Molecular Characterization of Cronobacter sakazakii Strains Isolated from Powdered Milk
Abstract
1. Introduction
2. Materials and Methods
2.1. Strains Used in This Study
2.2. Isolation and Primary Species Identification of Cronobacter Sakazakii Isolates
2.3. Whole-Genome Sequencing (WGS) Data Analysis
2.4. O-Serotype Determination Analysis
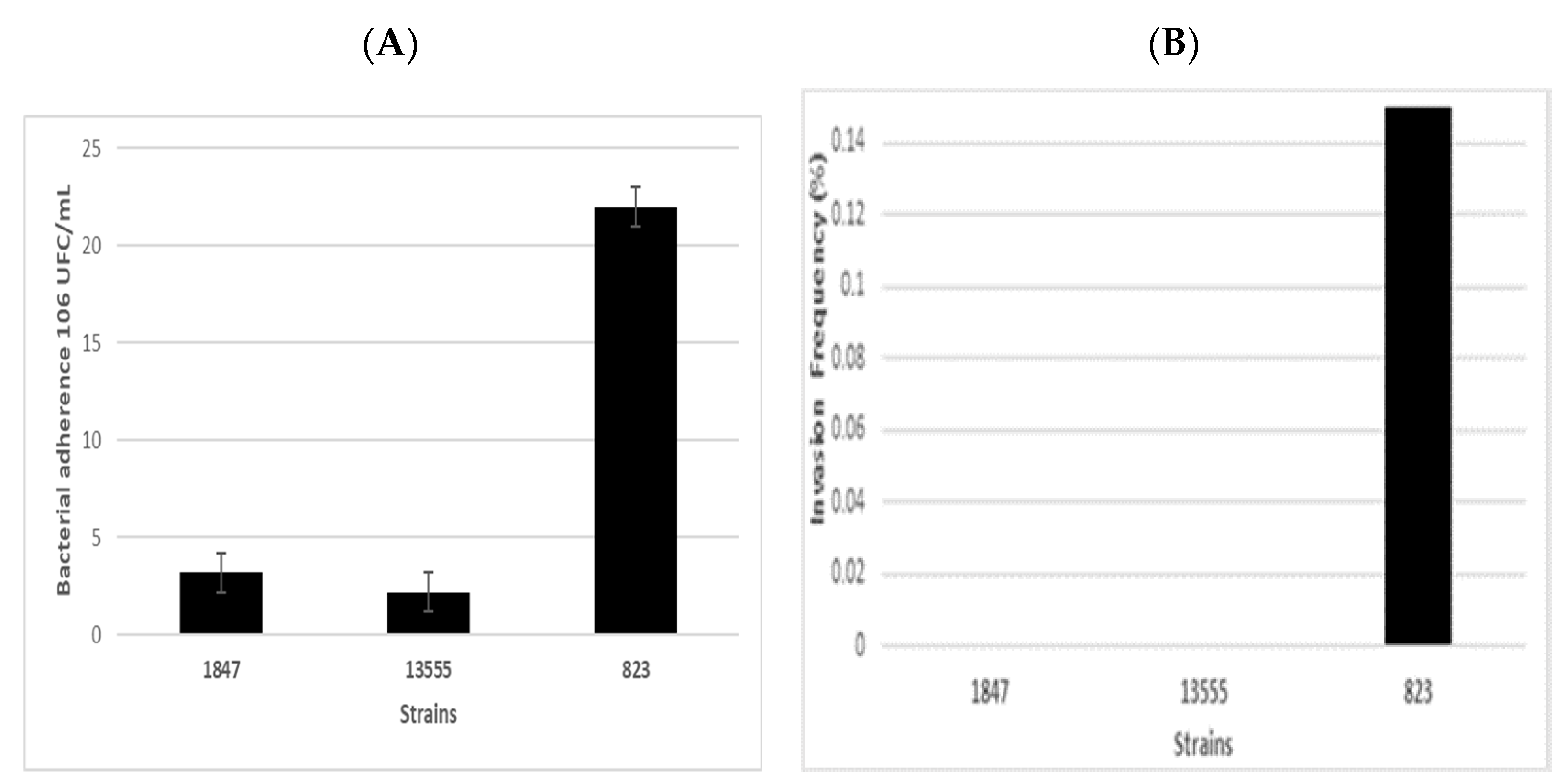
2.5. Adherence Assay
2.6. Invasion Assay
2.7. In Vitro Virulence Gene Detection
2.8. Antibiotic Resistance Profile
2.9. In Silico Detection of Virulence and Antibiotic Resistance Genes from Whole Genome Sequencing (WGS) Data
2.10. Plasmid Detection
2.11. Profiling of CRISPR-Cas Loci
3. Results and Discussion
4. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Iversen, C.; Lehner, A.; Mullane, N.; Bidlas, E.; Cleenwerck, I.; Marugg, J.; Fanning, S.; Stephan, R.; Joosten, H. The taxonomy of Enterobacter sakazakii: Proposal of a new genus Cronobacter gen. nov.and descriptions of Cronobacter sakazakii comb. nov. Cronobacter sakazakii subsp. sakazakii, comb. nov., Cronobacter sakazakii subsp. malonaticus subsp. nov., Cronobacter turicensis sp. nov., Cronobacter muytjensii sp. nov., Cronobacter dublinensis sp. nov.and Cronobacter genomospecies 1. BMC Evol. Biol. 2007, 7, 64–74. [Google Scholar]
- Iversen, C.; Mullane, N.; Mc Cardell, B.; Tall, B.; Lehner, A.; Fanning, S.; Stephan, R.; Joosten, H. Cronobacter gen. nov., a new genus to accommodate the biogroups of Enterobacter sakazakii, and proposal of Cronobacter sakazakii gen. nov.comb. nov., C. malonaticus sp. nov., C. turicensis sp. nov., C. muytjensii sp. nov., C. dublinensis sp. nov., Cronobacter genomospecies 1, and of three subspecies, C. dublinensis sp. nov.subsp. dublinensis subsp. nov., C. dublinensis sp. nov.subsp. lausannensis subsp. nov., and C. dublinensis sp. nov.subsp. lactaridi subsp. nov. Int. J. Syst. Evol. Microbiol. 2008, 58, 1442–1447. [Google Scholar]
- Joseph, S.; Cetinkaya, E.; Drahovska, H.; Levican, A.; Figueras, M.; Forsythe, S. Cronobacter condimenti sp. Nov., isolated from spiced meat, and Cronobacter universalis sp. Nov., a species designation for Cronobacter sp. Geneomoespecies 1, recovered from a leg infection, water and food ingredients. Int. J. Syst. Evol. Microbiol. 2012, 62, 1277–1283. [Google Scholar] [CrossRef]
- Holý, O.; Petrželová, J.; Hanulík, V.; Chromá, M.; Matoušková, I.; Forsythe, S. Epidemiology of Cronobacter spp. isolates from patients admitted to the Olomouc University Hospital (Czech Republic). Epidemiol. Mikrobiol. Imunol. 2014, 63, 69–72. [Google Scholar] [PubMed]
- Holý, O.; Forsythe, S. Cronobacter spp. as emerging causes of healthcare-associated infection. J. Hosp. Infect. 2014, 86, 169–177. [Google Scholar] [CrossRef]
- Forsythe, S.J. Updates on the Cronobacter Genus. Annu. Rev. Food Sci. Technol. 2018, 25, 23–44. [Google Scholar] [CrossRef] [PubMed]
- Bowen, A.; Braden, C. Invasive Enterobacter sakazakii disease in infants. Emerg. Infect. Dis. 2006, 12, 1185–1189. [Google Scholar] [CrossRef] [PubMed]
- Stoll, B.J.; Hansen, N.; Fanaroff, A.; Lemons, J.A. Enterobacter sakazakii is a rare cause of neonatal septicemia or meningitis in VLBW infants. J. Pediatr. 2004, 144, 821–823. [Google Scholar] [PubMed]
- Hunter, C.J.; Bean, J.F. Cronobacter: An emerging opportunistic pathogen associated with neonatal meningitis, sepsis and necrotizing enterocolitis. J. Perinatol. 2013, 33, 581–585. [Google Scholar] [CrossRef]
- FAO; WHO. Enterobacter Sakazakii (Cronobacter spp) in Powdered Follow-Up Formulae; Microbiological Risk Assessment Series; WHO Press Publisher: Italy, Rome, 2008; Volume 15, pp. 1–105. [Google Scholar]
- Baumgartner, A.; Grand, M.; Liniger, M.; Iversen, C. Detection and frequency of Cronobacter spp. (Enterobacter sakazakii) in different categories of ready-to-eat foods other than infant formula. Int. J. Food Microbiol. 2009, 136, 189–192. [Google Scholar] [CrossRef]
- Kalyantanda, G.; Shumyak, L.; Archibald, L.K. Cronobacter species contamination of powdered infant formula and the implications for neonatal health. Front. Pediatr. 2015, 3, 56. [Google Scholar] [CrossRef] [PubMed]
- Caubilla-Barron, J.; Forsythe, S. Dry stress and survival time of Enterobacter sakazakii and other Enterobacteriaceae in dehydrated powdered infant formula. J. Food Prot. 2007, 70, 2111–2117. [Google Scholar] [CrossRef] [PubMed]
- Chap, J.; Jackson, P.; Siqueira, R.; Gaspar, N.; Quintas, C.; Park, J.; Osaili, T.; Shaker, R.; Jaradat, Z.; Hartantyo, S.; et al. International survey of Cronobacter sakazakii and other Cronobacter spp. in follow up formulas and infant foods. Int. J. Food Microbiol. 2009, 136, 185–188. [Google Scholar] [CrossRef] [PubMed]
- Siqueira, R.F.; da Silva, N.; Junqueira, V.; Kajsik, M.; Forsythe, S.; Pereira, J. Screening for Cronobacter species in powdered and reconstituted infant formulas and from equipment used in formula preparation in maternity hospitals. Ann. Nut. Met. 2013, 63, 62–68. [Google Scholar] [CrossRef] [PubMed]
- Parra, J.; Oliveras, L.; Rodriguez, A.; Riffo, F.; Jackson, E.; Forsythe, S. Riesgo por Cronobacter sakazakii en leches en polvo para la nutrición de lactantes. Rev. Chil. Nut. 2015, 42, 83–89. [Google Scholar] [CrossRef]
- Parra-Flores, J.; Maury-Sintjago, E.; Rodriguez-Fernández, A.; Acuña, S.; Cerda, F.; Aguirre, J.; Holý, O. Microbiological quality of powdered infant formula in Latin America. J. Food. Prot. 2020, 83, 534–541. [Google Scholar] [CrossRef]
- Molloy, C.; Cagney, C.; O’Brien, S.; Iversen, C.; Fanning, S.; Duffy, G. Surveillance and characterization by Pulsed-Field Gel Electrophoresis of Cronobacter spp in farming and domestic environments, food production animals and retails foods. Int. J. Food Microbiol. 2009, 136, 198–238. [Google Scholar] [CrossRef]
- Carvalho, G.; Calarga, A.; Teodoro, J.; Queiroz, M.; Astudillo-Trujillo, C.; Levy, C.; Brocchi, M.; Kabuki, D. Isolation, comparison of identification methods and antibiotic resistance of Cronobacter spp. in infant foods. Food Res. Int. 2020, 137, 109643. [Google Scholar] [CrossRef]
- Parra-Flores, J.; Aguirre, J.; Juneja, V.; Jackson, E.; Cruz, A.; Silva, J.; Forsythe, S. Virulence and Antibiotic Resistance Profiles of Cronobacter sakazakii and Enterobacter spp. Involved in the Diarrheic Hemorrhagic Outbreak in Mexico. Front. Microbiol. 2018, 9, 2206. [Google Scholar] [CrossRef]
- Fei, P.; Jiang, Y.; Yuan, X.; Yang, T.; Chen, J.; Wang, Z.; Kang, H.; Forsythe, S. Antibiotic and Desiccation Resistance of Cronobacter sakazakii and C. malonaticus Isolates from Powdered Infant Formula and Processing Environments. Front. Microbiol. 2017, 8, 316. [Google Scholar] [CrossRef]
- Cruz, A.; Xicohtencatl, J.; Gonzalez, B.; Bobadilla, M.; Eslava, C.; Rosas, I. Virulence traits in Cronobacter species isolated from different sources. Can. J. Microbiol. 2011, 7, 735–744. [Google Scholar] [CrossRef] [PubMed]
- Holý, O.; Cruz-Cordova, A.; Xicohtencatl-Cortés, J.; Hochel, I.; Parra-Flores, J.; Petrzelova, J.; Facevicova, K.; Forsythe, S.; Alsonosi, A. Occurrence of virulence factors in Cronobacter sakazakii and Cronobacter malonaticus originated from clinical samples. Microb. Pathog. 2019, 127, 250–256. [Google Scholar] [CrossRef] [PubMed]
- Hamby, S.; Joseph, S.; Forsythe, S.; Chuzhanova, N. In Silico identification of pathogenic strains of Cronobacter from biochemical data reveals association of inositol fermentation with pathogenicity. BMC Microbiol. 2011, 11, 204–213. [Google Scholar] [CrossRef] [PubMed]
- Townsend, S.; Hurrell, E.; Forsythe, S. Virulence studies of Enterobacter sakazakii isolates associated with a neonatal intensive care unit outbreak. BMC Microbiol. 2008, 8, 64. [Google Scholar] [CrossRef] [PubMed]
- Franco, A.; Kothary, M.; Gopinath, G.; Jarvis, K.; Grim, C.; Hu, L.; Datta, A.; McCardell, B.A.; Tall, B.D. Cpa, the outer membrane protease of Cronobacter sakazakii, activates plasminogen and mediates resistance to serum bactericidal activity. Infect. Immun. 2011, 79, 1578–1587. [Google Scholar] [CrossRef] [PubMed]
- Cruz-Córdova, A.; Rocha-Ramírez, L.; Ochoa, S.; Gónzalez-Pedrajo, B.; Espinosa, N.; Eslava, C.; Hernández-Chiñas, U.; Mendoza-Hernández, G.; Rodríguez-Leviz, A.; Valencia-Mayoral, P.; et al. Flagella from five Cronobacter species induce pro-inflammatory cytokines in macrophage derivatives from human monocytes. PLoS ONE 2012, 7, e52091. [Google Scholar] [CrossRef]
- Kim, K.; Kim, K.; Choi, J.; Lim-Jeong, A.; Lee, J.; Hwang, S.; Ryu, S. Outer Membrane Proteins A (OmpA) and X (OmpX) Are Essential for Basolateral Invasion of Cronobacter sakazakii. Appl. Environ. Microbiol. 2010, 76, 5188–5198. [Google Scholar] [CrossRef]
- Ogrodzki, P.; Forsythe, S. Capsular profiling of the Cronobacter genus and the association of specific Cronobacter sakazakii and C. malonaticus capsule types with neonatal meningitis and necrotizing enterocolitis. BMC Genom. 2015, 16, 758. [Google Scholar] [CrossRef]
- Masood, N.; Moore, K.; Farbos, A.; Hariri, S.; Block, C.; Paszkiewicz, K.; Dickins, B.; McNally, A.; Forsythe, S. Draft Genome Sequence of a Meningitic Isolate of Cronobacter sakazakii Clonal Complex 4, Strain 8399. Genome Announc. 2013, 1, e00833-13. [Google Scholar] [CrossRef]
- Ogrodzki, P.; Forsythe, S.J. DNA-Sequence Based Typing of the Cronobacter Genus Using MLST, CRISPR-cas Array and Capsular Profiling. Front. Microbiol. 2017, 8, 1875. [Google Scholar] [CrossRef]
- Jolany vangah, S.; Katalani, C.; Boone, H.; Abbas Hajizade, A.; Ahmadian, G. CRISPR-Based Diagnosis of Infectious and Noninfectious Diseases. Biol. Proced. Online 2020, 22, 22. [Google Scholar] [CrossRef] [PubMed]
- Baldwin, A.; Loughlin, M.; Caubilla-Barron, J.; Kucerova, E.; Manning, G.; Dowson, C.; Forsythe, S. Multilocus sequence typing of Cronobacter sakazakii and Cronobacter malonaticus reveals stable clonal structures with clinical significance, which do not correlate with biotypes. BMC Microbiol. 2009, 9, 223. [Google Scholar] [CrossRef] [PubMed]
- Forsythe, S.J.; Dickins, B.; Jolley, K.A. Cronobacter, the emergent bacterial pathogen Enterobacter sakazakii comes of age; MLST and whole genome sequence analysis. BMC Genom. 2014, 15, 1121. [Google Scholar] [CrossRef] [PubMed]
- Grad, Y.; Lipsitch, M. Epidemiologic data and pathogen genome sequences: A powerful synergy for public health. Genome Biol. 2014, 15, 538. [Google Scholar] [CrossRef]
- Iversen, C.; Forsythe, S.J. Isolation of Enterobacter sakazakii and other Enterobacteriaceae from powdered infant formula milk and related products. Food Microbiol. 2004, 21, 771–776. [Google Scholar] [CrossRef]
- Lepuschitz, S.; Sorschag, S.; Springer, B.; Allerberger, F.; Ruppitsch, W. Draft genome sequence of carbapenemase-producing Serratia marcescens isolated from a patient with chronic obstructive pulmonary disease. Genome Announc. 2017, 5, e01288-17. [Google Scholar] [CrossRef]
- Lepuschitz, S.; Ruppitsch, W.; Pekard-Amenitsch, S.; Forsythe, S.J.; Cormican, M.; Mach, R.L.; Piérard, D.; Allerberger, F.; the EUCRONI Study Group. Multicenter Study of Cronobacter sakazakii Infections in Humans, Europe, 2017. Emerg. Infect. Dis. 2019, 25, 515–522. [Google Scholar] [CrossRef]
- Bankevich, A.; Nurk, S.; Antipov, D.; Gurevich, A.A.; Dvorkin, M.; Kulikov, A.S.; Lesin, V.M.; Nikolenko, S.; Pham, S.; Prjibelski, A.; et al. SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 2012, 19, 455–477. [Google Scholar] [CrossRef]
- Ruppitsch, W.; Pietzka, A.; Prior, K.; Bletz, S.; Fernandez, H.L.; Allerberger, F.; Harmsen, D.; Mellmann, A. Defining and evaluating a core genome MLST scheme for whole genome sequence-based typing of Listeria monocytogenes. J. Clin. Microbiol. 2015, 53, 2869–2876. [Google Scholar] [CrossRef]
- Jia, B.; Raphenya, A.R.; Alcock, B.; Waglechner, N.; Guo, P.; Tsang, K.; Lago, B.; Dave, B.; Pereira, S.; Sharma, A.; et al. CARD 2017: Expansion and model-centric curation of the comprehensive antibiotic resistance database. Nucleic Acids Res. 2017, 45, D566–D573. [Google Scholar] [CrossRef]
- Carattoli, A.; Zankari, E.; García-Fernández, A.; Voldby-Larsen, M.; Lund, O.; Villa, L.; Møller-Aarestrup, F.; Hasman, H. In silico detection and typing of plasmids using PlasmidFinder and plasmid multilocus sequence typing. Antimicrob. Agents Chemother. 2014, 58, 3895–3903. [Google Scholar] [CrossRef] [PubMed]
- Couvin, D.; Bernheim, A.; Toffano-Nioche, C.; Touchon, M.; Michalik, J.; Néron, B.; Rocha, E.; Vergnaud, G.; Gautheret, D.; Pourcel, C. CRISPRCasFinder, an update of CRISRFinder, includes a portable version, enhanced performance and integrates search for Cas proteins. Nucleic Acids Res. 2018, 46, 246–251. [Google Scholar] [CrossRef]
- Ge, R.; Mai, G.; Wang, P.; Zhou, M.; Luo, Y.; Cai, Y.; Zhou, F. CRISPRdigger: Detecting CRISPRs with better direct repeat annotations. Sci. Rep. 2016, 6, 32942. [Google Scholar] [CrossRef] [PubMed]
- Joseph, S.; Forsythe, S. Insights into the emergent bacterial pathogen Cronobacter spp., generated by multilocus sequence typing and analysis. Front. Food Microbiol. 2012, 3, 397. [Google Scholar] [CrossRef] [PubMed]
- Dugan, K. Advances in Understanding Bacterial Pathogenesis Gained from Whole-Genome Sequencing and Phylogenetics. Cell Host Microbe 2016, 19, 599–610. [Google Scholar]
- Hunter, C.J.; Singamsetty, V.K.; Chokshi, N.K.; Boyle, P.; Camerini, V.; Grishin, A.V.; Upperman, J.S.; Ford, H.R.; Prasadarao, N.V. Enterobacter sakazakii enhances epithelial cell injury by inducing apoptosis in a rat model of necrotizing enterocolitis. J Infect. Dis. 2008, 198, 586–593. [Google Scholar] [CrossRef]
- Mange, J.P.; Stephan, R.; Borel, L.; Wild, P.; Kim, K.S.; Pospischil, A.; Lenher, A. Adhesive properties of Enterobacter sakazakii to human epithelial and brain microvascular endothelial cells. BMC Microbiol. 2006, 6, 58–68. [Google Scholar] [CrossRef]
- Townsend, S.; Hurrell, E.; Gonzalez-Gomez, I.; Lowe, J.; Frye, J.; Forsythe, S.; Badger, J. Enterobacter sakazakii invades brain capillary endothelial cells, persists in human macrophages influencing cytokine secretion and induces severe brain pathology in the neonatal rat. Microbiology 2007, 153, 3538–3547. [Google Scholar] [CrossRef]
- Ye, Y.; Li, H.; Ling, N.; Han, Y.; Wu, Q.; Xu, X.; Jiao, R.; Gao, J. Identification of potential virulence factors of Cronobacter sakazakii isolates by comparative proteomic analysis. Int. J. Food Microbiol. 2016, 217, 182–188. [Google Scholar] [CrossRef]
- Quintero-Villegas, M.; Wittke, A.; Hutkins, R. Adherence Inhibition of Cronobacter sakazakii to Intestinal Epithelial Cells by Lactoferrin. Curr. Microbiol. 2014, 69, 574–579. [Google Scholar] [CrossRef]
- Alsonosi, A.; Holý, O.; Forsythe, S. Characterization of the pathogenicity of clinical Cronobacter malonaticus strains based on the tissue culture investigations. Antonie Leeuwenhoek. 2019, 112, 435–450. [Google Scholar] [CrossRef] [PubMed]
- Wu, H.; Andrew, H.-J.; Wang, A.; Jennings, M. Discovery of virulence factors of pathogenic bacteria. Curr. Opin. Chem. Biol. 2008, 12, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Papadopoulos, D.; Schneider, D.; Meier-Eiss, J.; Arber, W.; Lenski, R.; Blot, M. Genomic evolution during a 10,000-generation experiment with bacteria. Proc. Natl. Acad. Sci. USA 1999, 96, 3807–3812. [Google Scholar] [CrossRef] [PubMed]
- Nair, M.K.M.; Venkitanarayanan, K. Role of bacterial OmpA and host citoskeleton in the invasion of human intestinal epithelial cells by Enterobacter sakazakii. Pediatr. Res. 2007, 62, 664–669. [Google Scholar] [CrossRef] [PubMed]
- Jang, H.; Chase, H.R.; Gangiredla, J.; Grim, C.J.; Patel, I.R.; Kothary, M.H.; Jackson, S.; Mammel, M.K.; Carter, L.; Negrete, F.; et al. Analysis of the molecular diversity among Cronobacter species isolated from filth flies using targeted PCR, pan genomic DNA microarray, and whole genome sequencing analyses. Front. Microbiol. 2020, 11, 561204. [Google Scholar] [CrossRef]
- Baida, G.E.; Kuzmin, N.P. Mechanism of action of hemolysin III from Bacillus cereus. Biochim. Biophys. Acta 1996, 1284, 22–124. [Google Scholar] [CrossRef]
- Chen, Y.C.; Chang, M.C.; Chuang, Y.C.; Jeang, C.L. Characterization and virulence of hemolysin III from Vibrio vulnificus. Curr. Microbiol. 2004, 49, 175–179. [Google Scholar] [CrossRef]
- Himelright, I.; Harris, E.; Lorch, V.; Anderson, M. Enterobacter sakazakii infections associated with the use of powdered infant formula—Tennessee, 2001. J. Am. Med. Assoc. 2002, 287, 2204–2205. [Google Scholar]
- Abreu, A.; Bueris, V.; Porangaba, T.; Sircili, M.; Navarro-Garcia, F.; Elias, W. Autotransporter Protein-Encoding Genes of Diarrheagenic Escherichia coli Are Found in both Typical and Atypical Enteropathogenic E. coli Strains. Appl. Environ. Microbiol. 2013, 79, 411–414. [Google Scholar] [CrossRef]
- Proudy, I.; Bouglé, D.; Coton, E.; Coton, M.; Leclercq, R.; Vergnaud, M. Genotypic characterization of Enterobacter sakazakii isolates by PFGE, BOX-PCR and sequencing of the fliC gene. J. Appl. Microbiol. 2008, 104, 26–34. [Google Scholar] [CrossRef]
- Hoeflinger, J.L.; Miller, M.J. Cronobacter sakazakii ATCC 29544 Autoaggregation requires FliC flagellation, not motility. Front. Microbiol. 2017, 8, 301. [Google Scholar] [CrossRef] [PubMed]
- Aldubyan, M.; Almami, I.; Benslimane, F.; Alsonosi, A.; Forsythe, S. Comparative Outer Membrane Protein Analysis of High and Low-Invasive Strains of Cronobacter malonaticus. Front. Microbiol. 2017, 8, 2268. [Google Scholar] [CrossRef] [PubMed]
- Dingle, T.; Mulvey, G.; Armstrong, G. Mutagenic analysis of the Clostridium difficile flagellar proteins, FliC and FliD, and their contribution to virulence in hamsters. Infect. Immun. 2011, 79, 4061–4067. [Google Scholar] [CrossRef]
- Ye, Y.; Zhang, X.; Zhang, M.; Ling, N.; Zeng, H.; Gao, J.; Jiao, R.; Wu, Q.; Zhang, J. Potential factors involved in virulence of Cronobacter sakazakii isolates by comparative transcriptome analysis. J. Dairy Sci. 2017, 100, 8826–8837. [Google Scholar] [CrossRef]
- Kim, K.; Jang, S.; Kim, S.; Park, J.; Heu, S.; Ryu, S. Prevalence and genetic diversity of Enterobacter sakazakii in ingredients of infant foods. Int. J. Food Microbiol. 2008, 122, 196–203. [Google Scholar] [CrossRef] [PubMed]
- Chon, J.; Song, K.; Kim, S.; Hyeon, J.; Seo, K. Isolation and characterization of Cronobacter from desiccated foods in Korea. J. Food Sci. 2012, 77, 354–358. [Google Scholar] [CrossRef] [PubMed]
- Parra-Flores, J.; Arvizu, S.; Silva, J.; Fernández, E. Two cases of hemorrhagic diarrhea caused by Cronobacter sakazakii in hospitalized nursing infants associated with the consumption of powdered infant formula. J. Food Prot. 2011, 74, 2177–2181. [Google Scholar] [CrossRef]
- Aly, M.A.; Domig, K.J.; Kneifel, W.; Reimhult, E. Whole Genome Sequencing-Based Comparison of Food Isolates of Cronobacter sakazakii. Front. Microbiol. 2019, 10, 1464. [Google Scholar] [CrossRef]
- Touze, T.; Eswaran, J.; Bokma, E.; Koronakis, E.; Hughes, C.; Koronakis, V. Interactions underlying assembly of the Escherichia coli AcrAB-TolC multidrug efflux system. Mol. Microbiol. 2004, 53, 697–706. [Google Scholar] [CrossRef]
- Kucerova, E.; Clifton, S.W.; Xia, X.Q.; Long, F.; Porwollik, S.; Fulton, L.; Feng, D.; Wollam, A.; Shah, N.; Bhonogiri, V.; et al. Genome sequence of Cronobacter sakazakii BAA-894 and comparative genomic hybridization analysis with other Cronobacter species. PLoS ONE 2010, 5, e9556. [Google Scholar] [CrossRef]
- El-Sharoud, W.; O’Brien, S.; Negredo, C.; Iversen, C.; Fanning, S.; Healy, B. Characterization of Cronobacter recovered from dried milk and related products. BMC Microbiol. 2009, 9, 9. [Google Scholar] [CrossRef] [PubMed]
- Carroll, L.; Gaballa, A.; Guldimann, C.; Sullivan, G.; Henderson, L.; Wiedmann, M. Identification of Novel Mobilized Colistin Resistance Gene mcr-9 in a Multidrug-Resistant, Colistin-Susceptible Salmonella enterica Serotype Typhimurium Isolate. mBio 2019, 10, e00853-19. [Google Scholar] [CrossRef] [PubMed]
- Kieffer, N.; Royer, G.; Decousser, J.-W.; Bourrel, A.-S.; Palmieri, M.; Ortiz De La Rosa, J.-M.; Jacquier, H.; Denamur, E.; Nordmann, P.; Poirel, L. mcr-9, an Inducible Gene Encoding an Acquired Phosphoethanolamine Transferase in Escherichia coli, and Its Origin. Antimicrob. Agents Chemother. 2019, 63, e00965-19. [Google Scholar] [CrossRef] [PubMed]
- Yuan, Y.; Li, Y.; Wang, G.; Li, C.; Xiang, L.; She, J.; Yang, Y.; Zhong, F.; Zhang, L. Coproduction of MCR-9 and NDM-1 By Colistin-Resistant Enterobacter hormaechei Isolated from Bloodstream Infection. Infect. Drug Resist. 2019, 12, 2979–2985. [Google Scholar] [CrossRef] [PubMed]
- Cheng, Y.; Chen, Y.; Liu, Y.; Guo, Y.; Zhou, Y.; Xiao, T.; Zhang, S.; Xu, H.; Chen, Y.; Shan, T.; et al. Identification of novel tetracycline resistance gene tet(X14) and its co-occurrence with tet(X2) in a tigecycline-resistant and colistin-resistant Empedobacter stercoris. Emerg. Microbes Infect. 2020, 9, 1843–1852. [Google Scholar] [CrossRef] [PubMed]
- Müller, A.; Hächler, H.; Stephan, R.; Lehner, A. Presence of AmpC beta-lactamases, CSA-1, CSA-2, CMA-1, and CMA-2 conferring an unusual resistance phenotype in Cronobacter sakazakii and Cronobacter malonaticus. Microb. Drug Resist. 2014, 20, 275–280. [Google Scholar] [CrossRef]
- Zhou, K.; Zhou, Y.; Zhang, C.; Song, J.; Cao, X.; Yu, X.; Shen, P.; Xiao, Y. Dissemination of a ‘rare’ extended-spectrum β-lactamase gene blaSFO-1 mediated by epidemic clones of carbapenemase-producing Enterobacter hormaechei in China. Int. J. Antimicrob. Agents 2020, 56, 106079. [Google Scholar] [CrossRef]
- Eshwar, A.K.; Tall, B.D.; Gangiredla, J.; Gopinath, G.R.; Patel, I.R.; Neuhauss, S.; Stephan, R.; Lehner, A. Linking Genomo- and Pathotype: Exploiting the Zebrafish Embryo Model to Investigate the Divergent Virulence Potential among Cronobacter spp. PLoS ONE 2016, 11, e0158428. [Google Scholar] [CrossRef]
- Makarova, K.S.; Koonin, E.V. Annotation and Classification of CRISPR-Cas Systems. Methods Mol. Biol. 2015, 1311, 47–75. [Google Scholar]
- Ogrodzki, P.; Forsythe, J. CRISPR–cas loci profiling of Cronobacter sakazakii pathovars. Future Microbiol. 2016, 11, 1507–1519. [Google Scholar] [CrossRef]
- Zeng, H.; Li, C.; He, W.; Zhang, J.; Chen, M.; Lei, T.; Wu, H.; Ling, N.; Cai, S.; Wang, J.; et al. Cronobacter sakazakii, Cronobacter malonaticus, and Cronobacter dublinensis Genotyping Based on CRISPR Locus Diversity. Front. Microbiol. 2019, 10, 1989. [Google Scholar] [CrossRef] [PubMed]
- Zeng, H.; Zhang, J.; Li, C.; Xie, T.; Ling, N.; Wu, Q.; Ye, Y. The driving force of prophages and CRISPR-Cas system in the evolution of Cronobacter sakazakii. Sci. Rep. 2017, 7, 40206. [Google Scholar] [CrossRef] [PubMed]
- Zeng, H.; Zhang, J.; Wu, Q.; He, W.; Wu, H.; Ye, Y.; Li, C.; Ling, N.; Chen, M.; Wang, J.; et al. Reconstituting the History of Cronobacter Evolution Driven by Differentiated CRISPR Activity. Appl. Environ. Microbiol. 2018, 84, e00267-18. [Google Scholar] [CrossRef] [PubMed]
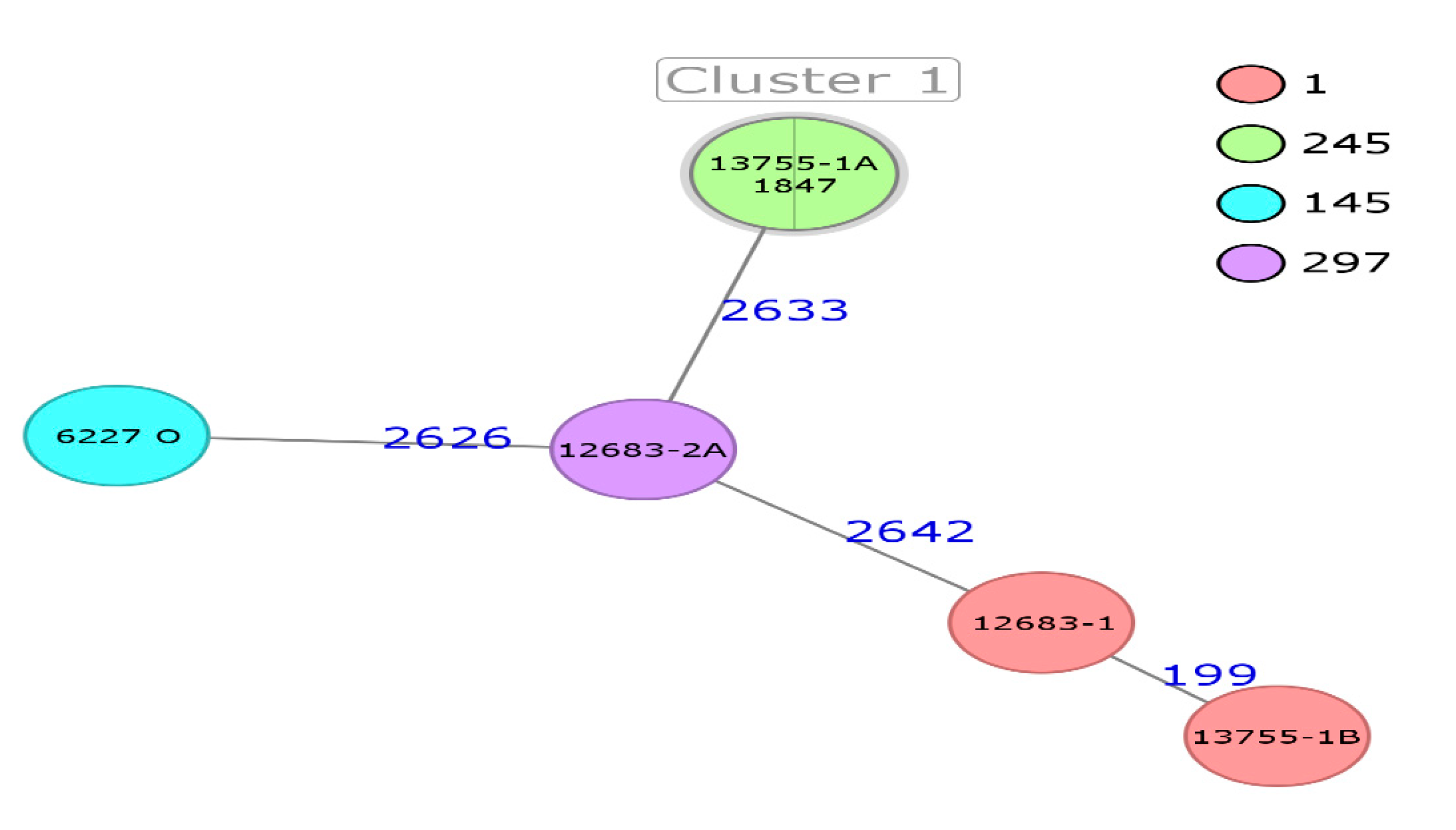
| Strain | NGS ID | Identification | ST | Source | Country | Year of Isolation | Manufacturer |
|---|---|---|---|---|---|---|---|
| * 1847 | 510284-18 | C. sakazakii | 245 | Powdered milk | Czech Republic | 2010 | A |
| 13755-1A | 510192-19 | C. sakazakii | 245 | Powdered milk | Czech Republic | 2014 | B |
| 13755-1B | 510193-19 | C. sakazakii | 1 | Powdered milk | Czech Republic | 2014 | B |
| 12683-1 | 510285-19 | C. sakazakii | 1 | Powdered milk | Czech Republic | 2014 | B |
| 12683-2A | 510194-19 | C. sakazakii | 297 | Powdered milk | Czech Republic | 2014 | B |
| 6227 | 510555-19 | C. sakazakii | 145 | Powdered milk | Czech Republic | 2014 | C |
| C. sakazakii Strain (ST) | Gene | |||||
|---|---|---|---|---|---|---|
| hlyA | ompA | aut | fliC | inv | cpa | |
| 13755-1B (1) | − | + | + | + | − | − |
| 1847 (245) | + | + | + | + | − | − |
| 823 (ATCC BAA-894) (1) control strain | + | + | + | + | + | + |
| Virulence Gene | Function | 1847 (ST245) | 13755-1A (ST245) | 13755-1B (ST1) | 12683-1 (ST1) | 12683-2A (ST 297) | 6227 (ST145) |
|---|---|---|---|---|---|---|---|
| flgB | motility | + | + | + | + | + | + |
| flgK | flagellar hook-associated protein 1 | + | + | + | + | + | + |
| flgL | flagellar hook-associated protein 3 | + | + | + | + | + | + |
| flgM | negative regulator of flagellin synthesis | + | + | + | + | + | + |
| flgN | flagella synthesis FlgN protein | + | + | + | + | + | + |
| flhD | flagellar hook-associated protein 2 | + | + | + | + | + | + |
| fliA | flagellar operon FliA | + | + | + | + | + | + |
| fliC | flagellin | + | + | + | + | + | + |
| fliD | flagellar hook-associated protein 2 | + | + | + | + | + | + |
| fliR | flagellar biosynthetic FliR protein | + | + | + | + | + | + |
| fliT | flagella FliT protein | + | + | + | + | + | + |
| fliZ | FliZ protein | + | + | + | + | + | + |
| lolA | outer membrane lipoprotein carrier protein | + | + | + | + | + | + |
| motB | chemotaxis MotA protein | + | + | − | + | + | + |
| ompW | transmembrane transport | + | + | + | + | + | + |
| sdiA | LuxR family transcriptional regulator | + | + | + | + | + | + |
| slyB | outer membrane lipoprotein SlyB | + | + | − | + | + | + |
| tolC | outer membrane channel protein | + | + | + | + | + | + |
| MsgA | survival in macrophage | + | + | + | + | + | + |
| MviN | protective immunity and colonization in Salmonella | + | + | + | + | + | + |
| cpa | plasminogen activator | − | − | + | + | + | + |
| hha | hemolysin expression modulating protein | + | + | + | + | + | + |
| hly III | hemolysin III | + | + | + | + | + | + |
| ompA | adhesion cell; cell death induction; biofilm formation | + | + | + | + | + | + |
| ompX | adhesion cell | + | + | + | + | + | + |
| blc | outer membrane lipoprotein | + | + | + | + | + | + |
| cheR | chemotaxis protein methyltransferase | + | + | − | + | + | + |
| cheY | response regulator of chemotaxis family | + | + | + | + | + | + |
| labp | epithelial cell invasion and lipid A production by LpxA | + | − | + | − | − | − |
| Best Hit Antibiotic Resistance Ontology (ARO) | Drug Class | Resistance Mechanism | 1847 (ST245) | 13755-1A (ST245) | 13755-1B (ST1) | 12683-1 (ST1) | 12683-2A (ST297) | 6227 (ST145) |
|---|---|---|---|---|---|---|---|---|
| CRP | fluoroquinolone antibiotic; macrolide antibiotic; penam | antibiotic efflux | + | + | + | + | + | + |
| marR | monobactam; triclosan; rifamycin antibiotic; penem; cephamycin; fluoroquinolone antibiotic; penam; phenicol antibiotic; glycylcycline; tetracycline antibiotic; cephalosporin; carbapenem | antibiotic efflux; reduced antibiotic permeability | + | + | + | + | + | + |
| H-NS | fluoroquinolone antibiotic; macrolide antibiotic; penam; tetracycline antibiotic; cephalosporin; cephamycin | antibiotic efflux | + | + | + | + | + | + |
| EF-Tu | elfamycin antibiotic | antibiotic target alteration | + | + | + | + | + | + |
| marA | fluoroquinolone antibiotic; triclosan; rifamycin antibiotic; penam; phenicol antibiotic; glycylcycline; tetracycline antibiotic; cephalosporin | antibiotic target alteration; antibiotic efflux | + | + | + | + | + | + |
| mrB | fluoroquinolone antibiotic | antibiotic efflux | + | + | + | + | + | + |
| emrR | fluoroquinolone antibiotic | antibiotic efflux | + | + | + | + | + | + |
| adeF | tetracycline antibiotic; fluoroquinolone antibiotic | antibiotic efflux | + | + | + | + | + | + |
| msbA | nitroimidazole antibiotic | antibiotic efflux | + | + | + | + | + | + |
| vgaC | streptogramin antibiotic; pleuromutilin antibiotic | antibiotic target protection | + | + | − | − | + | − |
| GlpT | fosfomycin | antibiotic target alteration | + | + | + | + | + | + |
| PBP3 | penam; cephalosporin; cephamycin; monobactam; carbapenem | antibiotic target alteration | + | + | + | + | + | + |
| Strains (ST) | Plasmids | Accession Number | Function |
|---|---|---|---|
| 1847 (1); 13755-1A (245) | IncFII(pECLA) | CP001919 | Antibiotic resistance |
| 12683-2A (297) | IncFIB(pCTU3) | FN543096 |
| Strains | Sequence Type (ST) | Operon Structure Type | Number of CRISPR Arrays per Strain | Maximum Number of Spacers per Strain | Sequences with Cas Cluster | Repeat Consensus/cas Genes |
|---|---|---|---|---|---|---|
| 1847 | 245 | Type I-E Cas | 29/7 | 32/9 | 1 | GTTCACTGCCGTACAGGCAGCTTAGAAA/CTGTTCCCCGCGCGAGCGGGGATAAACCG/ Cas3_0_I, Cse1_0_IE, Cse2_0_IE, Cas7_0_IE, Cas5_0_IE, Cas6_0_IE, Cas1_0_IE, Cas2_0_IE. |
| 13755-1A | 245 | Type I-F Cas Type I-E Cas | 10 18 | 9 17 | 1 1 | GTTCACTGCCGTACAGGCAGCTTAGAAA/ Cas1_0_IF, Cas3-Cas2_0_IF, Cas6_0_IF, Csy1_0_IF, Csy2_0_IF, Csy3_0_IF. CGGTTTATCCCCGCTCGCGCGGGGAACAC/ Cas3_0_I, Cse1_0_IE, Cse2_0_IE, Cas7_0_IE, Cas5_0_IE, Cas6_0_IE, Cas1_0_IE, Cas2_0_IE. |
| 13755-1B | 1 | Type I-E Cas Type I-F Cas | 22/30 14 | 24/32 16 | 1 1 | CTGTTCCCCGCGCGAGCGGGGATAAACCG/GTGTTCCCCGCGCGAGCGGGGATAAACCG/ Cas3_0_I, Cse1_0_IE, Cse2_0_IE, Cas7_0_IE, Cas5_0_IE, Cas6_0_IE, Cas1_0_IE, Cas2_0_IE. GTTCACTGCCGTACAGGCAGCTTAGAAA/ Cas1_0_IF, Cas3-Cas2_0_IF, Cas6_0_IF, Csy1_0_IF, Csy2_0_IF, Csy3_0_IF. |
| 12683-2A | 297 | Type I-F Cas Type I-E Cas | 49 20/58 | 53 22/62 | 1 1 | TTTCTAAGCTGCCTGTACGGCAGTGAAC/ Cas1_0_IF, Cas3-Cas2_0_IF, Cas6_0_IF, Csy1_0_IF, Csy2_0_IF, Csy3_0_IF. CTGTTCCCCGCGCGAGCGGGGATAAACCG/GTGTTCCCCGCGCGAGCGGGGATAAACCG/ Cas3_0_I, Cse1_0_IE, Cse2_0_IE, Cas7_0_IE, Cas5_0_IE, Cas6_0_IE, Cas1_0_IE, Cas2_0_IE. |
| 12683-1 | 1 | Type I-E Cas Type I-F Cas | 28/31 14 | 29/32 15 | 1 1 | CTGTTCCCCGCGCGAGCGGGGATAAACCG/GTGTTCCCCGCGCGAGCGGGGATAAACCG/ Cas3_0_I, Cse1_0_IE, Cse2_0_IE, Cas7_0_IE, Cas5_0_IE, Cas6_0_IE, Cas1_0_IE, Cas2_0_IE. GTTCACTGCCGTACAGGCAGCTTAGAAA/ Cas1_0_IF, Cas3-Cas2_0_IF, Cas6_0_IF, Csy1_0_IF, Csy2_0_IF, Csy3_0_IF. |
| 6227 | 145 | Type I-E Cas | 15/13 | 16/12 | 1 | CGGTTTATCCCCGCTCGCGCGGGGAACGG/GTGTTCCCCGCGCGAGCGGGGATAAACCG// Cas1_0_IE, Cas2_0_IE, Cas5_0_IE, Cas6_0_IE, Cas7_0_IE, Cse1_0_IE, Cse2_0_IE. |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Holý, O.; Parra-Flores, J.; Lepuschitz, S.; Alarcón-Lavín, M.P.; Cruz-Córdova, A.; Xicohtencatl-Cortes, J.; Mancilla-Rojano, J.; Ruppitsch, W.; Forsythe, S. Molecular Characterization of Cronobacter sakazakii Strains Isolated from Powdered Milk. Foods 2021, 10, 20. https://doi.org/10.3390/foods10010020
Holý O, Parra-Flores J, Lepuschitz S, Alarcón-Lavín MP, Cruz-Córdova A, Xicohtencatl-Cortes J, Mancilla-Rojano J, Ruppitsch W, Forsythe S. Molecular Characterization of Cronobacter sakazakii Strains Isolated from Powdered Milk. Foods. 2021; 10(1):20. https://doi.org/10.3390/foods10010020
Chicago/Turabian StyleHolý, Ondrej, Julio Parra-Flores, Sarah Lepuschitz, María Paula Alarcón-Lavín, Ariadnna Cruz-Córdova, Juan Xicohtencatl-Cortes, Jetsi Mancilla-Rojano, Werner Ruppitsch, and Stephen Forsythe. 2021. "Molecular Characterization of Cronobacter sakazakii Strains Isolated from Powdered Milk" Foods 10, no. 1: 20. https://doi.org/10.3390/foods10010020
APA StyleHolý, O., Parra-Flores, J., Lepuschitz, S., Alarcón-Lavín, M. P., Cruz-Córdova, A., Xicohtencatl-Cortes, J., Mancilla-Rojano, J., Ruppitsch, W., & Forsythe, S. (2021). Molecular Characterization of Cronobacter sakazakii Strains Isolated from Powdered Milk. Foods, 10(1), 20. https://doi.org/10.3390/foods10010020








