Synthesis, Reactivity Studies, and Cytotoxicity of Two trans-Iodidoplatinum(II) Complexes. Does Photoactivation Work?
Abstract
1. Introduction
2. Results
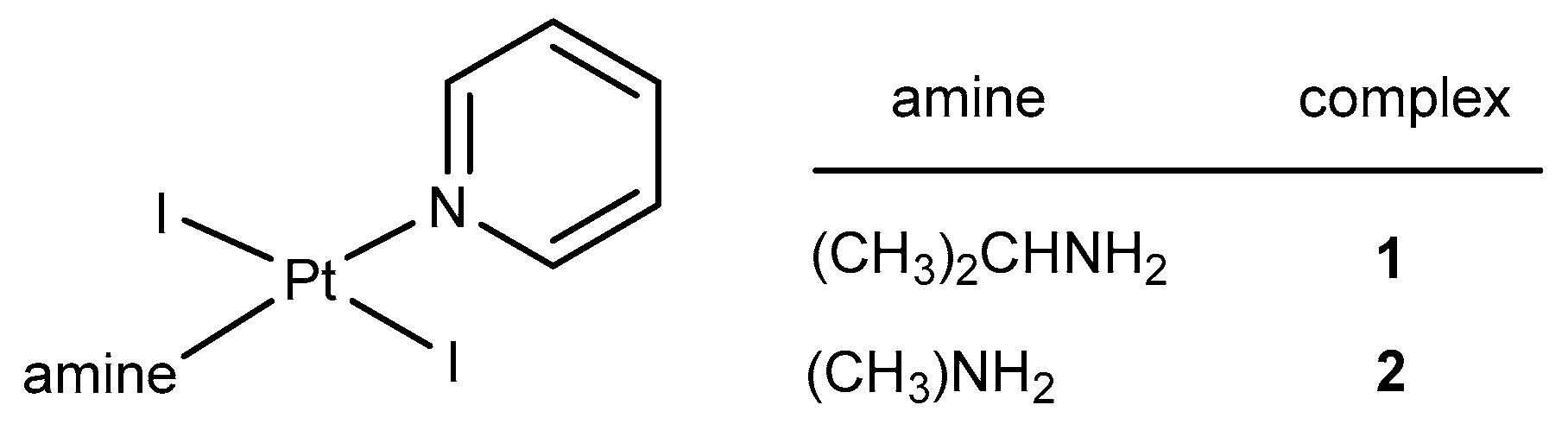
2.1. Synthesis and Characterization
2.2. Cytotoxicity of trans-[PtI2(amine)(py)] Complexes
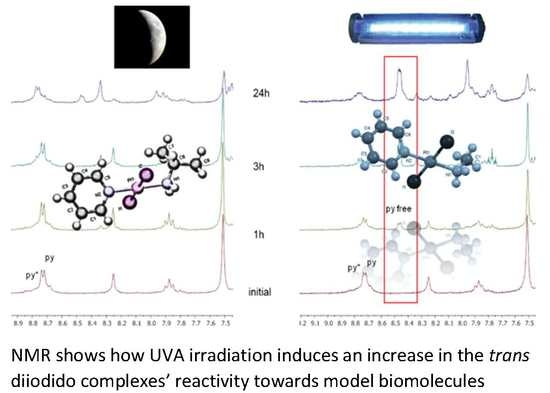
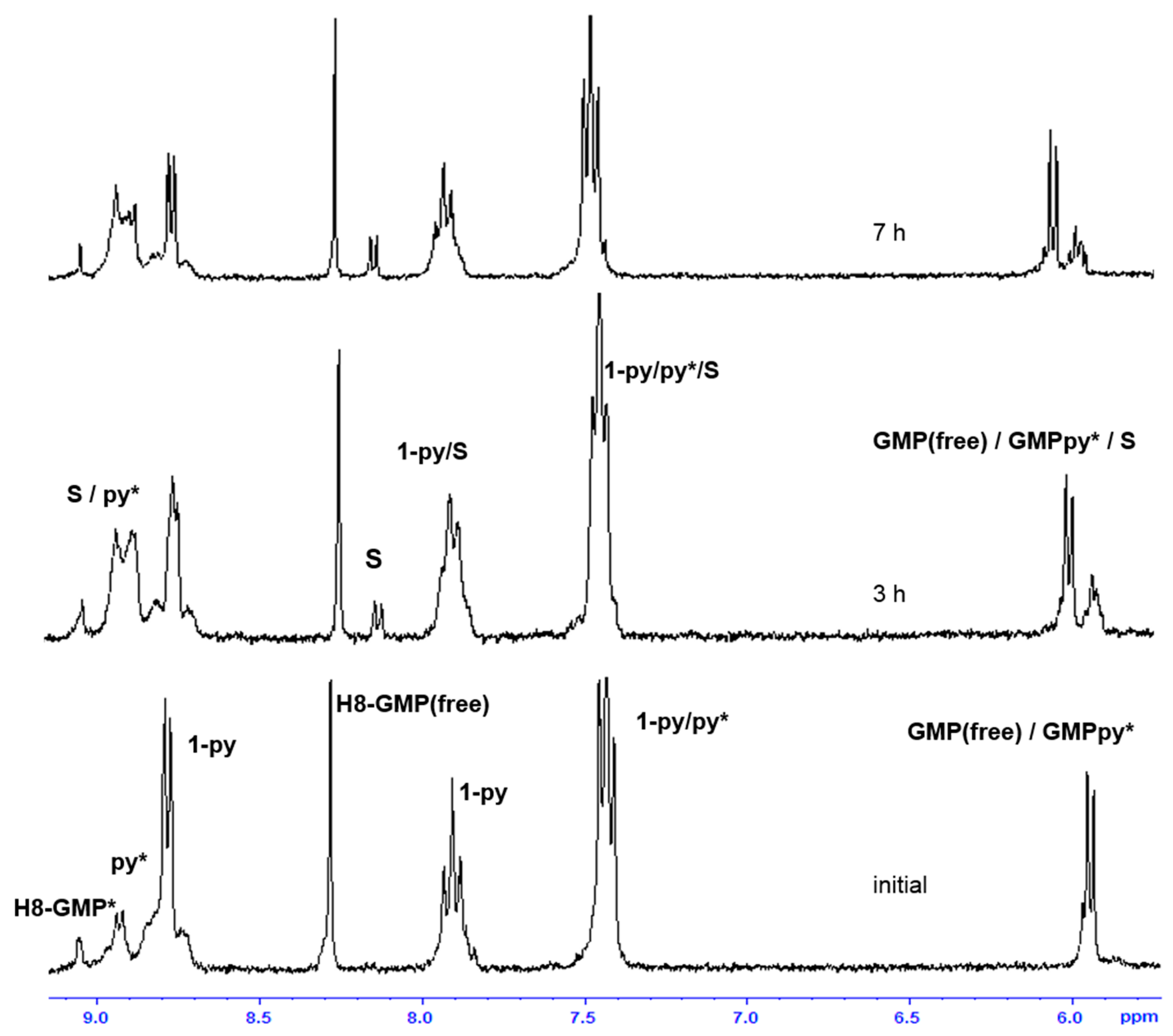
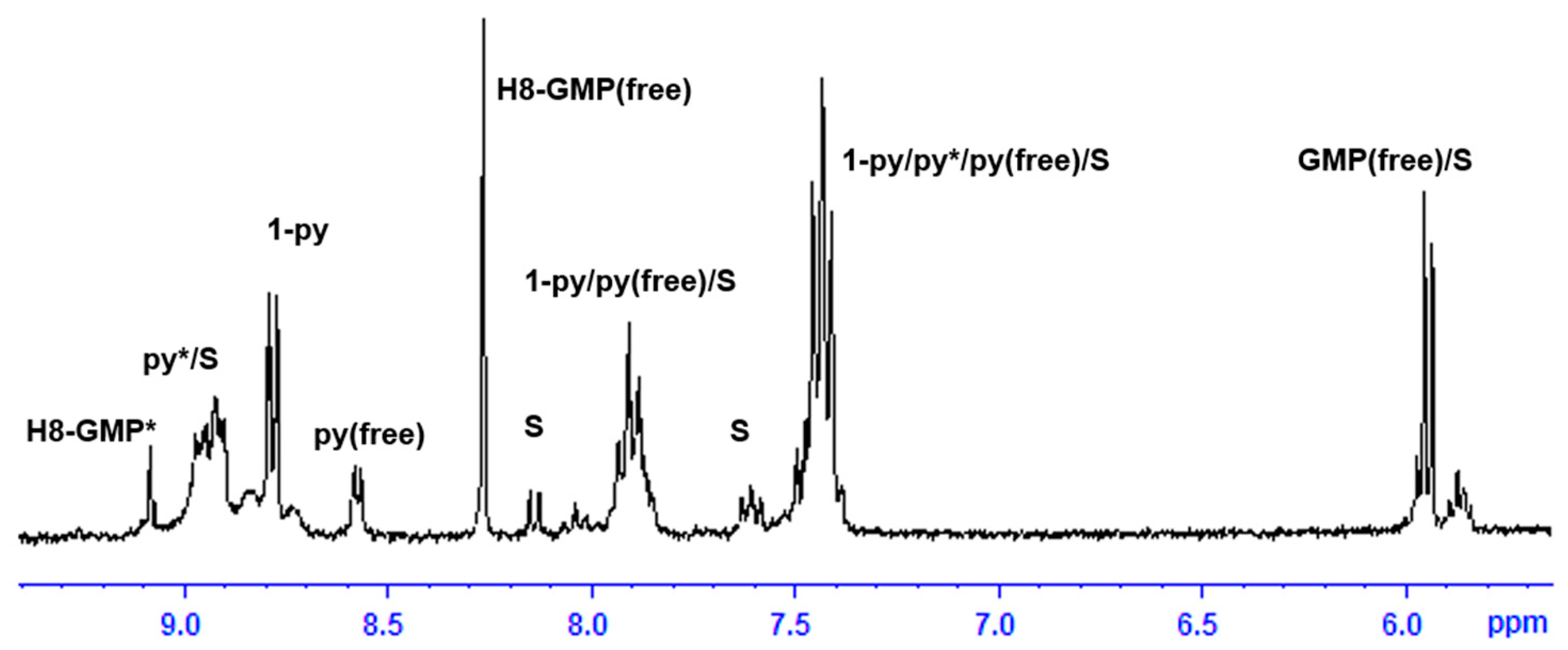
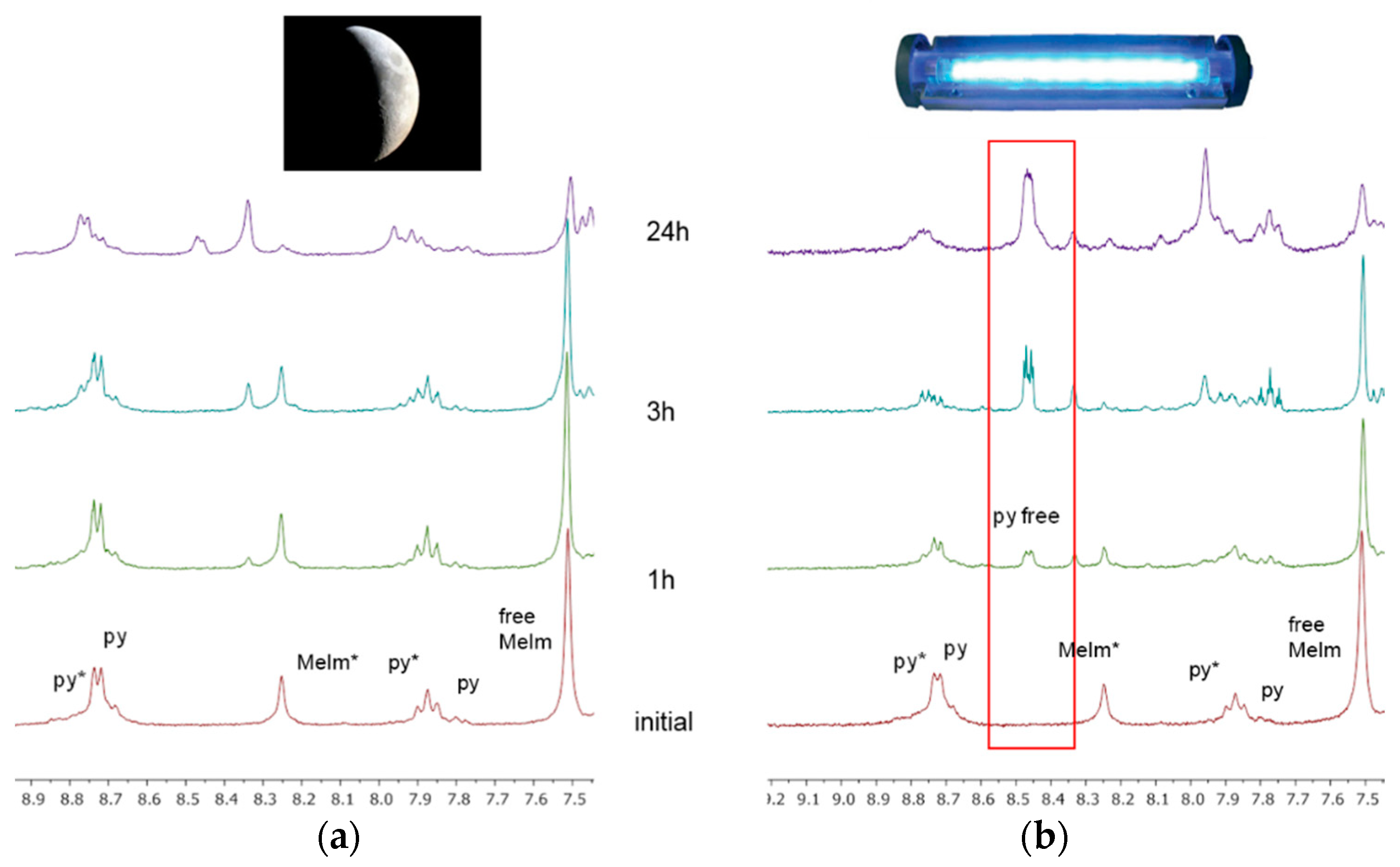
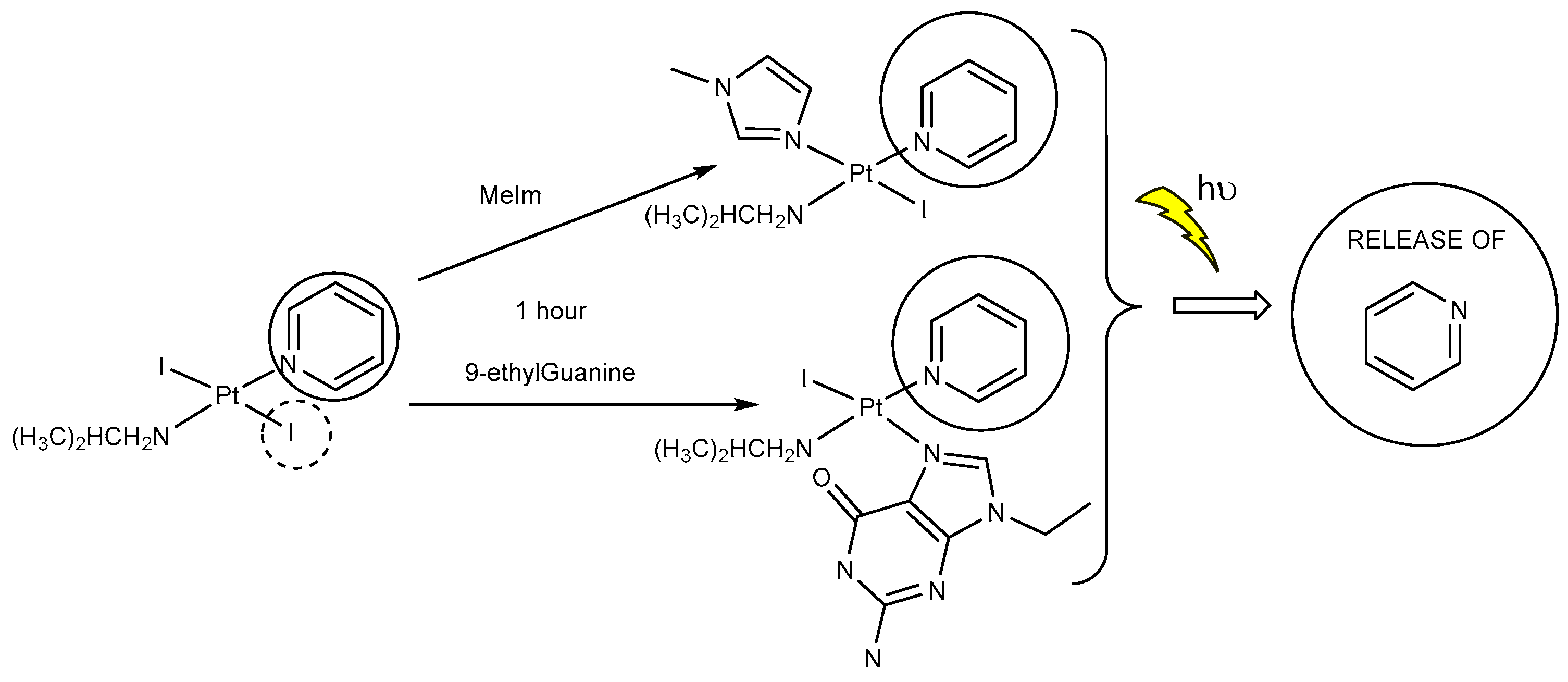
2.3. Reactivity of trans-[PtI2(amine)(py)] Complexes with 5′-GMP and MeIm
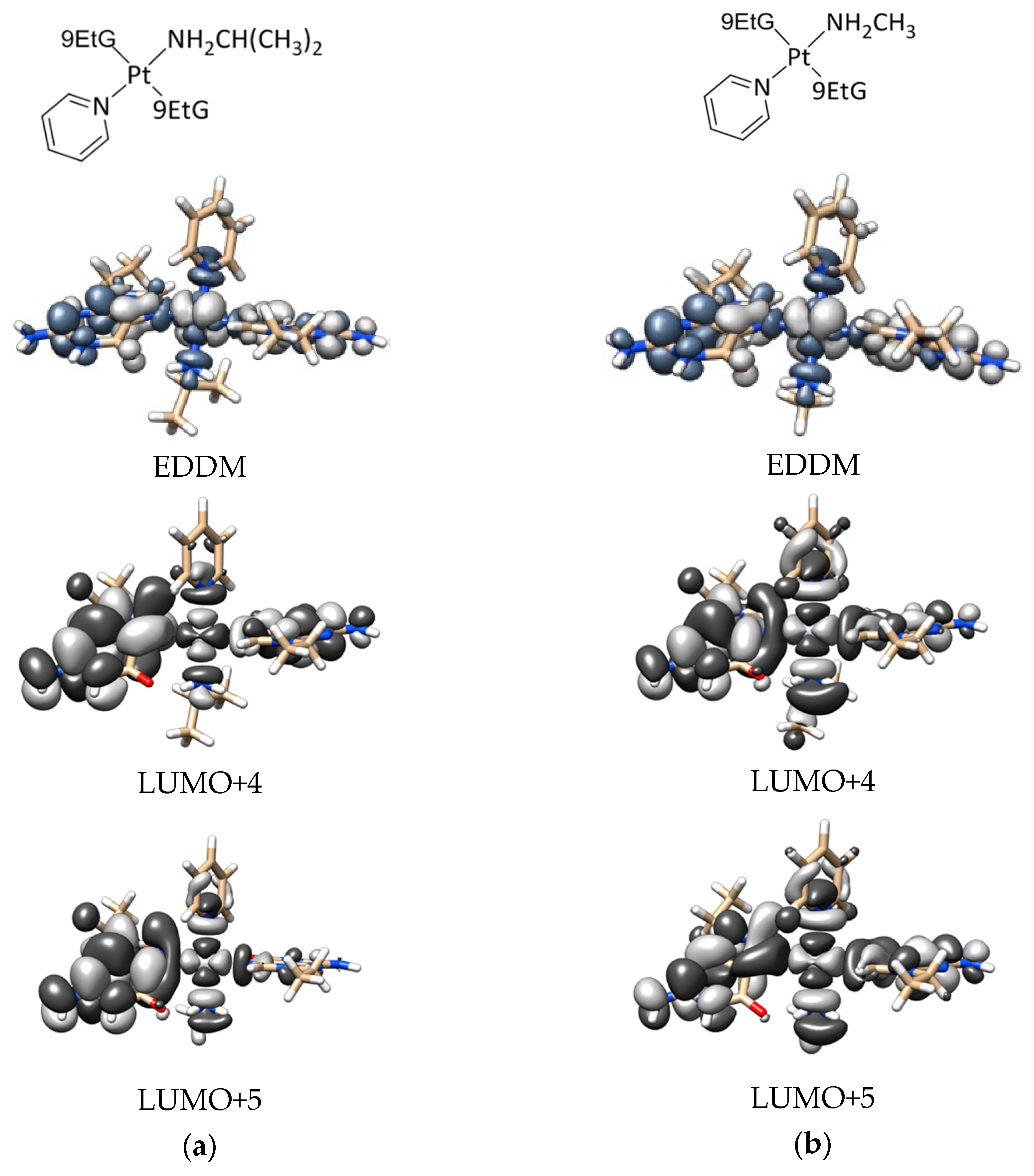
2.4. DFT Calculations
3. Discussion
- The interaction of these complexes with 5′-GMP shows the formation of the expected DNA adduct after hydrolysis of the iodido complexes as reported for similar compounds with no release of the spectator ligands.
- However, once we irradiate the samples, we can only observe a promotion of the reactivity in the first hour, and then the release of the pyridine ligand becomes clear at longer reaction times under irradiation.
- This release of the spectator ligand, enhanced by irradiation, has not been observed studying trans isomers with iodido ligands versus DNA [6], but only with cis isomers and versus proteins [5]. In addition, when the results of the interaction of compounds 1 and 2 with MeIm are evaluated, they are very similar to the results obtained with DNA.
- Our interpretation of the data is that after irradiation, the reactivity of complexes 1 and 2 is enhanced, forming active species. However, at longer reaction times, the compounds lose the pyridine, affording new adducts and species that are more similar to those reported from the cytotoxic compounds than their original aqua species/adducts.
- DFT calculations justify the quicker formation of the bis/monoadduct species (Figure 6) along the first hour, but once the irradiation applies for longer periods of time, the pyridine is released and the compound is no longer the structure proposed but similar to those bearing only aliphatic amines [12].
4. Materials and Methods
4.1. Method for the Synthesis of trans-[PtI2(amine)(pyridine)]
4.2. Nuclear Magnetic Resonance Spectroscopy
4.3. Irradiation
4.4. X-ray Diffraction
4.5. Cytotoxicity
4.6. DFT Calculations
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Johnstone, T.C.; Suntharalingam, K.; Lippard, S.J. The Next Generation of Platinum Drugs: Targeted Pt(II) Agents, Nanoparticle Delivery, and Pt(IV) Prodrugs. Chem. Rev. 2016, 116, 3436–3486. [Google Scholar] [CrossRef] [PubMed]
- Medrano, A.; Dennis, S.M.; Alvarez-Valdes, A.; Perles, J.; McGregor Mason, T.; Quiroga, A.G. Synthesis, cytotoxicity, DNA interaction and cell cycle studies of trans-diiodophosphine Pt(II) complexes. Dalton Trans. 2015, 44, 3557–3562. [Google Scholar] [CrossRef] [PubMed]
- Quiroga, A.G. Understanding trans platinum complexes as potential antitumor drugs beyond targeting DNA. J. Inorg. Biochem. 2012, 114, 106–112. [Google Scholar] [CrossRef] [PubMed]
- Messori, L.; Casini, A.; Gabbiani, C.; Michelucci, E.; Cubo, L.; Rios-Luci, C.; Padron, J.M.; Navarro-Ranninger, C.; Quiroga, A.G. Cytotoxic Profile and Peculiar Reactivity with Biomolecules of a Novel "Rule-Breaker" Iodidoplatinum(II) Complex. ACS Med. Chem. Lett. 2010, 1, 381–385. [Google Scholar] [CrossRef] [PubMed]
- Messori, L.; Cubo, L.; Gabbiani, C.; Alvarez-Valdes, A.; Michelucci, E.; Pieraccini, G.; Rios-Luci, C.; Leon, L.G.; Padron, J.M.; Navarro-Ranninger, C.; et al. Reactivity and Biological Properties of a Series of Cytotoxic PtI2(amine)2 Complexes, Either cis or trans Configured. Inorg. Chem. 2012, 51, 1717–1726. [Google Scholar] [CrossRef] [PubMed]
- Parro, T.; Medrano, M.A.; Cubo, L.; Munoz-Galvan, S.; Carnero, A.; Navarro-Ranninger, C.; Quiroga, A.G. The second generation of iodido complexes: trans-[Ptl2(amine)(amine’)] bearing different aliphatic amines. J. Inorg. Biochem. 2013, 127, 182–187. [Google Scholar] [CrossRef] [PubMed]
- Starha, P.; Vanco, J.; Travnicek, Z.; Hosek, J.; Klusakova, J.; Dvorak, Z. Platinum(II) Iodido Complexes of 7-Azaindoles with Significant Antiproliferative Effects: An Old Story Revisited with Unexpected Outcomes. PLoS ONE 2016, 11, e0165062. [Google Scholar] [CrossRef]
- Cubo, L.; Pizarro, A.M.; Quiroga, A.G.; Salassa, L.; Navarro-Ranninger, C.; Sadler, P.J. Photoactivation of trans diamine platinum complexes in aqueous solution and effect on reactivity towards nucleotides. J. Inorg. Biochem. 2010, 104, 909–918. [Google Scholar] [CrossRef]
- Presa, A.; Brissos, R.F.; Caballero, A.B.; Borilovic, I.; Korrodi-Gregório, L.; Pérez-Tomás, R.; Roubeau, O.; Gamez, P. Photoswitching the Cytotoxic Properties of Platinum(II) Compounds. Angew. Chem. Int. Ed. 2014, 54, 4561–4565. [Google Scholar] [CrossRef]
- Quental, L.; Raposinho, P.; Mendes, F.; Santos, I.; Navarro-Ranninger, C.; Alvarez-Valdes, A.; Huang, H.; Chao, H.; Rubbiani, R.; Gasser, G.; et al. Combining imaging and anticancer properties with new heterobimetallic Pt(II)/M(I) (M = Re, 99 mTc) complexes. Dalton Trans. 2017, 46, 14523–14536. [Google Scholar] [CrossRef]
- Frei, A.; Rubbiani, R.; Tubafard, S.; Blacque, O.; Anstaett, P.; FelgentrÃger, A.; Maisch, T.; Spiccia, L.; Gasser, G. Synthesis, Characterization, and Biological Evaluation of New Ru(II) Polypyridyl Photosensitizers for Photodynamic Therapy. J. Med. Chem. 2014, 57, 7280–7292. [Google Scholar] [CrossRef] [PubMed]
- Navas, F.; Perfahl, S.; Garino, C.; Salassa, L.; Novakova, O.; Navarro-Ranninger, C.; Bednarski, P.J.; Malina, J.; Quiroga, A.N.G. Increasing DNA reactivity and in vitro antitumor activity of trans diiodido Pt(II) complexes with UVA light. J. Inorg. Biochem. 2015, 153, 211–218. [Google Scholar] [CrossRef] [PubMed]
- Aris, S.M.; Farrell, N.P. Towards Antitumor Active trans-Platinum Compounds13. Eur. J. Inorg. Chem. 2009, 2009, 1293–1302. [Google Scholar] [CrossRef]
- Farrell, N.P. Multi-platinum anti-cancer agents. Substitution-inert compounds for tumor selectivity and new targets. Chem. Soc. Rev. 2015, 44, 8773–8785. [Google Scholar] [CrossRef] [PubMed]
- Brabec, V. DNA Modifications by antitumor platinum and ruthenium compounds: Their recognition and repair. In Progress in Nucleic Acid Research and Molecular Biology; Academic Press: New York, NY, USA, 2002; Volume 71, pp. 1–68. [Google Scholar]
- Mellish, K.J.; Qu, Y.; Scarsdale, N.; Farrell, N. Effect of Geometric Isomerism in Dinuclear Platinum Antitumour Complexes on the Rate of Formation and Structure of Intrastrand Adducts with Oligonucleotides. Nucleic Acids Res. 1997, 25, 1265–1271. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Woods, J.A.; Farrer, N.J.; Robinson, K.S.; Pracharova, J.; Kasparkova, J.; Novakova, O.; Li, H.; Salassa, L.; Pizarro, A.M.; et al. Diazido Mixed-Amine Platinum(IV) Anticancer Complexes Activatable by Visible-Light Form Novel DNA Adducts. Chem. A Eur. J. 2013, 19, 9578–9591. [Google Scholar] [CrossRef] [PubMed]
- Velders, A.H.; Quiroga, A.G.; Haasnoot, J.G.; Reedijk, J. Orientation- and Temperature-Dependent Rotational Behavior of Imidazole Ligands (L) in β-[Ru(azpy)2(L)2](PF6)2 Complexes. Eur. J. Inorg. Chem. 2003, 2003, 713–719. [Google Scholar] [CrossRef]
- Perez, J.M.; Montero, E.I.; Gonzalez, A.M.; Solans, X.; Font-Bardia, M.; Fuertes, M.A.; Alonso, C.; Navarro-Ranninger, C. X-ray Structure of Cytotoxic trans-[PtCl2(dimethylamine)(isopropylamine)]: Interstrand Cross-Link Efficiency, DNA Sequence Specificity, and Inhibition of the B–Z Transition. J. Med. Chem. 2000, 43, 2411–2418. [Google Scholar] [CrossRef]
- Rochon, F.D.; Buculei, V. Multinuclear NMR study and crystal structures of complexes of the types cis- and trans-Pt(amine)2I2. Inorg. Chim. Acta 2004, 357, 2218–2230. [Google Scholar] [CrossRef]
- Thiele, G.; Wagner, D. Über die Reaktion von Platiniodiden mit Pyridin und über die Molekül- und Kristallstruktur von trans-Diiodobis(pyridin)platin(II). Chem. Ber. 1978, 111, 3162–3170. [Google Scholar] [CrossRef]
- Berger, I.; Nazarov, A.A.; Hartinger, C.G.; Groessl, M.; Valiahdi, S.-M.; Jakupec, M.A.; Keppler, B.K. A glucose derivative as natural alternative to the cyclohexane-1,2-diamine ligand in the anticancer drug oxaliplatin? ChemMedChem 2007, 2, 505–514. [Google Scholar] [CrossRef] [PubMed]
- Tessier, C.; Rochon, F.D. Multinuclear NMR study and crystal structures of complexes of the types cis- and trans-Pt(Ypy)2X2, where Ypy=pyridine derivative and X=Cl and I. Inorg. Chim. Acta 1999, 295, 25–38. [Google Scholar] [CrossRef]
- Rochon, F.D.; Tessier, C. Pt(II) compounds with sulfoxide ligands and crystal structures of complexes of the types I(R2SO)Pt(μ-I)2Pt(R2SO)I and trans-Pt(R2SO)(L)X2 (L = amine, pyridine and pyrimidine). Inorg. Chim. Acta 2008, 361, 2591–2600. [Google Scholar] [CrossRef]
- Frisch, M.J.; Schlegel, G.E.; Scuseria, M.A.; Robb, J.R.; Cheeseman, G.; Scalmani, V.; Barone, B.; Mennucci, G.A.; Petersson, H.; Nakatsuji, M.; et al. Gaussian 09, Revision B.01; Gaussian, Inc.: England, UK, 2009. [Google Scholar]
- Stratmann, R.E.; Gustavo, E.S.; Michael, J.F. An efficient implementation of time-dependent density-functional theory for the calculation of excitation energies of large molecules. J. Chem. Phys. 1998, 109, 8218–8224. [Google Scholar] [CrossRef]
- Mark, E.C.; Christine, J.; Kim, C.C.; Dennis, R.S. Molecular excitation energies to high-lying bound states from time-dependent density-functional response theory: Characterization and correction of the time-dependent local density approximation ionization threshold. J. Chem. Phys. 1998, 108, 4439–4449. [Google Scholar] [CrossRef]
- Perdew, J.P.; Burke, K.; Ernzerhof, M. Generalized Gradient Approximation Made Simple. Phys. Rev. Lett. 1996, 77, 3865–3868. [Google Scholar] [CrossRef] [PubMed]
- Yanai, T.; Tew, D.P.; Handy, N.C. A new hybrid exchange-correlation functional using the Coulomb-attenuating method (CAM-B3LYP). Chem. Phys. Lett. 2004, 393, 51–57. [Google Scholar] [CrossRef]
- Miertu, S.; Scrocco, E.; Tomasi, J. Electrostatic interaction of a solute with a continuum. A direct utilizaion of AB initio molecular potentials for the prevision of solvent effects. Chem. Phys. 1981, 55, 117–129. [Google Scholar] [CrossRef]
- O’Boyle, N.M.; Tenderholt, A.L.; Langner, K.M. cclib: A library for package-independent computational chemistry algorithms. J. Comput. Chem. 2008, 29, 839–845. [Google Scholar] [CrossRef] [PubMed]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF ChimeraA visualization system for exploratory research and analysis. J. Comput. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef]
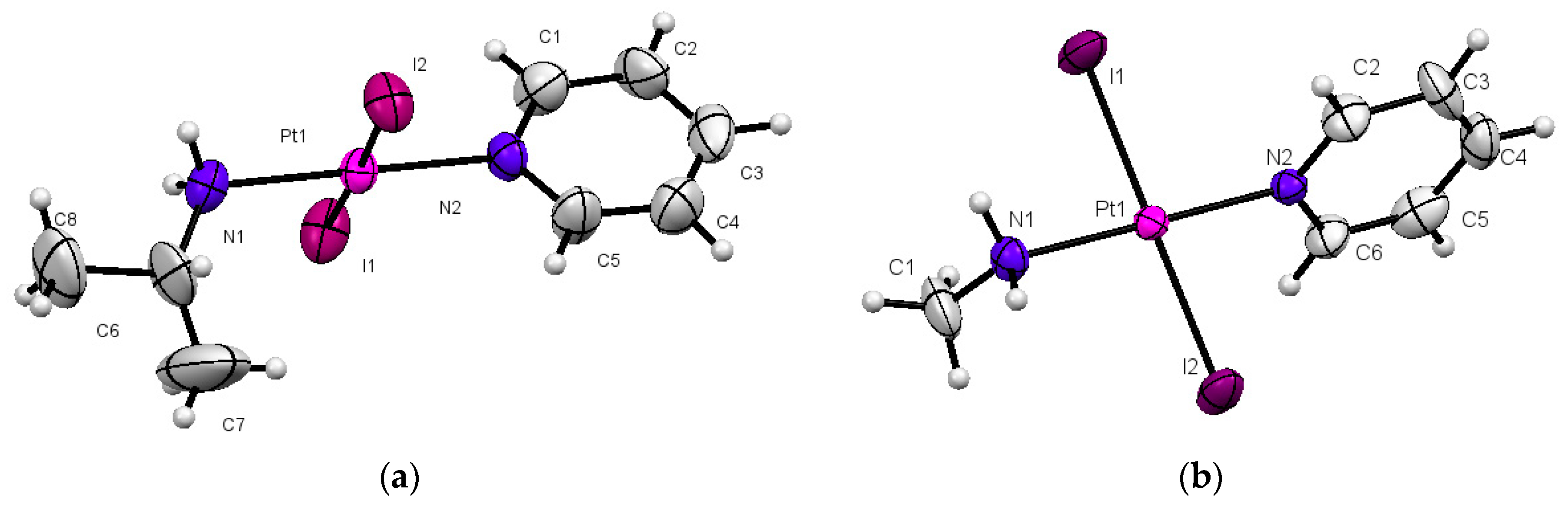
| Distances | Complex 1 | Complex 2 |
| Pt–I1 | 2.5962(9) | 2.5906(11) |
| Pt–I2 | 2.5920(8) | 2.59861(19) |
| Pt–N1 | 2.050(9) | 2.077(13) |
| Pt–N2 | 2.014(8) | 2.025(13) |
| Angles | Complex 1 | Complex 2 |
| N2–Pt–N1 | 179.24(4) | 179.3(5) |
| N1–Pt–I1 | 90.1(3) | 90.2(4) |
| N1–Pt–I2 | 90.0(3) | 89.1(4) |
| N2–Pt–I1 | 89.7(3) | 90.1(4) |
| N2–Pt–I2 | 90.02(3) | 90.6(4) |
| I1–Pt–I2 | 179.05(3) | 177.24(4) |
| Cell line | Complex 1 | Complex 2 | Cisplatin |
|---|---|---|---|
| SAOS-2 | 32.9 (22.6) * | 53.7(15.8) | 5.9(1.5) |
| A375 | 18.9(3.4) | 27.9(5.1) | 9.2(1.7) |
| T-47D | 30.9(5.6) | 45.0(2.5) | 10.2(3.4) |
| HCT116++ | 18.1(3.6) | 29.8(4.4) | 8.3(2.7) |
| HCT116−− | 56.4(10.3) | 63.4(17.3) | 62.2(14.3) |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cubo, L.; Parro, T.; Carnero, A.; Salassa, L.; Matesanz, A.I.; Quiroga, A.G. Synthesis, Reactivity Studies, and Cytotoxicity of Two trans-Iodidoplatinum(II) Complexes. Does Photoactivation Work? Inorganics 2018, 6, 127. https://doi.org/10.3390/inorganics6040127
Cubo L, Parro T, Carnero A, Salassa L, Matesanz AI, Quiroga AG. Synthesis, Reactivity Studies, and Cytotoxicity of Two trans-Iodidoplatinum(II) Complexes. Does Photoactivation Work? Inorganics. 2018; 6(4):127. https://doi.org/10.3390/inorganics6040127
Chicago/Turabian StyleCubo, Leticia, Thalia Parro, Amancio Carnero, Luca Salassa, Ana I. Matesanz, and Adoracion G. Quiroga. 2018. "Synthesis, Reactivity Studies, and Cytotoxicity of Two trans-Iodidoplatinum(II) Complexes. Does Photoactivation Work?" Inorganics 6, no. 4: 127. https://doi.org/10.3390/inorganics6040127
APA StyleCubo, L., Parro, T., Carnero, A., Salassa, L., Matesanz, A. I., & Quiroga, A. G. (2018). Synthesis, Reactivity Studies, and Cytotoxicity of Two trans-Iodidoplatinum(II) Complexes. Does Photoactivation Work? Inorganics, 6(4), 127. https://doi.org/10.3390/inorganics6040127