Comprehensive Classification and Regression Modeling of Wine Samples Using 1H NMR Spectra
Abstract
1. Introduction
2. Materials and Methods
2.1. Samples
2.2. Reference Measurements
2.3. 1H NMR Analysis and Spectral Preprocessing
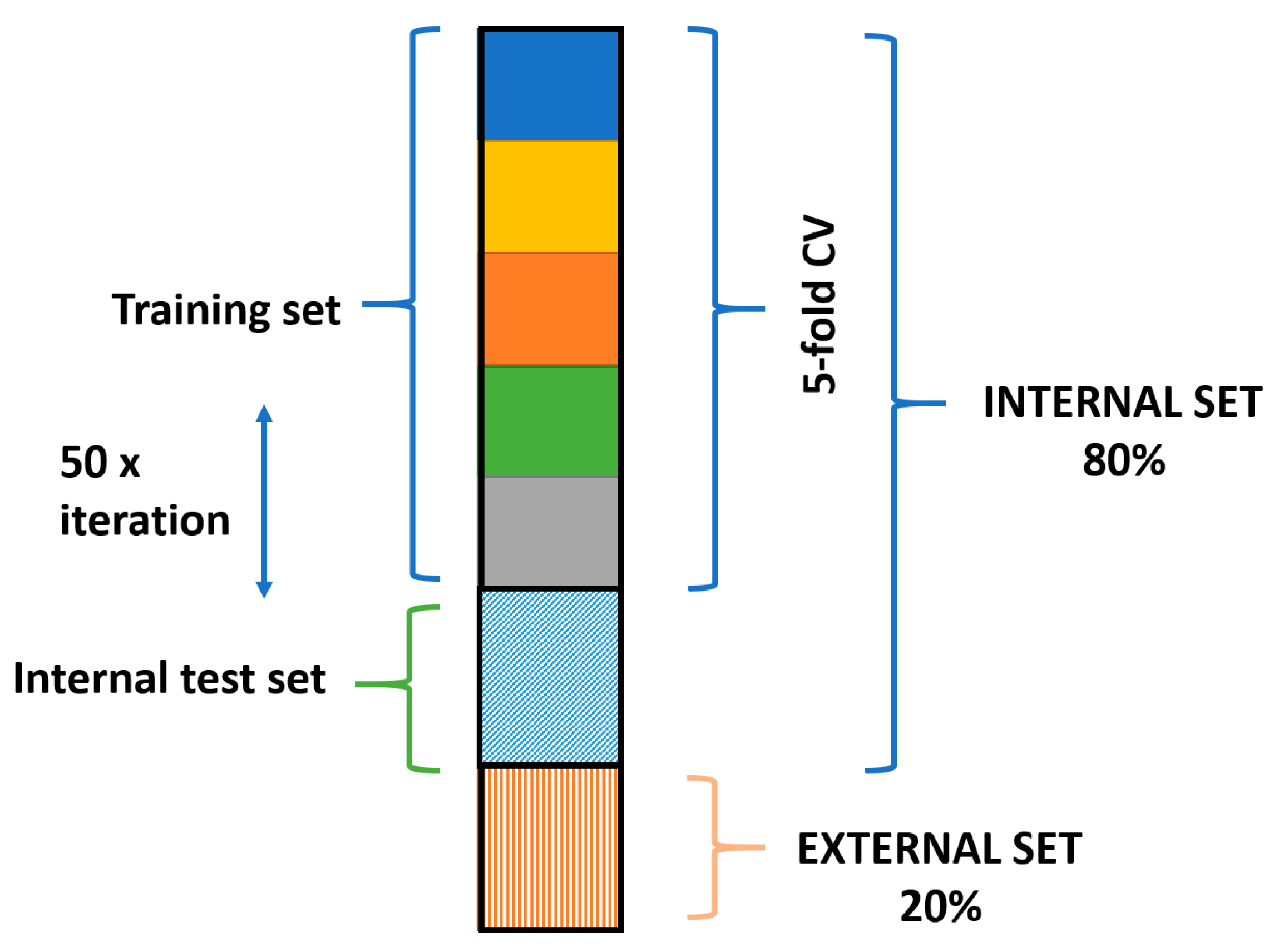
2.4. Chemometric Analysis
3. Results and Discussion
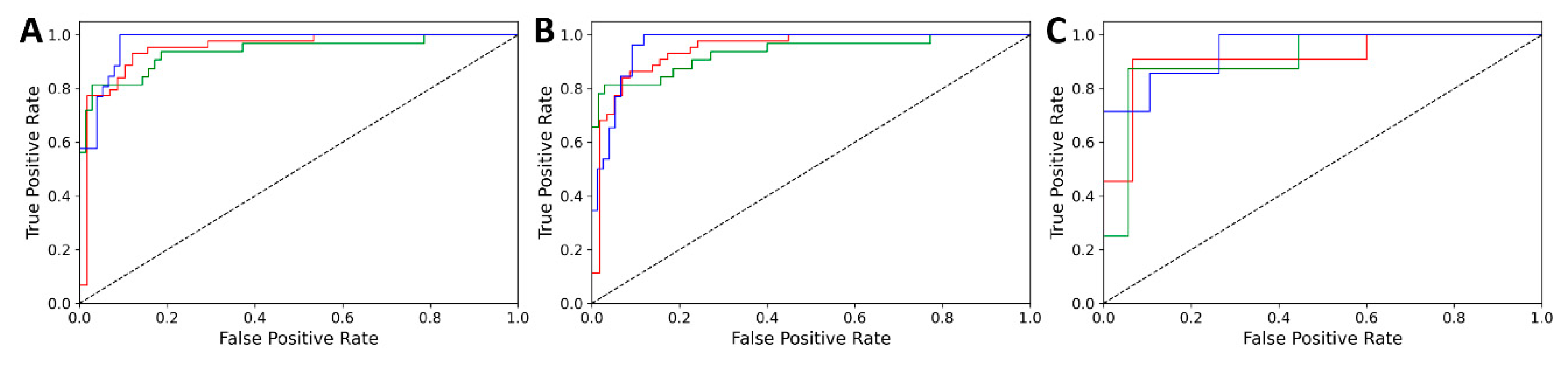
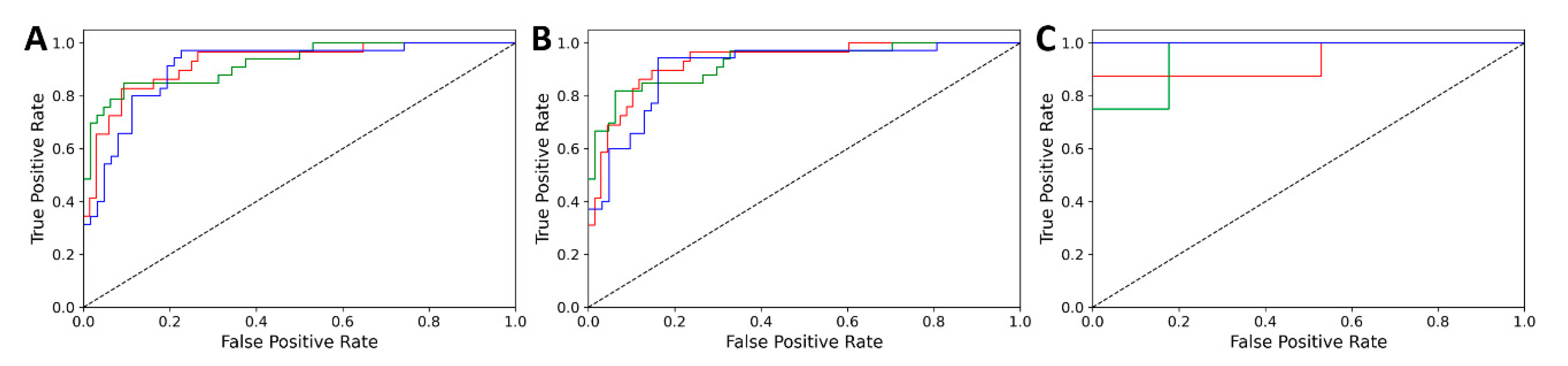
3.1. Classification Results
3.2. PLS Regression Results
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Correia, L.; Gouveia, S.; Martins, P. The European wine export cycle. Wine Econ. Policy 2019, 8, 91–101. [Google Scholar] [CrossRef]
- Pereira, G.E.; Gaudillere, J.P.; Van Leeuwen, C.; Hilbert, G.; Lavialle, O.; Maucourt, M.; Deborde, C.; Moing, A.; Rolin, D. 1H NMR and chemometrics to characterize mature grape berries in four wine-growing areas in Bordeaux, France. J. Agric. Food Chem. 2005, 53, 6382–6389. [Google Scholar] [CrossRef] [PubMed]
- Son, H.S.; Hwang, G.S.; Kim, K.M.; Ahn, H.J.; Park, W.M.; Van Den Berg, F.; Hong, Y.S.; Lee, C.H. Metabolomic studies on geographical grapes and their wines using 1H NMR analysis coupled with multivariate statistics. J. Agric. Food Chem. 2009, 57, 1481–1490. [Google Scholar] [CrossRef] [PubMed]
- Saurina, J. Characterization of wines using compositional profiles and chemometrics. TrAC Trends Anal. Chem. 2010, 29, 234–245. [Google Scholar] [CrossRef]
- Gougeon, L.; da Costa, G.; Guyon, F.; Richard, T. 1H NMR metabolomics applied to Bordeaux red wines. Food Chem. 2019, 301, 125257. [Google Scholar] [CrossRef] [PubMed]
- Hong, Y.S. NMR-based metabolomics in wine science. Magn. Reson. Chem. 2011, 49. [Google Scholar] [CrossRef]
- López-Rituerto, E.; Savorani, F.; Avenoza, A.; Busto, J.H.; Peregrina, J.M.; Engelsen, S.B. Investigations of la Rioja terroir for wine production using 1H NMR metabolomics. J. Agric. Food Chem. 2012, 60, 3452–3461. [Google Scholar] [CrossRef]
- Cassino, C.; Tsolakis, C.; Bonello, F.; Gianotti, V.; Osella, D. Wine evolution during bottle aging, studied by 1H NMR spectroscopy and multivariate statistical analysis. Food Res. Int. 2019, 116, 566–577. [Google Scholar] [CrossRef]
- Fan, S.; Zhong, Q.; Fauhl-Hassek, C.; Pfister, M.K.H.; Horn, B.; Huang, Z. Classification of Chinese wine varieties using 1H NMR spectroscopy combined with multivariate statistical analysis. Food Control 2018, 88, 113–122. [Google Scholar] [CrossRef]
- Brescia, M.A.; Košir, I.J.; Caldarola, V.; Kidrič, J.; Sacco, A. Chemometric classification of Apulian and Slovenian wines using 1H NMR and ICP-OES together with HPICE data. J. Agric. Food Chem. 2003, 51, 21–26. [Google Scholar] [CrossRef]
- Jiménez-Carvelo, A.M.; González-Casado, A.; Bagur-González, M.G.; Cuadros-Rodríguez, L. Alternative data mining/machine learning methods for the analytical evaluation of food quality and authenticity—A review. Food Res. Int. 2019, 122, 25–39. [Google Scholar] [CrossRef] [PubMed]
- López-Rituerto, E.; Cabredo, S.; López, M.; Avenoza, A.; Busto, J.H.; Peregrina, J.M. A thorough study on the use of quantitative 1H NMR in Rioja red wine fermentation processes. J. Agric. Food Chem. 2009, 57, 2112–2118. [Google Scholar] [CrossRef] [PubMed]
- Li, B.Q.; Xu, M.L.; Wang, X.; Zhai, H.L.; Chen, J.; Liu, J.J. An approach to the simultaneous quantitative analysis of metabolites in table wines by 1H NMR self-constructed three-dimensional spectra. Food Chem. 2017, 216, 52–59. [Google Scholar] [CrossRef] [PubMed]
- Larsen, F.H.; Van Den Berg, F.; Engelsen, S.B. An exploratory chemometric study of 1H NMR spectra of table wines. J. Chemom. 2006, 20, 198–208. [Google Scholar] [CrossRef]
- Monakhova, Y.B.; Godelmann, R.; Hermann, A.; Kuballa, T.; Cannet, C.; Schäfer, H.; Spraul, M.; Rutledge, D.N. Synergistic effect of the simultaneous chemometric analysis of 1H NMR spectroscopic and stable isotope (SNIF-NMR, 18O, 13C) data: Application to wine analysis. Anal. Chim. Acta 2014, 833, 29–39. [Google Scholar] [CrossRef]
- Mascellani, A.; Hoca, G.; Babisz, M.; Krska, P.; Kloucek, P.; Havlik, J. 1H NMR chemometric models for classification of Czech wine type and variety. Food Chem. 2021, 339, 127852. [Google Scholar] [CrossRef]
- Magdas, D.A.; Pirnau, A.; Feher, I.; Guyon, F.; Cozar, B.I. Alternative approach of applying 1H NMR in conjunction with chemometrics for wine classification. LWT 2019, 109, 422–428. [Google Scholar] [CrossRef]
- Godelmann, R.; Fang, F.; Humpfer, E.; Schütz, B.; Bansbach, M.; Schäfer, H.; Spraul, M. Targeted and nontargeted wine analysis by 1H NMR spectroscopy combined with multivariate statistical analysis. differentiation of important parameters: Grape variety, geographical origin, year of vintage. J. Agric. Food Chem. 2013, 61, 5610–5619. [Google Scholar] [CrossRef]
- Geana, E.I.; Popescu, R.; Costinel, D.; Dinca, O.R.; Ionete, R.E.; Stefanescu, I.; Artem, V.; Bala, C. Classification of red wines using suitable markers coupled with multivariate statistic analysis. Food Chem. 2016, 192, 1015–1024. [Google Scholar] [CrossRef]
- Košir, I.J.; Kocjančič, M.; Ogrinc, N.; Kidrič, J. Use of SNIF-NMR and IRMS in combination with chemometric methods for the determination of chaptalisation and geographical origin of wines (the example of Slovenian wines). Anal. Chim. Acta 2001, 429, 195–206. [Google Scholar] [CrossRef]
- Silvestri, M.; Elia, A.; Bertelli, D.; Salvatore, E.; Durante, C.; Li Vigni, M.; Marchetti, A.; Cocchi, M. A mid level data fusion strategy for the Varietal Classification of Lambrusco PDO wines. Chemom. Intell. Lab. Syst. 2014, 137, 181–189. [Google Scholar] [CrossRef]
- Godelmann, R.; Kost, C.; Patz, C.D.; Ristow, R.; Wachter, H. Quantitation of compounds in wine using 1H NMR spectroscopy: Description of the method and collaborative study. J. AOAC Int. 2016, 99, 1295–1304. [Google Scholar] [CrossRef] [PubMed]
- Chen, T.; Guestrin, C. XGBoost: A Scalable Tree Boosting Sysytem. In Proceedings of the 22nd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining, San Francisco, CA, USA, 13–17 August 2016; pp. 785–794. [Google Scholar] [CrossRef]
- Friedman, J.H. Greedy Function Approximation: A Gradient Boosting Machine. Ann. Stat. 2001, 29, 1189–1232. [Google Scholar] [CrossRef]
- Hastie, T.; Tibshirani, R.; Friedman, J. Boosting and Additive Trees. In Elements of Statistical Learning. Data Mining, Inference, Prediction; Springer: New York, NY, USA, 2009; pp. 337–387. [Google Scholar]
- Filzmoser, P.; Liebmann, B.; Varmuza, K. Repeated double cross validation. J. Chemom. 2009, 23, 160–171. [Google Scholar] [CrossRef]
- Rácz, A.; Bajusz, D.; Fodor, M.; Héberger, K. Comparison of classification methods with “n-class” receiver operating characteristic curves: A case study of energy drinks. Chemom. Intell. Lab. Syst. 2016, 151, 34–43. [Google Scholar] [CrossRef][Green Version]
- Geladi, P.; Kowalski, B.R. Partial least-squares regression: A tutorial. Anal. Chim. Acta 1986, 185, 1–17. [Google Scholar] [CrossRef]
- Leardi, R.; Lupiáñez González, A. Genetic algorithms applied to feature selection in PLS regression: How and when to use them. Chemom. Intell. Lab. Syst. 1998, 41, 195–207. [Google Scholar] [CrossRef]
- Boulton, R.B.; Singleton, V.L.; Bisson, L.F.; Kunkee, R.E.; Boulton, R.B.; Singleton, V.L.; Bisson, L.F.; Kunkee, R.E. The Role of Sulfur Dioxide in Wine. In Principles and Practices of Winemaking; Springer: Boston, MA, USA, 1999; pp. 448–473. [Google Scholar]
- Tao, J.; Dykes, S.I.; Kilmartin, P.A. Effect of SO2 concentration on polyphenol development during red wine micro-oxygenation. J. Agric. Food Chem. 2007, 55, 6104–6109. [Google Scholar] [CrossRef]

| Type of Analysis | Method | Features | Number of Samples | Analytical Method | Reference |
|---|---|---|---|---|---|
| regression | PLS | ethanol, glycerol, lactic acid, methanol and malic acid | 40 | 1H NMR | [14] |
| regression | Tchebichef moment-PLS | glycerol, ethanol, lactic acid, malic acid, methanol | 40 | 1H NMR 3D spectra | [13] |
| classification | LDA, PLS-DA, FDA, ICA | geographical origin, red wine varieties, year of vintage | 718 | 1H NMR and stable isotopes | [15] |
| classification | RF | white wine varieties | 679 | 1H NMR | [16] |
| classification | LDA | wine varieties, geographical origin | 107 | 1H NMR | [17] |
| classification | LDA, MANOVA | wine varieties, year of vintage, geographical origin | 579 | 1H NMR | [18] |
| classification | LDA | wine varieties, year of vintage | 56 | HPLC, Isotopic analysis, 1H NMR, 13C NMR | [19] |
| classification | PCA, SOM, CA | addition of beet or cane sugar, geographical origin | 50 | SNIF-NMR, Isotopic ratio | [20] |
| classification | PLS-DA | wine varieties | 58 | HPLC, EEM, 1H NMR | [21] |
| Type of Method | Model | Training Set | Test Set |
|---|---|---|---|
| PLS Regression | Density | 278 | 71 |
| Total alcohol concentration | 299 | 71 | |
| Total sugar concentration | 71 | 7 | |
| Total SO2 concentration | 250 | 55 | |
| Classification | White wine varieties | 102 | 26 |
| Red wine varieties | 97 | 25 |
| Dataset | Validation | Accuracy | AUC |
|---|---|---|---|
| White | CV | 0.833 | 0.952 |
| Internal test | 0.814 | 0.948 | |
| External test | 0.808 | 0.922 | |
| Red | CV | 0.814 | 0.920 |
| Internal test | 0.804 | 0.921 | |
| External test | 0.800 | 0.965 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Barátossy, G.; Berinkeiné Donkó, M.; Csikorné Vásárhelyi, H.; Héberger, K.; Rácz, A. Comprehensive Classification and Regression Modeling of Wine Samples Using 1H NMR Spectra. Foods 2021, 10, 64. https://doi.org/10.3390/foods10010064
Barátossy G, Berinkeiné Donkó M, Csikorné Vásárhelyi H, Héberger K, Rácz A. Comprehensive Classification and Regression Modeling of Wine Samples Using 1H NMR Spectra. Foods. 2021; 10(1):64. https://doi.org/10.3390/foods10010064
Chicago/Turabian StyleBarátossy, Gábor, Mária Berinkeiné Donkó, Helga Csikorné Vásárhelyi, Károly Héberger, and Anita Rácz. 2021. "Comprehensive Classification and Regression Modeling of Wine Samples Using 1H NMR Spectra" Foods 10, no. 1: 64. https://doi.org/10.3390/foods10010064
APA StyleBarátossy, G., Berinkeiné Donkó, M., Csikorné Vásárhelyi, H., Héberger, K., & Rácz, A. (2021). Comprehensive Classification and Regression Modeling of Wine Samples Using 1H NMR Spectra. Foods, 10(1), 64. https://doi.org/10.3390/foods10010064







