Complexation of 26-Mer Amylose with Egg Yolk Lipids with Different Numbers of Tails Using a Molecular Dynamics Simulation
Abstract
:1. Introduction
2. Simulation Methods
3. Results and Discussion
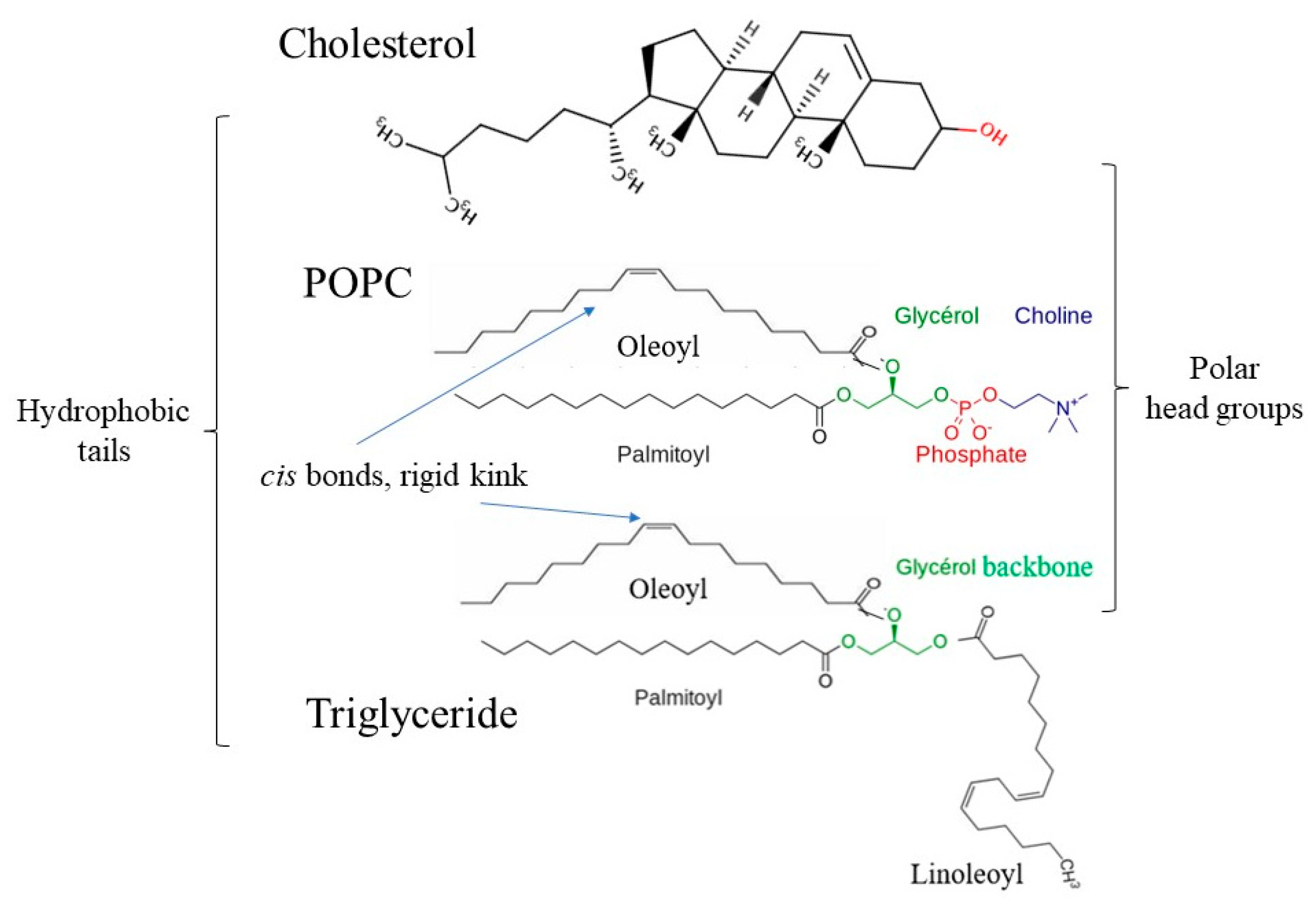
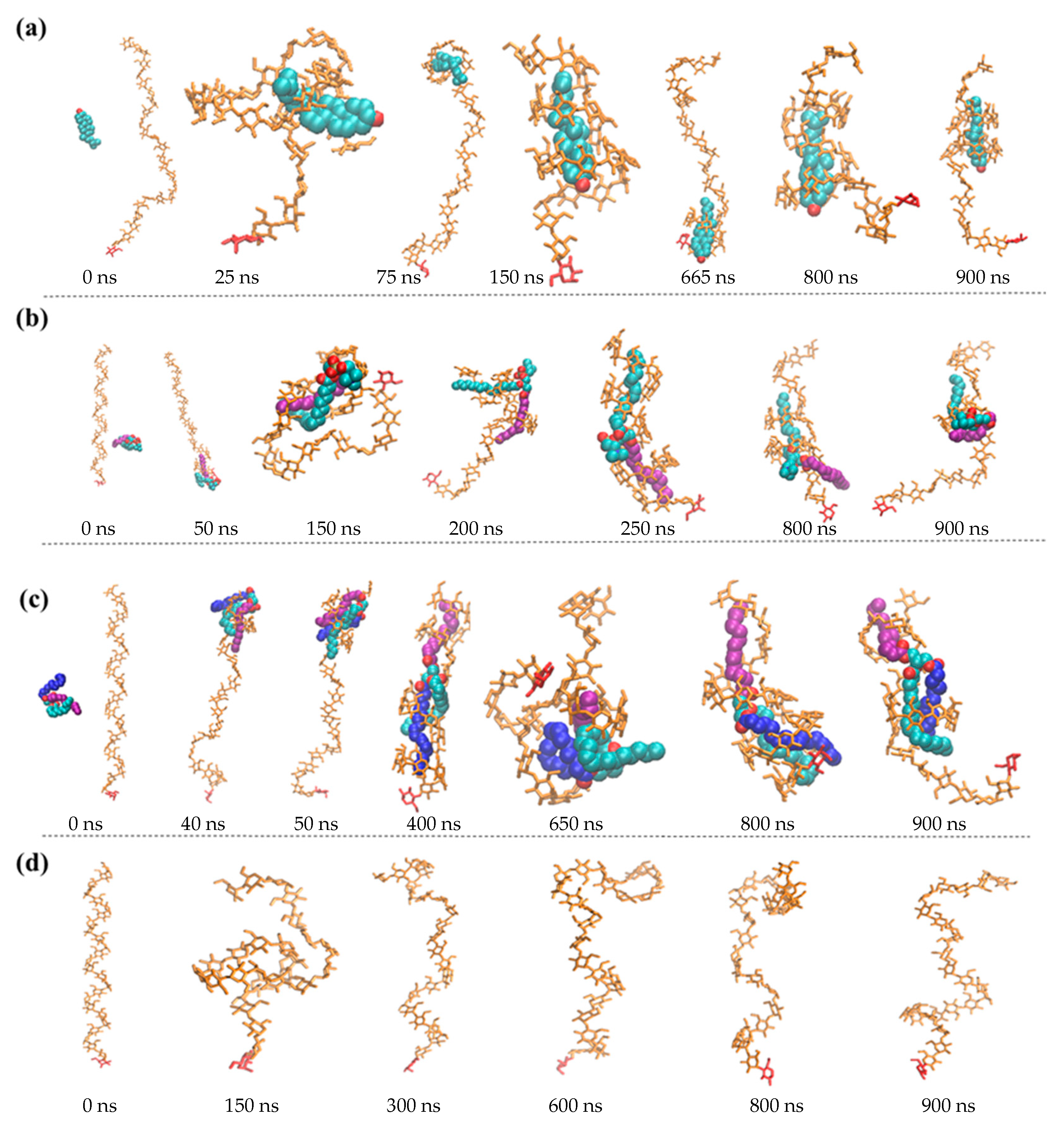
3.1. Structure
3.2. RMSD, Radium of Gyration, and Number of H-Bonds
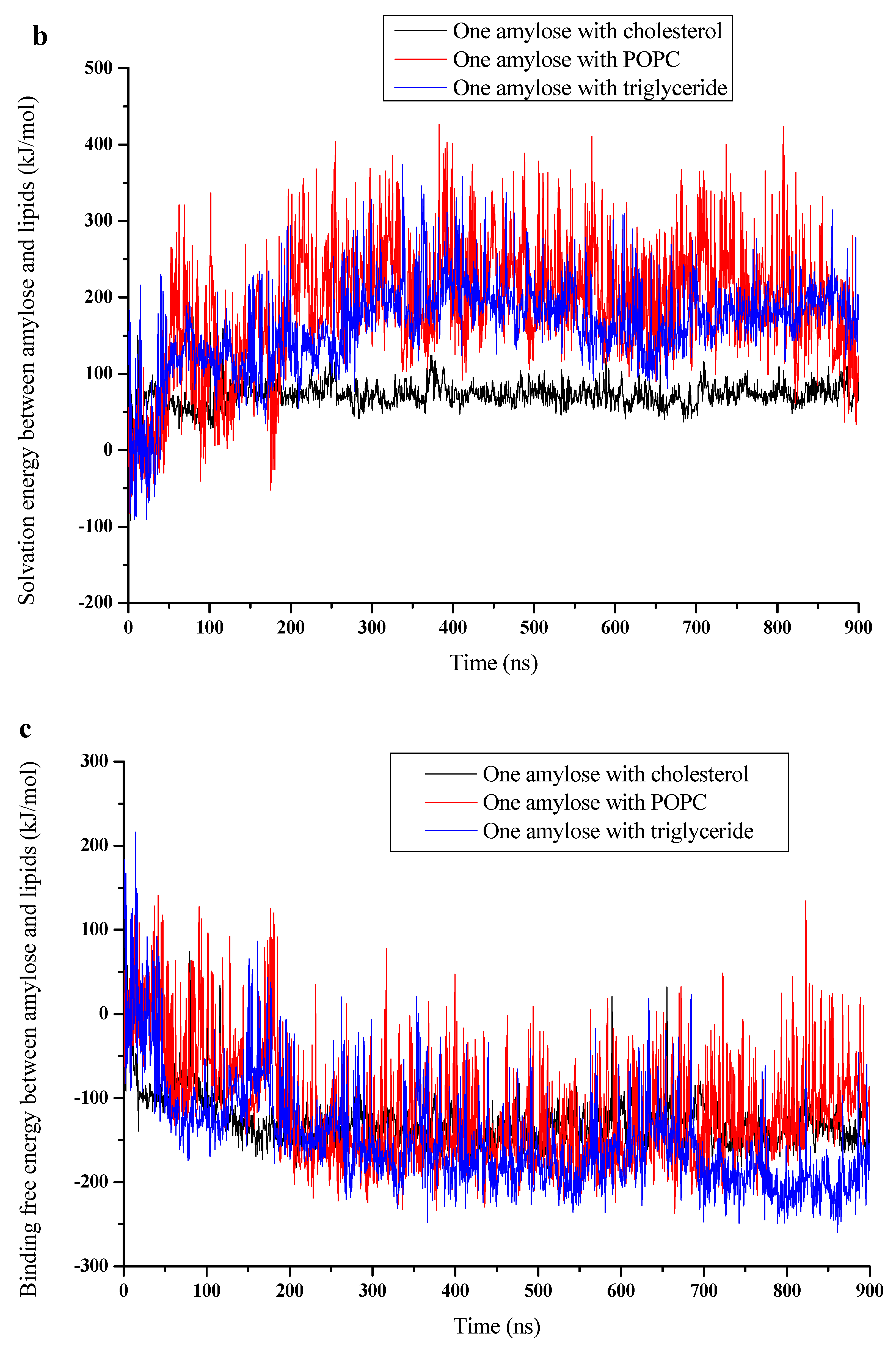
3.3. Binding Energy
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| ALC | amylose-lipid complex |
| DP | degree of polymerization |
| DPPC | dipalmitoylphosphatidylcholine |
| MD | molecular dynamics |
| POPC | 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine |
| RMSD | root mean square deviation |
References
- Obiro, W.C.; Ray, S.S.; Emmambux, M.N. V-amylose structural characteristics, methods of preparation, significance, and potential applications. Food Rev. Int. 2012, 28, 412–438. [Google Scholar] [CrossRef] [Green Version]
- Agyei-Amponsah, J.; Macakova, L.; DeKock, H.L.; Emmambux, M.N. Sensory, tribological, and rheological profiling of “clean label” starch–lipid complexes as fat replacers. Starch-Stärke 2019, 71, 1800340. [Google Scholar] [CrossRef]
- Panyoo, A.E.; Emmambux, M.N. Amylose-lipid complex production and potential health benefits: A mini-review. Starch-Stärke 2017, 69, 1600203. [Google Scholar] [CrossRef]
- Carlson, T.L.-G.; Larsson, K.; Dinh-Nguyen, N.; Krog, N. Study of the amylose-monoglyceride complex by raman-spectroscopy. Starch-Stärke 1979, 31, 222–224. [Google Scholar] [CrossRef]
- Godet, M.C.; Tran, V.; Delage, M.M.; Buleon, A. Molecular modeling of the specific interactions involved in the amylose complexation by fatty-acids. Int. J. Biol. Macromol. 1993, 15, 11–16. [Google Scholar] [CrossRef]
- Kawada, J.; Marchessault, R.H. Solid state NMR and X-ray studies on amylose complexes with small organic molecules. Starch-Stärke 2004, 56, 13–19. [Google Scholar] [CrossRef]
- Heinemann, C.; Zinsli, M.; Renggli, A.; Escher, F.; Conde-Petit, W. Influence of amylose-flavor complexation on build-up and breakdown of starch structures in aqueous food model systems. LWT-Food Sci. Technol. 2005, 38, 885–894. [Google Scholar] [CrossRef]
- Whittam, M.A.; Orford, P.D.; Ring, S.G.; Clark, S.A.; Parker, M.L.; Cairns, P.; Miles, M.J. Aqueous dissolution of crystalline and amorphous amylose-alcohol complexes. Int. J. Biol. Macromol. 1989, 11, 339–344. [Google Scholar] [CrossRef]
- Nuessli, J.; Putaux, J.L.; Le Bail, P.; Buleon, A. Crystal structure of amylose complexes with small ligands. Int. J. Biol. Macromol. 2003, 33, 227–234. [Google Scholar] [CrossRef]
- Rondeau-Mouro, C.; Le Bail, P.; Buleon, A. Structural investigation of amylose complexes with small ligands: Inter- or intra-helical associations? Int. J. Biol. Macromol. 2004, 34, 309–315. [Google Scholar] [CrossRef]
- Biliaderis, C.G.; Page, C.M.; Slade, L.; Sirett, R.R. Thermal behavior of amylose-lipid complexes. Carbohydr. Polym. 1985, 5, 367–389. [Google Scholar] [CrossRef]
- Cheng, L.; Feng, T.; Zhang, B.; Zhu, X.; Hamaker, B.; Zhang, H.; Campanella, O. A molecular dynamics simulation study on the conformational stability of amylose-linoleic acid complex in water. Carbohydr. Polym. 2018, 196, 56–65. [Google Scholar] [CrossRef]
- López, C.A.; de Vries, A.H.; Marrink, S.J. Amylose folding under the influence of lipids. Carbohydr. Res. 2012, 364, 1–7. [Google Scholar] [CrossRef] [Green Version]
- Putseys, J.A.; Lamberts, L.; Delcour, J.A. Amylose-inclusion complexes: Formation, identity and physico-chemical properties. J. Cereal Sci. 2010, 51, 238–247. [Google Scholar] [CrossRef]
- Oyeyinka, S.A.; Singh, S.; Venter, S.L.; Amonsou, E.O. Effect of lipid types on complexation and some physicochemical properties of bambara groundnut starch. Starch-Stärke 2017, 69, 1600158. [Google Scholar] [CrossRef]
- Tang, M.C.; Copeland, L. Analysis of complexes between lipids and wheat starch. Carbohydr. Polym. 2007, 67, 80–85. [Google Scholar] [CrossRef]
- Kawai, K.; Takato, S.; Sasaki, T.; Kajiwara, K. Complex formation, thermal properties, and in-vitro digestibility of gelatinized potato starch-fatty acid mixtures. Food Hydrocoll. 2012, 27, 228–234. [Google Scholar] [CrossRef]
- Bhosale, R.G.; Ziegler, G.R. Preparation of spherulites from amylose-palmitic acid complexes. Carbohydr. Polym. 2010, 80, 53–64. [Google Scholar] [CrossRef]
- Derycke, V.; Vandeputte, G.E.; Vermeylen, R.; De Man, W.; Goderis, B.; Koch, M.; Delcour, J.A. Starch gelatinization and amylose-lipid interactions during rice parboiling investigated by temperature resolved wide angle X-ray scattering and differential scanning calorimetry. J. Cereal Sci. 2005, 42, 334–343. [Google Scholar] [CrossRef]
- Eliasson, A.C.; Finstad, H.; Ljunger, G. A study of starch-lipid interactions for some native and modified maize starches. Starch-Stärke 1988, 40, 95–100. [Google Scholar] [CrossRef]
- Godet, M.C.; Bizot, H.; Buleon, A. Crystallization of amylose-fatty acid complexes prepared with different amylose chain lengths. Carbohydr. Polym. 1995, 27, 47–52. [Google Scholar] [CrossRef]
- Yotsawimonwat, S.; Sriroth, K.; Kaewvichit, S.; Piyachomkwan, K.; Jane, J.L.; Sirithunyalug, J. Effect of ph on complex formation between debranched waxy rice starch and fatty acids. Int. J. Biol. Macromol. 2008, 43, 94–99. [Google Scholar] [CrossRef]
- Gunaratne, A.; Corke, H. Effect of hydroxypropyl beta-cyclodextrin on physical properties and transition parameters of amylose-lipid complexes of native and acetylated starches. Food Chem. 2008, 108, 14–22. [Google Scholar] [CrossRef]
- Guraya, H.S.; Kadan, R.S.; Champagne, E.T. Effect of rice starch-lipid complexes on in vitro digestibility, complexing index, and viscosity. Cereal Chem. 1997, 74, 561–565. [Google Scholar] [CrossRef] [Green Version]
- Eliasson, A.-C.; Carlson, T.L.-G.; Larsson, K. Some effects of starch lipids on the thermal and rheological properties of wheat starch. Starch-Stärke 1981, 33, 130–134. [Google Scholar] [CrossRef]
- Mira, I.; Persson, K.; Villwock, V.K. On the effect of surface active agents and their structure on the temperature-induced changes of normal and waxy wheat starch in aqueous suspension. Part I. Pasting and calorimetric studies. Carbohydr. Polym. 2007, 68, 665–678. [Google Scholar] [CrossRef]
- Czuchajowska, Z.; Sievert, D.; Pomeranz, Y. Enzyme-resistant starch. 4. Effects of complexing lipids. Cereal Chem. 1991, 68, 537–542. [Google Scholar]
- Gudmundsson, M. Effects of an added inclusion-amylose complex on the retrogradation of some starches and amylopectin. Carbohydr. Polym. 1992, 17, 299–304. [Google Scholar] [CrossRef]
- Sang, S.; Chen, Y.; Zhu, X.; Narsimhan, G.; Hu, Q.; Jin, Z.; Xu, X. Effect of egg yolk lipids on structure and properties of wheat starch in steamed bread. J. Cereal Sci. 2019, 86, 77–85. [Google Scholar] [CrossRef]
- Hess, B.; Kutzner, C.; van der Spoel, D.; Lindahl, E. Gromacs 4: Algorithms for highly efficient, load-balanced, and scalable molecular simulation. J. Chem. Theory Comput. 2008, 4, 435–447. [Google Scholar] [CrossRef] [Green Version]
- Lyu, Y.; Zhu, X.; Xiang, N.; Narsimhan, G. Molecular dynamics study of pore formation by melittin in a 1,2-dioleoyl-sn-glycero-3-phosphocholine and 1,2-di(9z-octadecenoyl)-sn-glycero-3-phospho-(1′-rac-glycerol) mixed lipid bilayer. Ind. Eng. Chem. Res. 2015, 54, 10275–10283. [Google Scholar] [CrossRef]
- Payne, M.C.; Teter, M.P.; Allan, D.C.; Arias, T.A.; Joannopoulos, J.D. Iterative minimization techniques for abinitio total-energy calculations—Molecular-dynamics and conjugate gradients. Rev. Mod. Phys. 1992, 64, 1045–1097. [Google Scholar] [CrossRef] [Green Version]
- Kumari, R.; Kumar, R.; Lynn, A. Open Source Drug Discovery Consortium. G_mmpbsa—A GROMACS tool for high-throughput MM-PBSA calculations. J. Chem. Inf. Model. 2014, 54, 1951–1962. [Google Scholar] [CrossRef] [PubMed]
- Immel, S.; Lichtenthaler, F.W. The hydrophobic topographies of amylose and its blue iodine complex. Starch-Stärke 2000, 52, 1–8. [Google Scholar] [CrossRef]
- Koneru, J.K.; Zhu, X.; Mondal, J. Quantitative assessment of the conformational heterogeneity in amylose across force fields. J. Chem. Theory Comput. 2019, 15, 6203–6212. [Google Scholar] [CrossRef]
- Saenger, W. The structure of the blue starch-iodine complex. Naturwissenschaften 1984, 71, 31–36. [Google Scholar] [CrossRef]
- Nuessli, J.; CondePetit, B.; Trommsdorff, U.R.; Escher, F. Influence of starch flavour interactions on rheological properties of low concentration starch systems. Carbohydr. Polym. 1995, 28, 167–170. [Google Scholar] [CrossRef]
- Heinemann, C.; Conde-Petit, B.; Nuessli, J.; Escher, F. Evidence of starch inclusion complexation with lactones. J. Agric. Food Chem. 2001, 49, 1370–1376. [Google Scholar] [CrossRef]
- Biliaderis, C.G.; Galloway, G. Crystallization behavior of amylose-V complexes—Structure property relationships. Carbohydr. Res. 1989, 189, 31–48. [Google Scholar] [CrossRef]
- Buleon, A.; Delage, M.M.; Brisson, J.; Chanzy, H. Single-crystals of V-amylose complexed with isopropanol and acetone. Int. J. Biol. Macromol. 1990, 12, 25–33. [Google Scholar] [CrossRef]
- Neszmelyi, A.; Laszlo, E.; Hollo, J. Biomolecular modeling—An interactive program for the visualization and modeling of carbohydrate (starch and oligosaccharide) complexes in solution. Starch-Stärke 1987, 39, 393–396. [Google Scholar] [CrossRef]
- Yamashita, Y.H.; Ryugo, J.; Monobe, K. Electron-microscopic study on crystals of amylose V complexes. J. Electron Microsc. 1973, 22, 19–26. [Google Scholar]
- Chao, C.; Yu, J.; Wang, S.; Copeland, L.; Wang, S. Mechanisms underlying the formation of complexes between maize starch and lipids. J. Agric. Food Chem. 2018, 66, 272–278. [Google Scholar] [CrossRef] [PubMed]
- Raphaelides, S.; Karkalas, J. Thermal dissociation of amylose-fatty acid complexes. Carbohydr. Res. 1988, 172, 65–82. [Google Scholar] [CrossRef]
- Kong, L.; Bhosale, R.; Ziegler, G.R. Encapsulation and stabilization of beta-carotene by amylose inclusion complexes. Food Res. Int. 2018, 105, 446–452. [Google Scholar] [CrossRef] [PubMed]
- Karkalas, J.; Ma, S.; Morrison, W.R.; Pethrick, R.A. Some factors determining the thermal-properties of amylose inclusion complexes with fatty-acids. Carbohydr. Res. 1995, 268, 233–247. [Google Scholar] [CrossRef]
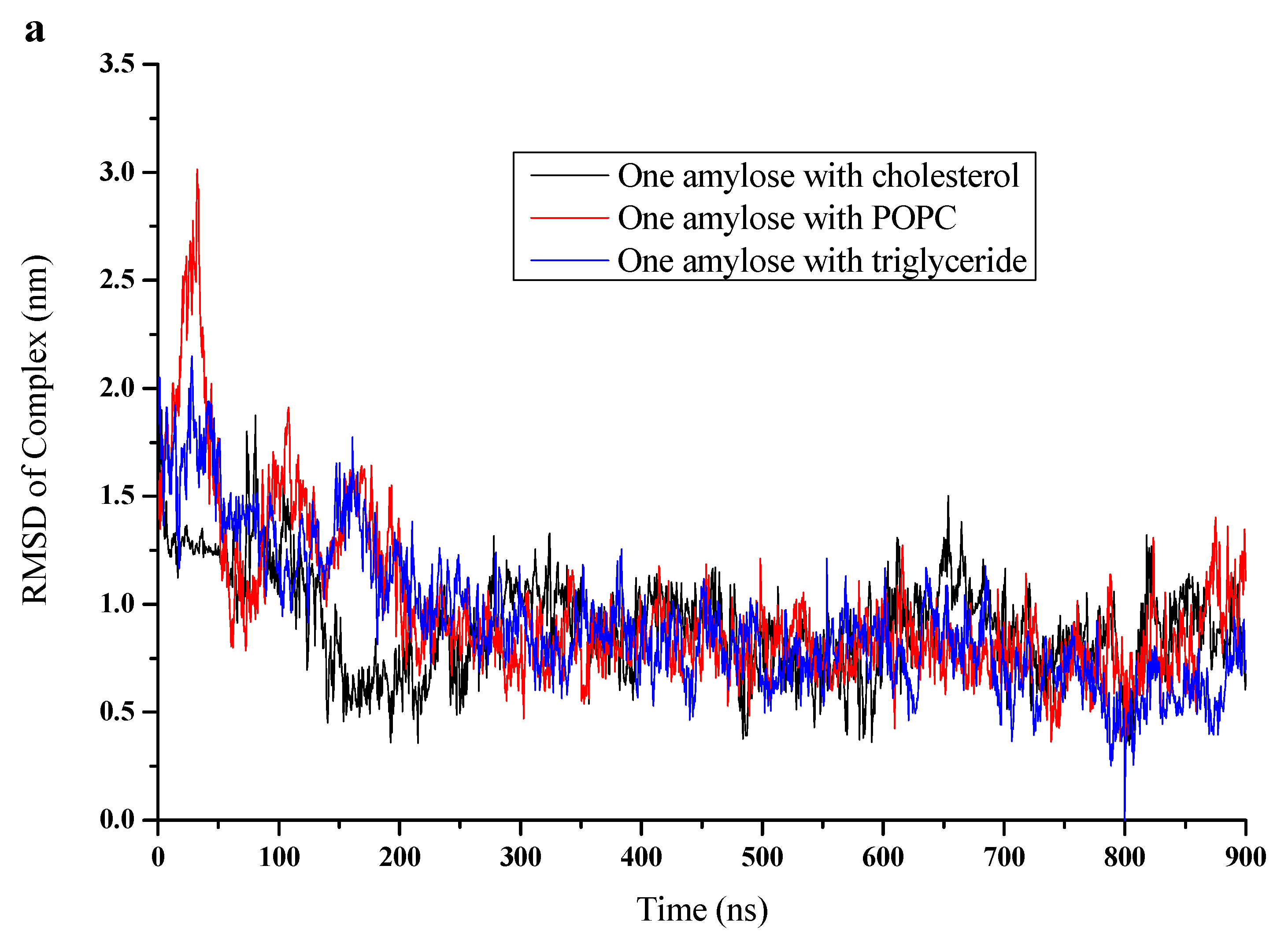
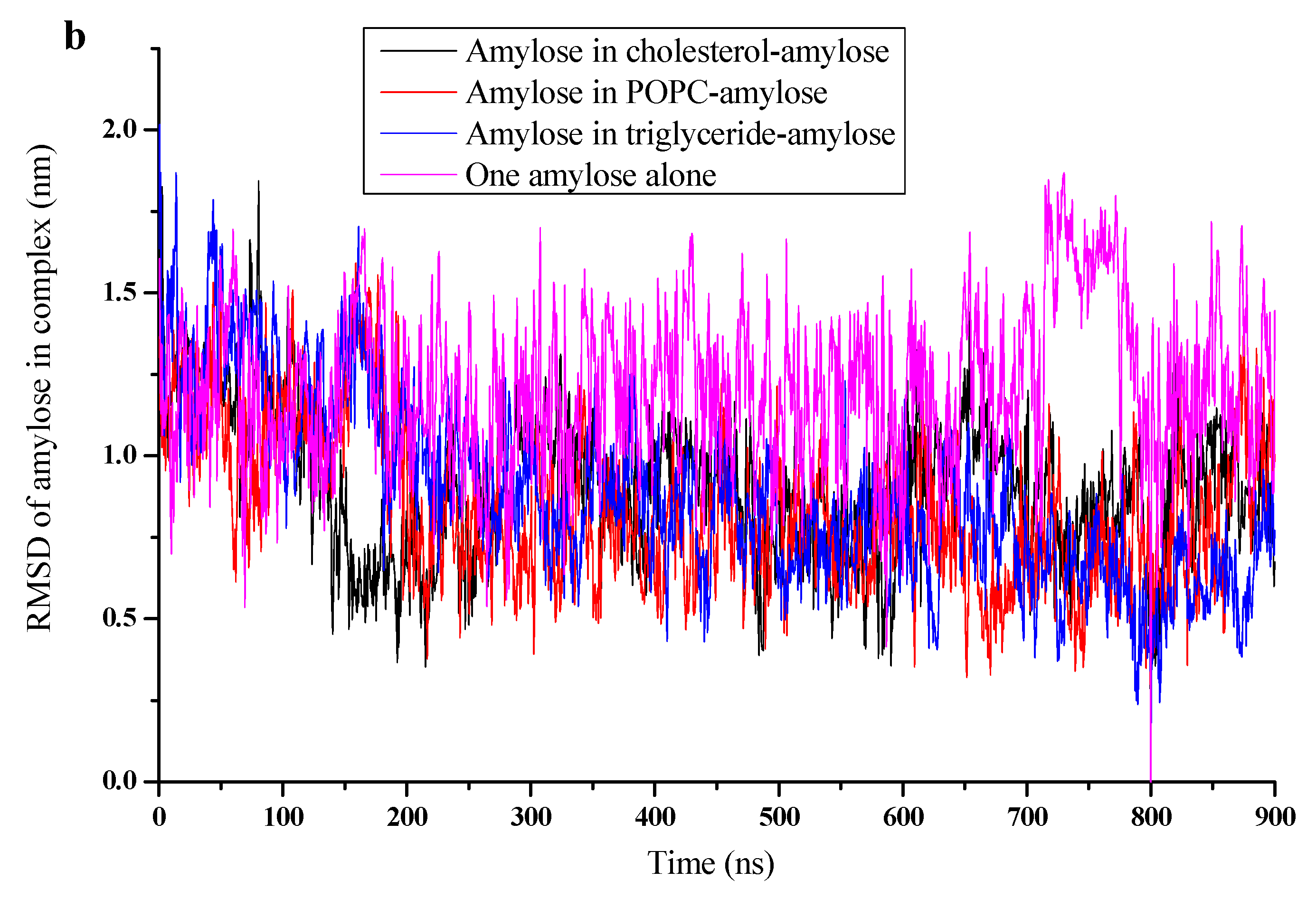


| Simulations | Box Side Length (Nm) | Water Molecule Number | RMSD of Complex (Nm) | RMSD Of Amylose (Nm) | Rg of Complex (Nm) | Rg of Amylose (Nm) | NO. of H-Bonds |
|---|---|---|---|---|---|---|---|
| Cholesterol | 11.6265 | 51,493 | 0.879 | 0.888 | 1.37 | 1.41 | 18 |
| POPC | 11.0265 | 43,296 | 0.862 | 0.829 | 1.55 | 1.6 | 16 |
| Triglyceride | 11.6265 | 51,462 | 0.593 | 0.614 | 1.25 | 1.3 | 21 |
| Amylose alone | 10.6265 | 38,877 | - | 1.19 | - | 1.75 | 10 |
| Helical Regions | Cholesterol | POPC | Triglyceride | |||
|---|---|---|---|---|---|---|
| Parameters | Hydrophobic Tail | Palmitoyl Tail | Oleoyl Tail | Palmitoyl Tail | Oleoyl and Linoleoyl Tails | |
| NO. of glucose units per turn | 8 | 6 | 6 | 6 | 8 | |
| Maximum NO. of glucose units in each helical region | 16 | 13 | 16 | 13 | 18 | |
| Pitch of helical regions (nm) | 0.992 | 0.877 | 0.789 | 0.774 | 1.051 | |
| Inner diameter of helical cavity (nm) | 0.927 | 0.696 | 0.871 | 0.802 | 1.207 | |
| Outer diameter of helical cavity (nm) | 1.744 | 1.080 | 1.249 | 1.314 | 1.530 | |
| Parameters | Van Der Waals (kJ/mol) | Electrostatic (kJ/mol) | MM Potential (kJ/mol) | Solvation (kJ/mol) | Binding Free Energy (kJ/mol) | |
|---|---|---|---|---|---|---|
| Simulations | ||||||
| Cholesterol | −207.62 | −3.88 | −211.5 | 76.6 | −134.89 | |
| POPC | −231.97 | −43.29 | −275.26 | 178.23 | −97.03 | |
| Triglyceride | −363.13 | −22.83 | −385.96 | 187.61 | −198.35 | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sang, S.; Xu, X.; Zhu, X.; Narsimhan, G. Complexation of 26-Mer Amylose with Egg Yolk Lipids with Different Numbers of Tails Using a Molecular Dynamics Simulation. Foods 2021, 10, 2355. https://doi.org/10.3390/foods10102355
Sang S, Xu X, Zhu X, Narsimhan G. Complexation of 26-Mer Amylose with Egg Yolk Lipids with Different Numbers of Tails Using a Molecular Dynamics Simulation. Foods. 2021; 10(10):2355. https://doi.org/10.3390/foods10102355
Chicago/Turabian StyleSang, Shangyuan, Xueming Xu, Xiao Zhu, and Ganesan Narsimhan. 2021. "Complexation of 26-Mer Amylose with Egg Yolk Lipids with Different Numbers of Tails Using a Molecular Dynamics Simulation" Foods 10, no. 10: 2355. https://doi.org/10.3390/foods10102355






