
| 9-Oxo-10(E),12(E)-octadecadienoic acid | C18H30O3 | 54232-58-5 | 2.618 |
| Cynaroside | C21H20O11 | 5373-11-5 | 2.382 |
| Phthalic anhydride | C8H4O3 | 85-44-9 | 1.956 |
| Hexadecanamide | C16H33NO | 629-54-9 | 1.883 |
| 3,4,5-tricaffeoylquinic acid | C34H30O15 | 86632-03-3 | 1.773 |
| Diisobutyl phthalate | C16H22O4 | 84-69-5 | 1.139 |
| Triethyl phosphate | C6H15O4P | 78-40-0 | 1.061 |
| ID | Title | x | y | n | N | Prot ID |
|---|---|---|---|---|---|---|
| 04614 | Renin-angiotensin system | 2 | 12 | 27 | 3214 | B2R941 P23946 |
| 00650 | Butanoate metabolism | 2 | 18 | 27 | 3214 | Q02338 Q08426 |
| 00380 | Tryptophan metabolism | 2 | 21 | 27 | 3214 | Q9BS61 Q08426 |
| 04640 | Hematopoietic cell lineage | 2 | 23 | 27 | 3214 | P17301 E7ESP4 |
| 00232 | Caffeine metabolism | 1 | 2 | 27 | 3214 | P47989 |
| 04611 | Platelet activation | 3 | 74 | 27 | 3214 | P17301 E7ESP4 O15264 |
| 04977 | Vitamin digestion and absorption | 1 | 4 | 27 | 3214 | E1B4S8 |
| 00310 | Lysine degradation | 2 | 36 | 27 | 3214 | Q9NVH6 Q08426 |
| 03320 | PPAR signaling pathway | 2 | 36 | 27 | 3214 | A0A140VKG0 Q08426 |
| 05412 | Arrhythmogenic right ventricular cardiomyopathy (ARVC) | 2 | 37 | 27 | 3214 | P17301 E7ESP4 |
| 04610 | Complement and coagulation cascades | 2 | 40 | 27 | 3214 | A0A0F7G8J1 Q19UG4 |
| 04975 | Fat digestion and absorption | 1 | 6 | 27 | 3214 | E1B4S8 |
| Prot ID | Description |
|---|---|
| B2R941 | cDNA, FLJ94198, highly similar to Homo sapiens carboxypeptidase A3 (mast cell) |
| P23946 | Chymase |
| Q02338 | D-beta-hydroxybutyrate dehydrogenase, mitochondrial |
| Q08426 | Peroxisomal bifunctional enzyme |
| Q9BS61 | Kynurenine 3-monooxygenase |
| Q08426 | Peroxisomal bifunctional enzyme |
| P17301 | Integrin alpha-2 |
| E7ESP4 | Integrin alpha-2 |
| P47989 | Xanthine dehydrogenase/oxidase |
| O15264 | Mitogen-activated protein kinase 13 |
| E1B4S8 | Apolipoprotein B (Fragment) |
| Q9NVH6 | Trimethyllysine dioxygenase |
| Q08426 | Peroxisomal bifunctional enzyme |
| A0A0F7G8J1 | Plasminogen |
| Q19UG4 | Christmas factor (Fragment) |
| Prot ID | KEGG Pathway |
|---|---|
| P17301 | Hematopoietic cell lineage * Platelet activation * Arrhythmogenic right ventricular cardiomyopathy (ARCV) ECM-receptor interaction * Hypertrophic cardiomyopathy (HCM) Dilated cardiomyopathy (DCM) Small-cell lung cancer * Proteoglycans in cancer * Focal adhesion * Phagosome * Regulation of actin cytoskeleton * PI3K-Akt signaling pathway * Human papillomavirus infection * Pathways in cancer * |
| E1B4S8 | Vitamin digestion and absorption Fat digestion and absorption Cholesterol metabolism * |
| P12830 | Apelin signaling pathway * Thyroid cancer * Rap1 signaling pathway * Bladder cancer * Endometrial cancer * Melanoma * Cell adhesion molecules (CAMs) * Pathogenic Escherichia coli infection * Gastric cancer * Adherens junction * Bacterial invasion of epithelial cells Pathways in cancer * Hippo signaling pathway * |
| Q19UG4 | Complement and coagulation cascades * |
| A0A0F7G8J1 | Complement and coagulation cascades * Neuroactive ligand-receptor interaction * Staphylococcus aureus infection * Influenza A |
| B2R941 | Renin-angiotensin system * Protein digestion and absorption * Pancreatic secretion * |
| P23946 | Renin-angiotensin system * |
| Q08426 | Butanoate metabolism Tryptophan metabolism * Lysine degradation * PPAR signaling pathway * Peroxisome * Beta-alanine metabolism * Propanoate metabolismCarbon metabolism * Metabolic pathways * Fatty acid degradation * Fatty acid metabolism * Valine, leucine, and isoleucine degradation * |
| P48163 | Carbon metabolism * Pyruvate metabolism Metabolic pathways * |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gao, J.; Wang, Y.; Lyu, B.; Chen, J.; Chen, G. Component Identification of Phenolic Acids in Cell Suspension Cultures of Saussureainvolucrata and Its Mechanism of Anti-Hepatoma Revealed by TMT Quantitative Proteomics. Foods 2021, 10, 2466. https://doi.org/10.3390/foods10102466
Gao J, Wang Y, Lyu B, Chen J, Chen G. Component Identification of Phenolic Acids in Cell Suspension Cultures of Saussureainvolucrata and Its Mechanism of Anti-Hepatoma Revealed by TMT Quantitative Proteomics. Foods. 2021; 10(10):2466. https://doi.org/10.3390/foods10102466
Chicago/Turabian StyleGao, Junpeng, Yi Wang, Bo Lyu, Jian Chen, and Guang Chen. 2021. "Component Identification of Phenolic Acids in Cell Suspension Cultures of Saussureainvolucrata and Its Mechanism of Anti-Hepatoma Revealed by TMT Quantitative Proteomics" Foods 10, no. 10: 2466. https://doi.org/10.3390/foods10102466
APA StyleGao, J., Wang, Y., Lyu, B., Chen, J., & Chen, G. (2021). Component Identification of Phenolic Acids in Cell Suspension Cultures of Saussureainvolucrata and Its Mechanism of Anti-Hepatoma Revealed by TMT Quantitative Proteomics. Foods, 10(10), 2466. https://doi.org/10.3390/foods10102466







