An Integrated Bile Acids Profile Determination by UHPLC-MS/MS to Identify the Effect of Bile Acids Supplement in High Plant Protein Diet on Common Carp (Cyprinus carpio)
Abstract
1. Introduction
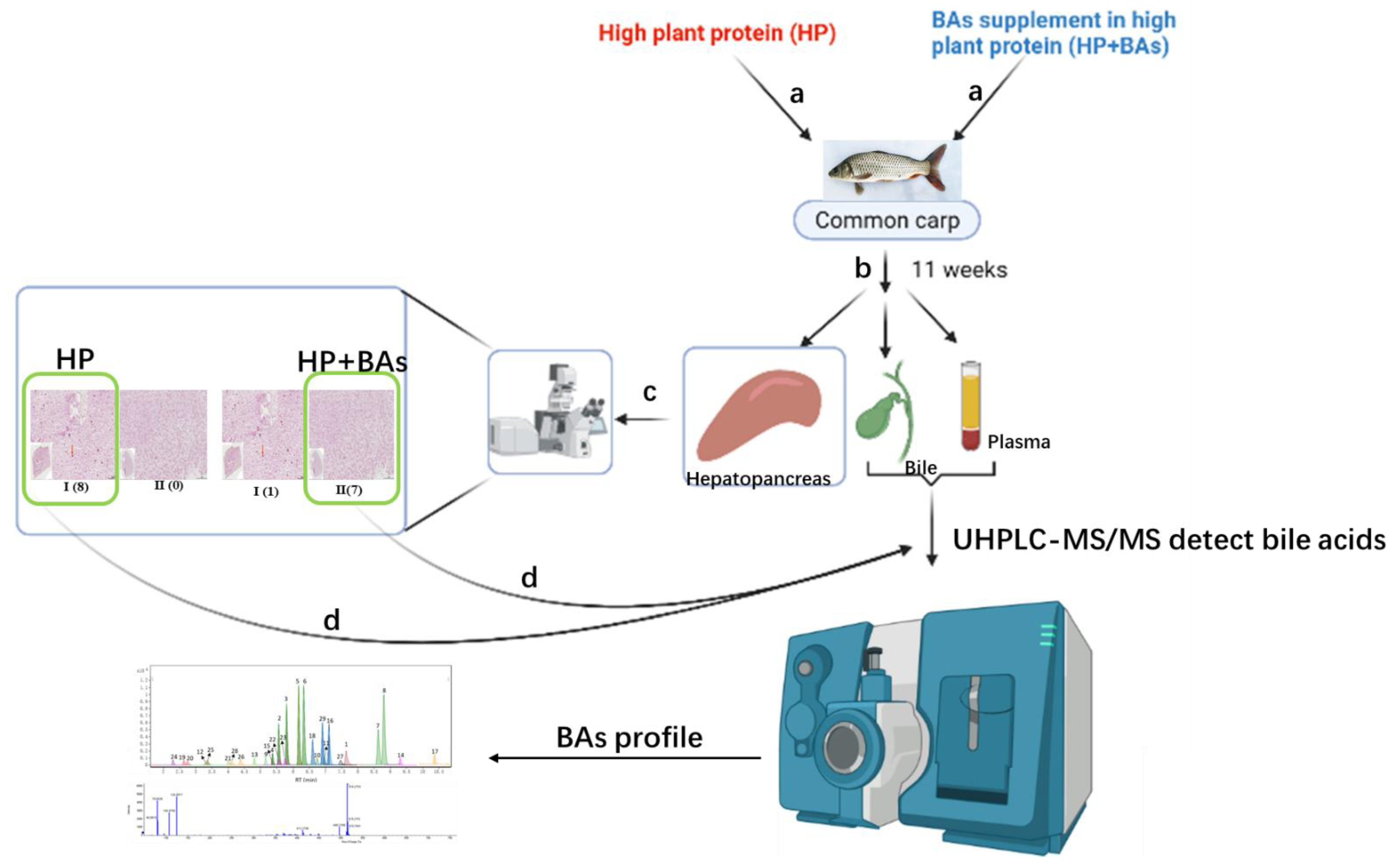
2. Materials and Methods
2.1. Chemicals and Reagents
2.2. Bile and Plasma Sampling
2.3. Plasma Biochemical Parameters
2.4. Histopathological Detections of Hepatopancreas Tissues
2.5. Bile Acids Quantitative Analysis
2.6. TβMCA and TωMCA Qualitative Analysis
2.7. Statistical Analyses
3. Results
3.1. Growth Performance
3.2. Hepatopancreas Histopathological Examination
3.3. Plasma Biochemical Parameters and Hepatic Glycogen
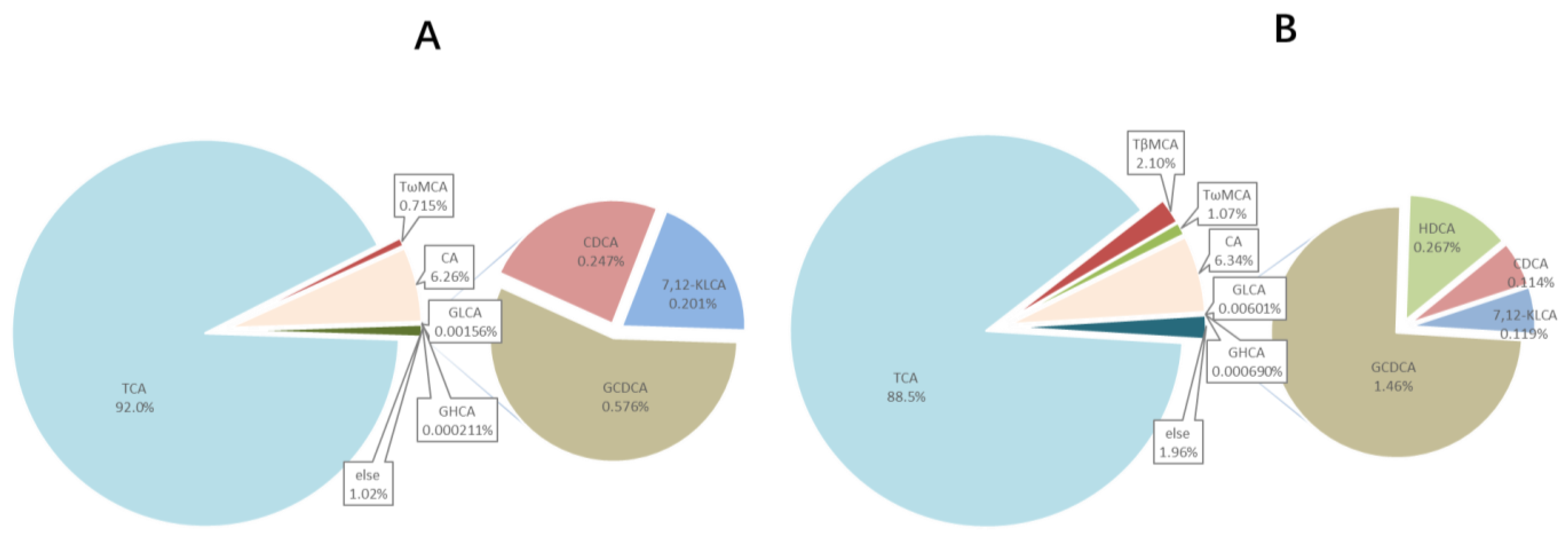
3.4. Bile Acids Profile of in Common Carp Bile and Plasma
3.5. Supplement BAs to High Plant Protein Feed Altered the BA profile
4. Discussion
4.1. BA Profile Changes Caused by Supplements of BAs
4.2. Supplement BAs Affected Common Carp BA Profile to Reduce Hepatopancreas Glycogen Accumulation and Alleviated Hepatopancreas Damage with a High Plant Protein Diet
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
References
- Lefebvre, P.; Cariou, B.; Lien, F.; Kuipers, F.; Staels, B. Role of Bile Acids and Bile Acid Receptors in Metabolic Regulation. Physiol. Rev. 2009, 89, 147–191. [Google Scholar] [CrossRef]
- Bachmann, V.; Kostiuk, B.; Unterweger, D.; Diaz-Satizabal, L.; Ogg, S.; Pukatzki, S. Bile Salts Modulate the Mucin-Activated Type VI Secretion System of Pandemic Vibrio cholerae. PLoS Negl. Trop. Dis. 2015, 9, e0004031. [Google Scholar] [CrossRef] [PubMed]
- Sun, J.; Cao, Z.; Smith, A.D.; Carlson, P.E., Jr.; Coryell, M.; Chen, H.; Beger, R.D. Bile Acid Profile and its Changes in Response to Cefoperazone Treatment in MR1 Deficient Mice. Metabolites 2020, 10, 127. [Google Scholar] [CrossRef] [PubMed]
- Ridlon, J.M.; Kang, D.-J.; Hylemon, P.B. Bile salt biotransformations by human intestinal bacteria. J. Lipid Res. 2006, 47, 241–259. [Google Scholar] [CrossRef] [PubMed]
- Begley, M.; Gahan, C.G.; Hill, C. The interaction between bacteria and bile. FEMS Microbiol. Rev. 2005, 29, 625–651. [Google Scholar] [CrossRef] [PubMed]
- Insull, W., Jr. Clinical utility of bile acid sequestrants in the treatment of dyslipidemia a scientific review. South. Med. J. 2006, 99, 257–273. [Google Scholar] [CrossRef]
- Staels, B.; Fonseca, V.A. Bile Acids and Metabolic Regulation: Mechanisms and clinical responses to bile acid sequestration. Diabetes Care 2009, 32, S237–S245. [Google Scholar] [CrossRef]
- Ferrell, J.M.; Chiang, J.Y.L. Understanding Bile Acid Signaling in Diabetes: From Pathophysiology to Therapeutic Targets. Diabetes Metab. J. 2019, 43, 257–272. [Google Scholar] [CrossRef]
- Lien, F.; Berthier, A.; Bouchaert, E.; Gheeraert, C.; Alexandre, J.; Porez, G.; Prawitt, J.; Dehondt, H.; Ploton, M.; Colin, S.; et al. Metformin interferes with bile acid homeostasis through AMPK-FXR crosstalk. J. Clin. Investig. 2014, 124, 1037–1051. [Google Scholar] [CrossRef]
- Chiang, J.Y.L. Bile Acid Metabolism and Signaling. Compr. Physiol. 2013, 3, 1191–1212. [Google Scholar] [CrossRef]
- Katsuma, S.; Hirasawa, A.; Tsujimoto, G. Bile acids promote glucagon-like peptide-1 secretion through TGR5 in a murine enteroendocrine cell line STC-1. Biochem. Biophys. Res. Commun. 2005, 329, 386–390. [Google Scholar] [CrossRef]
- Pols, T.W.; Nomura, M.; Harach, T.; Sasso, G.L.; Oosterveer, M.H.; Thomas, C.; Rizzo, G.; Gioiello, A.; Adorini, L.; Pellicciari, R.; et al. TGR5 Activation Inhibits Atherosclerosis by Reducing Macrophage Inflammation and Lipid Loading. Cell Metab. 2011, 14, 747–757. [Google Scholar] [CrossRef]
- Vallim, T.Q.D.A.; Tarling, E.J.; Edwards, P.A. Pleiotropic Roles of Bile Acids in Metabolism. Cell Metab. 2013, 17, 657–669. [Google Scholar] [CrossRef]
- Li, F.; Jiang, C.; Krausz, K.W.; Li, Y.; Albert, I.; Hao, H.; Fabre, K.M.; Mitchell, J.B.; Patterson, A.; Gonzalez, F.J. Microbiome remodelling leads to inhibition of intestinal farnesoid X receptor signalling and decreased obesity. Nat. Commun. 2013, 4, 2384. [Google Scholar] [CrossRef]
- Abdel-Latif, H.M.; Abdel-Daim, M.M.; Shukry, M.; Nowosad, J.; Kucharczyk, D. Benefits and applications of Moringa oleifera as a plant protein source in Aquafeed: A review. Aquaculture 2021, 547, 737369. [Google Scholar] [CrossRef]
- Wei, H.; Xing, S.; Chen, P.; Wu, X.; Gu, X.; Luo, L.; Liang, X.; Xue, M. Plant protein diet-induced hypoimmunity by affecting the spiral valve intestinal microbiota and bile acid enterohepatic circulation in Amur sturgeon (Acipenser schrenckii). Fish Shellfish Immunol. 2020, 106, 421–430. [Google Scholar] [CrossRef]
- Zhang, Y.; Chen, P.; Liang, X.F.; Han, J.; Wu, X.F.; Yang, Y.H.; Xue, M. Metabolic disorder induces fatty liver in Japanese seabass, Lateolabrax japonicas fed a full plant protein diet and regulated by cAMP-JNK/NF-kB-caspase signal pathway. Fish Shellfish Immunol. 2019, 90, 223–234. [Google Scholar] [CrossRef] [PubMed]
- Romano, N.; Kumar, V.; Yang, G.; Kajbaf, K.; Rubio, M.B.; Overturf, K.; Brezas, A.; Hardy, R. Bile acid metabolism in fish: Disturbances caused by fishmeal alternatives and some mitigating effects from dietary bile inclusions. Rev. Aquac. 2020, 12, 1792–1817. [Google Scholar] [CrossRef]
- Jin, M.; Pan, T.; Cheng, X.; Zhu, T.T.; Sun, P.; Zhou, F.; Ding, X.; Zhou, Q.-C. Effects of supplemental dietary l-carnitine and bile acids on growth performance, antioxidant and immune ability, histopathological changes and inflammatory response in juvenile black seabream (Acanthopagrus schlegelii) fed high-fat diet. Aquaculture 2019, 504, 199–209. [Google Scholar] [CrossRef]
- Kortner, T.M.; Penn, M.H.; Björkhem, I.; Måsøval, K.; Krogdahl, Å. Bile components and lecithin supplemented to plant based diets do not diminish diet related intestinal inflammation in Atlantic salmon. BMC Vet. Res. 2016, 12, 190. [Google Scholar] [CrossRef] [PubMed]
- Bathena, S.P.R.; Mukherjee, S.; Olivera, M.; Alnouti, Y. The profile of bile acids and their sulfate metabolites in human urine and serum. J. Chromatogr. B 2013, 942–943, 53–62. [Google Scholar] [CrossRef]
- Chen, C.; Hu, B.; Wu, T.; Zhang, Y.; Xu, Y.; Feng, Y.; Jiang, H. Bile acid profiles in diabetic (db/db) mice and their wild type littermates. J. Pharm. Biomed. Anal. 2016, 131, 473–481. [Google Scholar] [CrossRef]
- Buchinger, T.J.; Li, W.; Johnson, N.S. Bile Salts as Semiochemicals in Fish. Chem. Senses 2014, 39, 647–654. [Google Scholar] [CrossRef]
- Satoh, R.; Saito, T.; Ogata, H.; Ohsaki, A.; Iida, T.; Asahina, K.; Mitamura, K.; Ikegawa, S.; Hofmann, A.F.; Hagey, L.R. N-Methyltaurine N-acyl amidated bile acids and deoxycholic acid in the bile of angelfish (Pomacanthidae): A novel bile acid profile in Perciform fish. Steroids 2014, 80, 15–23. [Google Scholar] [CrossRef]
- Schmucker, A.K.; Johnson, N.S.; Bussy, U.; Li, K.; Galbraith, H.S.; Chung-Davidson, Y.; Li, W. American eels produce and release bile acid profiles that vary across life stage. J. Fish Biol. 2020, 96, 1024–1033. [Google Scholar] [CrossRef]
- Wang, H.; Yeh, C.-Y.; Li, K.; Chung-Davidson, Y.-W.; Li, W. An UPLC–MS/MS method for quantitative profiling of bile acids in sea lamprey plasma and tissues. J. Chromatogr. B 2015, 980, 72–78. [Google Scholar] [CrossRef]
- Xu, H.; Zhang, Q.; Kim, S.-K.; Liao, Z.; Wei, Y.; Sun, B.; Jia, L.; Chi, S.; Liang, M. Dietary taurine stimulates the hepatic biosynthesis of both bile acids and cholesterol in the marine teleost, tiger puffer (Takifugu rubripes). Br. J. Nutr. 2020, 123, 1345–1356. [Google Scholar] [CrossRef]
- Yao, T.; Wei, X.; Xue, M.; Sun, B.; Gu, X.; Zhang, S. Analysis of Bile Acid Profile in Bile and Plasma of Turbot. Chin. J. Anim. Nutr. 2020, 32, 5816–5826. [Google Scholar]
- Zhang, J.; Li, W.; Zhou, H.; Li, M.; Wang, M.; Wu, S. An analysis of Bile Acid composition in different tissues of Grass Carp (Ctenopharyngodon idellus). Acta Hydrobiol. Sin. 2017, 41, 479–482. [Google Scholar]
- Yao, T.; Gu, X.; Liang, X.; Fall, F.N.; Cao, A.; Zhang, S.; Guan, Y.; Sun, B.; Xue, M. Tolerance assessment of dietary bile acids in common carp (Cyprinus carpio L.) fed a high plant protein diet. Aquaculture 2021, 543, 737012. [Google Scholar] [CrossRef]
- Long, Y.; Li, X.; Li, F.; Ge, G.; Liu, R.; Song, G.; Li, Q.; Qiao, Z.; Cui, Z. Transcriptional Programs Underlying Cold Acclimation of Common Carp (Cyprinus carpio L.). Front. Genet. 2020, 11, 556418. [Google Scholar] [CrossRef] [PubMed]
- Yu, H.; Zhang, L.; Chen, P.; Liang, X.; Cao, A.; Han, J.; Wu, X.; Zheng, Y.; Qin, Y.; Xue, M. Dietary Bile Acids Enhance Growth, and Alleviate Hepatic Fibrosis Induced by a High Starch Diet via AKT/FOXO1 and cAMP/AMPK/SREBP1 Pathway in Micropterus salmoides. Front. Physiol. 2019, 10, 1430. [Google Scholar] [CrossRef]
- Gu, Y.; Wang, X.; Li, J.; Zhang, Y.; Zhong, H.; Liu, R.; Zhang, D.; Feng, Q.; Xie, X.; Hongmei, Z.; et al. Analyses of gut microbiota and plasma bile acids enable stratification of patients for antidiabetic treatment. Nat. Commun. 2017, 8, 1785. [Google Scholar] [CrossRef]
- Humbert, L.; Maubert, M.A.; Wolf, C.; Duboc, H.; Mahé, M.; Farabos, D.; Seksik, P.; Mallet, J.-M.; Trugnan, G.; Masliah, J.; et al. Bile acid profiling in human biological samples: Comparison of extraction procedures and application to normal and cholestatic patients. J. Chromatogr. B 2012, 899, 135–145. [Google Scholar] [CrossRef]
- Zheng, X.; Huang, F.; Zhao, A.; Lei, S.; Zhang, Y.; Xie, G.; Chen, T.; Qu, C.; Rajani, C.; Dong, B.; et al. Bile acid is a significant host factor shaping the gut microbiome of diet-induced obese mice. BMC Biol. 2017, 15, 120. [Google Scholar] [CrossRef]
- Staessen, T.W.O.; Verdegem, M.C.J.; Nederlof, M.A.J.; Eding, E.H.; Schrama, J.W. Effect of type of dietary non- protein energy source (starch vs. fat) on the body bile acid pool size and composition, faecal bile acid loss and bile acid synthesis in rainbow trout (Oncorhynchus mykiss). Aquac. Nutr. 2021, 27, 865–879. [Google Scholar] [CrossRef]
- Vessey, D.A. The biochemical basis for the conjugation of bile acids with either glycine or taurine. Biochem. J. 1978, 174, 621–626. [Google Scholar] [CrossRef]
- Hofmann, A.F.; Hagey, L.R.; Krasowski, M. Bile salts of vertebrates: Structural variation and possible evolutionary significance. J. Lipid Res. 2010, 51, 226–246. [Google Scholar] [CrossRef]
- Li, K.; Buchinger, T.J.; Bussy, U.; Fissette, S.D.; Johnson, N.S.; Li, W. Quantification of 15 bile acids in lake charr feces by ultra-high performance liquid chromatography–tandem mass spectrometry. J. Chromatogr. B 2015, 1001, 27–34. [Google Scholar] [CrossRef] [PubMed]
- Jurkowska, H.; Stipanuk, M.H.; Hirschberger, L.L.; Roman, H.B. Propargylglycine inhibits hypotaurine/taurine synthesis and elevates cystathionine and homocysteine concentrations in primary mouse hepatocytes. Amino Acids 2015, 47, 1215–1223. [Google Scholar] [CrossRef] [PubMed]
- Ridlon, J.M.; Wolf, P.G.; Gaskins, H.R. Taurocholic acid metabolism by gut microbes and colon cancer. Gut Microbes 2016, 7, 201–215. [Google Scholar] [CrossRef]
- Wahlström, A.; Sayin, S.I.; Marschall, H.-U.; Bäckhed, F. Intestinal Crosstalk between Bile Acids and Microbiota and Its Impact on Host Metabolism. Cell Metab. 2016, 24, 41–50. [Google Scholar] [CrossRef]
- DiMarzio, M.; Rusconi, B.; Yennawar, N.H.; Eppinger, M.; Patterson, A.D.; Dudley, E. GIdentification of a mouse Lactobacillus johnsonii strain with deconjugase activity against the FXR an-tagonist T-beta-MCA. PLoS ONE 2017, 12, e0183564. [Google Scholar] [CrossRef] [PubMed]
- Sun, L.; Xie, C.; Wang, G.; Wu, Y.; Wu, Q.; Wang, X.; Liu, J.; Deng, Y.; Xia, J.; Chen, B.; et al. Gut microbiota and intestinal FXR mediate the clinical benefits of metformin. Nat. Med. 2018, 24, 1919–1929. [Google Scholar] [CrossRef] [PubMed]
- Sayin, S.I.; Wahlström, A.; Felin, J.; Jäntti, S.; Marschall, H.-U.; Bamberg, K.; Angelin, B.; Hyötyläinen, T.; Oresic, M.; Bäckhed, F. Gut Microbiota Regulates Bile Acid Metabolism by Reducing the Levels of Tauro-beta-muricholic Acid, a Naturally Occurring FXR Antagonist. Cell Metab. 2013, 17, 225–235. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, S.; Fukami, T.; Masuo, Y.; Brocker, C.N.; Xie, C.; Krausz, K.W.; Wolf, C.R.; Henderson, C.J.; Gonzalez, F.J. Cyp2c70 is responsible for the species difference in bile acid metabolism between mice and humans. J. Lipid Res. 2016, 57, 2130–2137. [Google Scholar] [CrossRef]
- Bansal, M.; Fu, Y.; Alrubaye, B.; Abraha, M.; Almansour, A.; Gupta, A.; Liyanage, R.; Wang, H.; Hargis, B.; Sun, X. A secondary bile acid from microbiota metabolism attenuates ileitis and bile acid reduction in subclinical necrotic enteritis in chickens. J. Anim. Sci. Biotechnol. 2020, 11, 37. [Google Scholar] [CrossRef]
- Jia, W.; Rajani, C.; Zheng, X.; Jia, W. Hyocholic acid and glycemic regulation: Comments on ‘Hyocholic acid species improve glucose homeostasis through a distinct TGR5 and FXR signaling mechanism’. J. Mol. Cell Biol. 2021, 13, 460–462. [Google Scholar] [CrossRef]
- Cui, Y.; Xu, B.; Zhang, X.; He, Y.; Shao, Y.; Ding, M. Diagnostic and therapeutic profiles of serum bile acids in women with intrahepatic cholestasis of pregnancy-a pseudo-targeted metabolomics study. Clin. Chim. Acta 2018, 483, 135–141. [Google Scholar] [CrossRef]
- Wei, H.; Chen, P.; Liang, X.; Yu, H.; Wu, X.; Han, J.; Luo, L.; Gu, X.; Xue, M. Plant protein diet suppressed immune function by inhibiting spiral valve intestinal mucosal barrier integrity, anti-oxidation, apoptosis, autophagy and proliferation responses in amur sturgeon (Acipenser schrenckii). Fish Shellfish Immunol. 2019, 94, 711–722. [Google Scholar] [CrossRef]
- Urán, P.; Gonçalves, A.; Taverne-Thiele, J.; Schrama, J.; Verreth, J.; Rombout, J. Soybean meal induces intestinal inflammation in common carp (Cyprinus carpio L.). Fish Shellfish Immunol. 2008, 25, 751–760. [Google Scholar] [CrossRef]
- Kaushik, S.; Cravedi, J.; Lallès, J.-P.; Sumpter, J.; Fauconneau, B.; Laroche, M. Partial or total replacement of fish meal by soybean protein on growth, protein utilization, potential estrogenic or antigenic effects, cholesterolemia and flesh quality in rainbow trout, Oncorhynchus mykiss. Aquaculture 1995, 133, 257–274. [Google Scholar] [CrossRef]
- Escaffre, A.M.; Infante, J.L.Z.; Cahu, C.L.; Mambrini, M.; Bergot, P.; Kaushik, S.J. Nutritional value of soy protein concentrate for larvae of common carp (Cyprinus carpio). Aquaculture 1997, 153, 63–80. [Google Scholar] [CrossRef]
- Laubertová, L.; Koňariková, K.; Gbelcová, H.; Ďuračková, Z.; Žitňanová, I. Effect of walnut oil on hyperglycemia-induced oxidative stress and pro-inflammatory cytokines production. Eur. J. Nutr. 2015, 54, 291–299. [Google Scholar] [CrossRef]
- Knudsen, D.; Urán, P.; Arnous, A.; Koppe, A.W.; Frøkiær, H. Saponin-Containing Subfractions of Soybean Molasses Induce Enteritis in the Distal Intestine of Atlantic Salmon. J. Agric. Food Chem. 2007, 55, 2261–2267. [Google Scholar] [CrossRef] [PubMed]
- Matin, H.H.; Shariatmadari, F.; Torshizi, M.K.; Chiba, L. In vitro bile acid-binding capacity of dietary fibre sources and their effects with bile acid on broiler chicken performance and lipid digestibility. Br. Poult. Sci. 2016, 57, 348–357. [Google Scholar] [CrossRef]
- Lee, S.-O.; Simons, A.L.; Murphy, P.A.; Hendrich, S. Soyasaponins Lowered Plasma Cholesterol and Increased Fecal Bile Acids in Female Golden Syrian Hamsters. Exp. Biol. Med. 2005, 230, 472–478. [Google Scholar] [CrossRef]
- Sato, H.; Genet, C.; Strehle, A.; Thomas, C.; Lobstein, A.; Wagner, A.; Mioskowski, C.; Auwerx, J.; Saladin, R. Anti-hyperglycemic activity of a TGR5 agonist isolated from Olea europaea. Biochem. Biophys. Res. Commun. 2007, 362, 793–798. [Google Scholar] [CrossRef]
- Guo, C.; Chen, W.-D.; Wang, Y.-D. TGR5, Not Only a Metabolic Regulator. Front. Physiol. 2016, 7, 646. [Google Scholar] [CrossRef]
- Wang, Y.-D.; Chen, W.-D.; Yu, D.; Forman, B.M.; Huang, W. The G-protein-coupled bile acid receptor, Gpbar1 (TGR5), negatively regulates hepatic inflammatory response through antagonizing nuclear factor kappa light-chain enhancer of activated B cells (NF-kappaB) in mice. Hepatology 2011, 54, 1421–1432. [Google Scholar] [CrossRef]
- Keitel, V.; Donner, M.; Winandy, S.; Kubitz, R.; Häussinger, D. Expression and function of the bile acid receptor TGR5 in Kupffer cells. Biochem. Biophys. Res. Commun. 2008, 372, 78–84. [Google Scholar] [CrossRef]
- Häussinger, D.; Keitel, V. Role of TGR5 (GPBAR1) in Liver Disease. Semin. Liver Dis. 2018, 38, 333–339. [Google Scholar] [CrossRef]
- Li, Y.-F.; Wu, J.-S.; Li, Y.-Y.; Dai, Y.; Zheng, M.; Zeng, J.-K.; Wang, G.-F.; Wang, T.-M.; Li, W.-K.; Zhang, X.-Y.; et al. Chicken bile powder protects against α-naphthylisothiocyanate-induced cholestatic liver injury in mice. Oncotarget 2017, 8, 97137–97152. [Google Scholar] [CrossRef][Green Version]
- Holter, M.M.; Chirikjian, M.K.; Govani, V.N.; Cummings, B.P. TGR5 Signaling in Hepatic Metabolic Health. Nutrients 2020, 12, 2598. [Google Scholar] [CrossRef] [PubMed]
- Ide, T.; Kano, S.; Murata, M.; Yanagita, T.; Sugano, M. Dietary modifications of the biliary bile acid glycine:taurine ratio and activity of hepatic bile acid-CoA:amino acid N-acyltransferase (EC 2.3.1) in the rat. Br. J. Nutr. 1994, 72, 93–100. [Google Scholar] [CrossRef] [PubMed]
- Rivera, C.A.; Bradford, B.U.; Hunt, K.J.; Adachi, Y.; Schrum, L.W.; Koop, D.R.; Burchardt, E.-R.; Rippe, R.A.; Thurman, R.G. Attenuation of CCl4-induced hepatic fibrosis by GdCl3 treatment or dietary glycine. Am. J. Physiol. Gastrointest. Liver Physiol. 2001, 281, G200–G208. [Google Scholar] [CrossRef] [PubMed]
- Ma, K.; Saha, P.K.; Chan, L.; Moore, D.D. Farnesoid X receptor is essential for normal glucose homeostasis. J. Clin. Investig. 2006, 116, 1102–1109. [Google Scholar] [CrossRef]
- Van Dijk, T.H.; Grevhorst, A.; Oosterveer, M.H.; Bloks, V.W.; Staels, B.; Reijngoud, D.-J.; Kuipers, F. An increased flux through the glucose 6-phosphate pool in enterocytes delays glucose absorption in Fxr−/− mice. J. Biol. Chem. 2009, 284, 10315–10323. [Google Scholar] [CrossRef]
- Hui, S.; Liu, Y.; Chen, M.; Wang, X.; Lang, H.; Zhou, M.; Yi, L.; Mi, M. Capsaicin Improves Glucose Tolerance and Insulin Sensitivity Through Modulation of the Gut Microbiota-Bile Acid-FXR Axis in Type 2 Diabetic db/db Mice. Mol. Nutr. Food Res. 2019, 63, e1900608. [Google Scholar] [CrossRef]





| No. | Compounds | Retention Time | Transition | Fragmentor | CE | Linearity Range | Calibration Curves * | R2 | Internal Standard |
|---|---|---|---|---|---|---|---|---|---|
| (min) | (m/z) | (V) | (eV) | (μg/mL) | |||||
| 1 | CA | 6.339 | 407.3 -> 407.3 | 240 | 10 | 0.0025–5 | y = 13.798x | 0.9989 | CA -d4 |
| 2 | αMCA | 5.570 | 407.3 -> 407.3 | 240 | 10 | 0.001–5 | y = 100.08x | 0.9992 | βMCA -d5 |
| 3 | ωMCA | 5.384 | 407.3 -> 407.3 | 240 | 10 | 0.001–5 | y = 65.329x | 0.9982 | βMCA -d5 |
| 4 | HCA | 6.178 | 407.3 -> 407.3 | 240 | 10 | 0.001–5 | y = 11.074x | 0.9994 | βMCA -d5 |
| 5 | CDCA | 8.628 | 391.3 -> 391.3 | 230 | 10 | 0.001–5 | y = 39.802x | 0.9995 | CDCA-d4 |
| 6 | DCA | 8.805 | 391.3 -> 391.3 | 230 | 10 | 0.001–5 | y = 8.9922x | 0.9998 | DCA-d4 |
| 7 | HDCA | 7.110 | 391.3 -> 391.3 | 230 | 10 | 0.001–5 | y = 8.6365x | 0.9993 | DCA-d4 |
| 8 | 7,12KLCA | 7.641 | 389.3 -> 389.3 | 225 | 10 | 0.005–0.5 | y = 52.86x | 0.9934 | LCA-d4 |
| 9 | LCA | 10.367 | 375.3 -> 375.3 | 235 | 10 | 0.005–0.5 | y = 17.63x | 0.9972 | LCA-d4 |
| 10 | GCA | 5.169 | 464.3 -> 74.0 | 230 | 46 | 0.001–5 | y = 44.935x | 0.9926 | GCA-d4 |
| 11 | GHCA | 4.823 | 464.3 -> 74.0 | 230 | 46 | 0.001–5 | y = 118.29x | 0.9974 | GCDCA-d4 |
| 12 | GCDCA | 6.744 | 448.3 -> 74.0 | 220 | 40 | 0.001–5 | y = 27.884x | 0.9930 | GCDCA-d4 |
| 13 | GLCA | 9.304 | 432.3 -> 74.0 | 225 | 36 | 0.001–5 | y = 17.814x | 0.9996 | GLCA-d4 |
| 14 | TCA | 4.059 | 514.3 -> 79.9 | 300 | 77 | 0.001–5 | y = 32.025x + 0.2914 | 0.9989 | TCA-d5 |
| 15 | TβMCA | 2.787 | 514.3 -> 79.9 | 300 | 77 | 0.005–5 | y = 28.963x − 0.0743 | 0.9966 | TβMCA -d4 |
| 16 | TωMCA | 2.371 | 514.3 -> 79.9 | 300 | 77 | 0.005–5 | y = 28.963x − 0.0743 | 0.9966 | TβMCA -d4 |
| 17 | TCDCA | 5.572 | 498.3 -> 79.9 | 280 | 80 | 0.001–5 | y = 51.689x + 0.0782 | 0.9999 | TUDCA-d5 |
| 18 | TDCA | 5.753 | 498.3 -> 79.9 | 280 | 80 | 0.001–5 | y = 191.74x | 0.9950 | TDCA-d5 |
| 19 | THDCA | 4.412 | 498.3 -> 79.9 | 280 | 80 | 0.001–5 | y = 48.675x − 0.0104 | 0.9999 | TUDCA-d5 |
| 20 | TLCA | 7.463 | 482.3 -> 79.9 | 290 | 80 | 0.001–0.5 | y = 9.4573x | 0.9931 | LCA-d4 |
| 21 | βMCA | 5.812 | 407.3 -> 407.3 | 240 | 10 | 0.001–5 | y = 83.77x + 0.1057 | 0.9995 | βMCA -d5 |
| 22 | UDCA | 6.913 | 391.3 -> 391.3 | 230 | 10 | 0.001–5 | y = 61.645x + 1.3425 | 0.9991 | UDCA-d4 |
| 23 | MuroCA | 6.622 | 391.3 -> 391.3 | 230 | 10 | 0.0025–5 | y = 1.2135x + 0.0362 | 0.9993 | DCA-d4 |
| 24 | GDCA | 6.976 | 448.3 -> 74.0 | 220 | 40 | 0.001–5 | y = 7.6437x + 0.0541 | 0.9997 | GDCA-d4 |
| 25 | GUDCA | 5.387 | 448.3 -> 74.0 | 220 | 40 | 0.005–5 | y = 130.8x − 5.5375 | 0.9982 | GCDCA-d4 |
| 26 | GDHCA | 3.354 | 458.2 -> 74.0 | 205 | 36 | 0.001–5 | y = 88.496x + 4.2369 | 0.9973 | GCDCA-d4 |
| 27 | TαMCA | 2.657 | 514.3 -> 79.9 | 300 | 77 | 0.001–5 | y = 27.164x − 0.0574 | 0.9977 | TβMCA -d4 |
| 28 | THCA | 3.405 | 514.3 -> 79.9 | 300 | 77 | 0.0025–5 | y = 23.806x + 0.1518 | 0.9994 | TDCA-d5 |
| 29 | TUDCA | 4.151 | 498.3 -> 79.9 | 280 | 80 | 0.001–5 | y = 35.379x − 0.1025 | 0.9999 | TUDCA-d5 |
| 30 | TDHCA | 2.314 | 508.3 -> 79.9 | 285 | 72 | 0.001–5 | y = 15.83x + 0.1023 | 0.9997 | TUDCA-d5 |
| Feed Formulation | HP | HP+BAs |
|---|---|---|
| Fish meal a | 10.00 | 10.00 |
| Soybean meal | 18.00 | 18.00 |
| Cottonseed protein concentrated | 18.00 | 18.00 |
| Tapioca flour | 5.00 | 5.00 |
| Wheat flour | 39.80 | 39.74 |
| Soy oil | 4.00 | 4.00 |
| Vitamin and mineral premix b | 4.10 | 4.10 |
| Lecithin oi | 1.00 | 1.00 |
| DL-me | 0.10 | 0.10 |
| Total | 100 | 100 |
| Bile Acid (mg/kg) c | 0 | 600 |
| Analyzed nutrients compositions | ||
| Moisture | 8.24 | 9.11 |
| Crude protein | 30.47 | 29.15 |
| Crude lipid | 7.53 | 7.47 |
| Crude Ash | 7.25 | 7.31 |
| Gross energy (MJ/Kg) | 18.28 | 18.20 |
| HP | HP+BAs | |
|---|---|---|
| AST(U/L) | 103 ± 11.8 * | 62.0 ± 5.84 |
| ALT(U/L) | 73.7 ± 9.97 * | 46.5 ± 5.50 |
| TC (mmol/L) | 5.2 ± 0.308 | 5.3 ± 0.345 |
| BAs in Bile | HP | HP+BAs | |
|---|---|---|---|
| Free BAs | CA | 162.0 ± 51.0 | 149.7 ± 30.0 |
| HDCA | ND | 6.3 ± 2.4 | |
| LCA | ND | 3.8 × 10−2 ± 7.1 × 10−3 | |
| DCA | 7.4 × 10−4 ± 1.9 × 10−4 | ND | |
| CDCA | 6.4 ± 1.7 | 2.7 ± 0.7 | |
| 7,12-KLCA | 5.2 ± 1.0 | 2.8 ± 0.2 | |
| T-BAs | TCA | 2380.6 ± 356.4 | 2091.1 ± 262.4 |
| TCDCA | ND | ND | |
| TβMCA | ND | 49.6 ± 5.4 ** | |
| TωMCA | 18.5 ± 3.7 | 25.3 ± 2.5 | |
| G-BAs | GCA | ND | ND |
| GLCA | 4.0 × 10−2 ± 1.6 × 10−3 | 1.4 × 10−1 ± 1.2 × 10−2 ** | |
| GHCA | 5.5 × 10−3 ± 2.8 × 10−4 | 1.6 × 10−2 ± 2.7 × 10−3 ** | |
| GCDCA | 14.9 ± 0.9 | 34.5 ± 2.6 *** | |
| TBA | 2561.7 ± 346.0 | 2337.7 ± 286.9 |
| BAs in Plasma | HP | HP+BAs | |
|---|---|---|---|
| Free BAs | CA | ND | 5.7 × 10−3 ± 2.0 × 10−3 * |
| HDCA | 3.5 × 10−3 ± 2.1 × 10−3 | 5.0 × 10−3 ± 1.1 × 10−3 | |
| CDCA | ND | 2.0 × 10−3 ± 3.5 × 10−4 * | |
| 7,12-KLCA | ND | ND | |
| LCA | ND | 3.8 × 10−2 ± 7.06 × 10−3 * | |
| T-BAs | TCA | ND | ND |
| TCDCA | 1.1 × 10−1 ± 1.3 × 10−2 | 2.3 × 10−1 ± 7.4 × 10−2 | |
| TβMCA | ND | ND | |
| TωMCA | ND | ND | |
| G-BAs | GCA | ND | ND |
| GLCA | 8.5 × 10−5 ± 3.2 × 10−5 | 1.0 × 10−4 ± 3.10 × 10−8 | |
| GHCA | ND | ND | |
| GCDCA | ND | 3.7 × 10−3 ± 5.2 × 10−4 * | |
| TBA | 1.3 × 10−1 ± 1.9 × 10−2 | 3.2 × 10−1 ± 9.4 × 10−2 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wei, X.; Yao, T.; Fall, F.N.; Xue, M.; Liang, X.; Wang, J.; Du, W.; Gu, X. An Integrated Bile Acids Profile Determination by UHPLC-MS/MS to Identify the Effect of Bile Acids Supplement in High Plant Protein Diet on Common Carp (Cyprinus carpio). Foods 2021, 10, 2465. https://doi.org/10.3390/foods10102465
Wei X, Yao T, Fall FN, Xue M, Liang X, Wang J, Du W, Gu X. An Integrated Bile Acids Profile Determination by UHPLC-MS/MS to Identify the Effect of Bile Acids Supplement in High Plant Protein Diet on Common Carp (Cyprinus carpio). Foods. 2021; 10(10):2465. https://doi.org/10.3390/foods10102465
Chicago/Turabian StyleWei, Xian, Ting Yao, Fatou Ndoye Fall, Min Xue, Xiaofang Liang, Jie Wang, Wenlong Du, and Xu Gu. 2021. "An Integrated Bile Acids Profile Determination by UHPLC-MS/MS to Identify the Effect of Bile Acids Supplement in High Plant Protein Diet on Common Carp (Cyprinus carpio)" Foods 10, no. 10: 2465. https://doi.org/10.3390/foods10102465
APA StyleWei, X., Yao, T., Fall, F. N., Xue, M., Liang, X., Wang, J., Du, W., & Gu, X. (2021). An Integrated Bile Acids Profile Determination by UHPLC-MS/MS to Identify the Effect of Bile Acids Supplement in High Plant Protein Diet on Common Carp (Cyprinus carpio). Foods, 10(10), 2465. https://doi.org/10.3390/foods10102465








