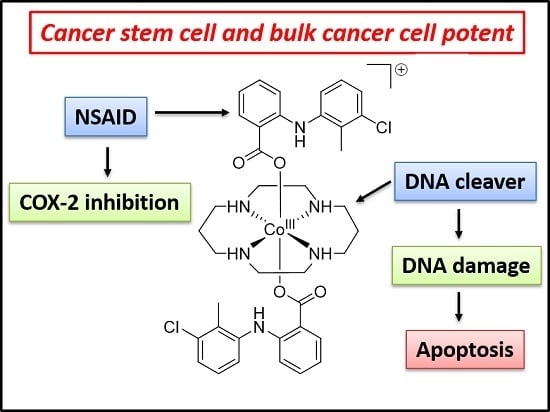
A Cancer Stem Cell Potent Cobalt(III)–Cyclam Complex Bearing Two Tolfenamic Acid Moieties
Abstract
:1. Introduction
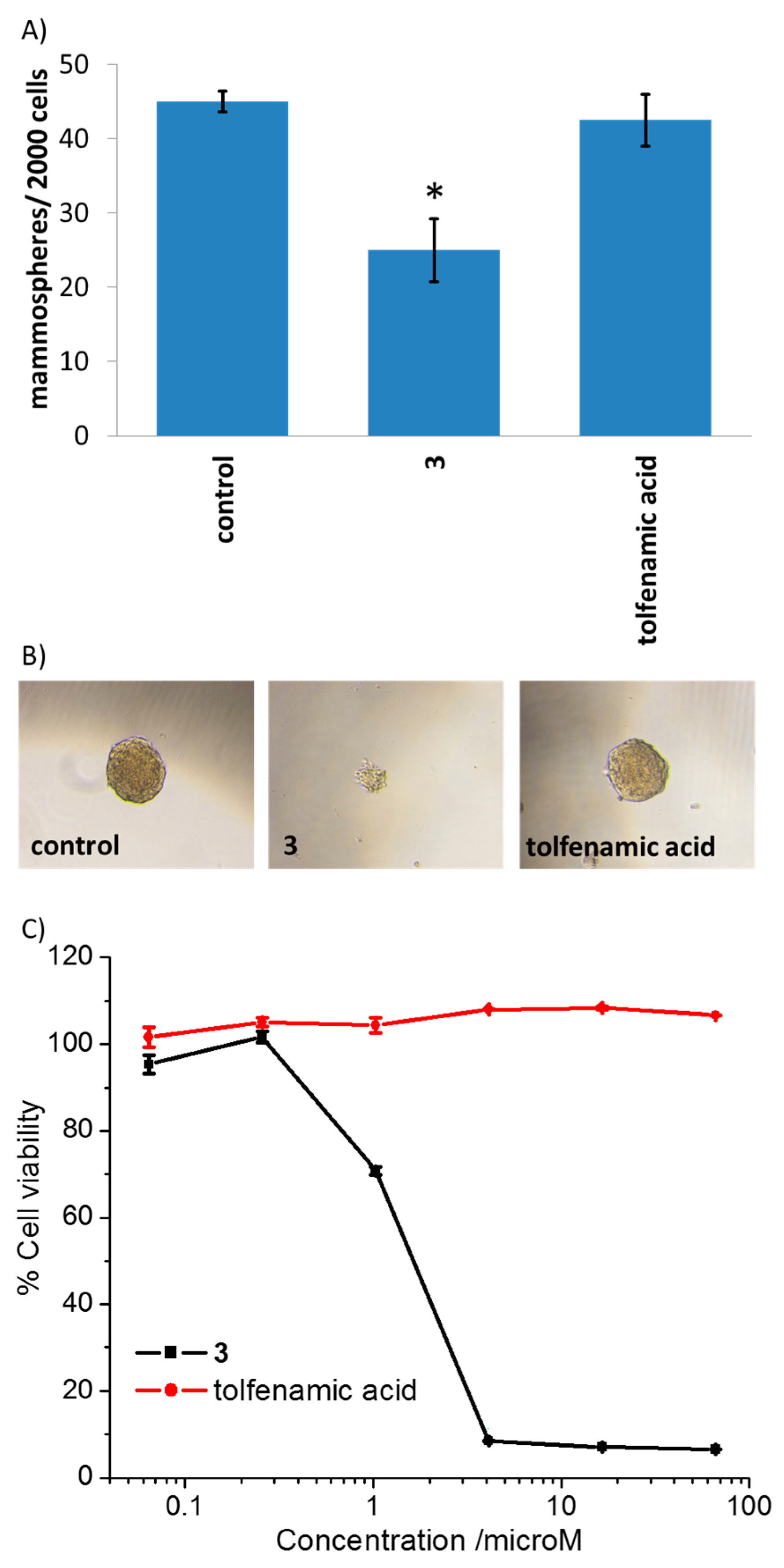
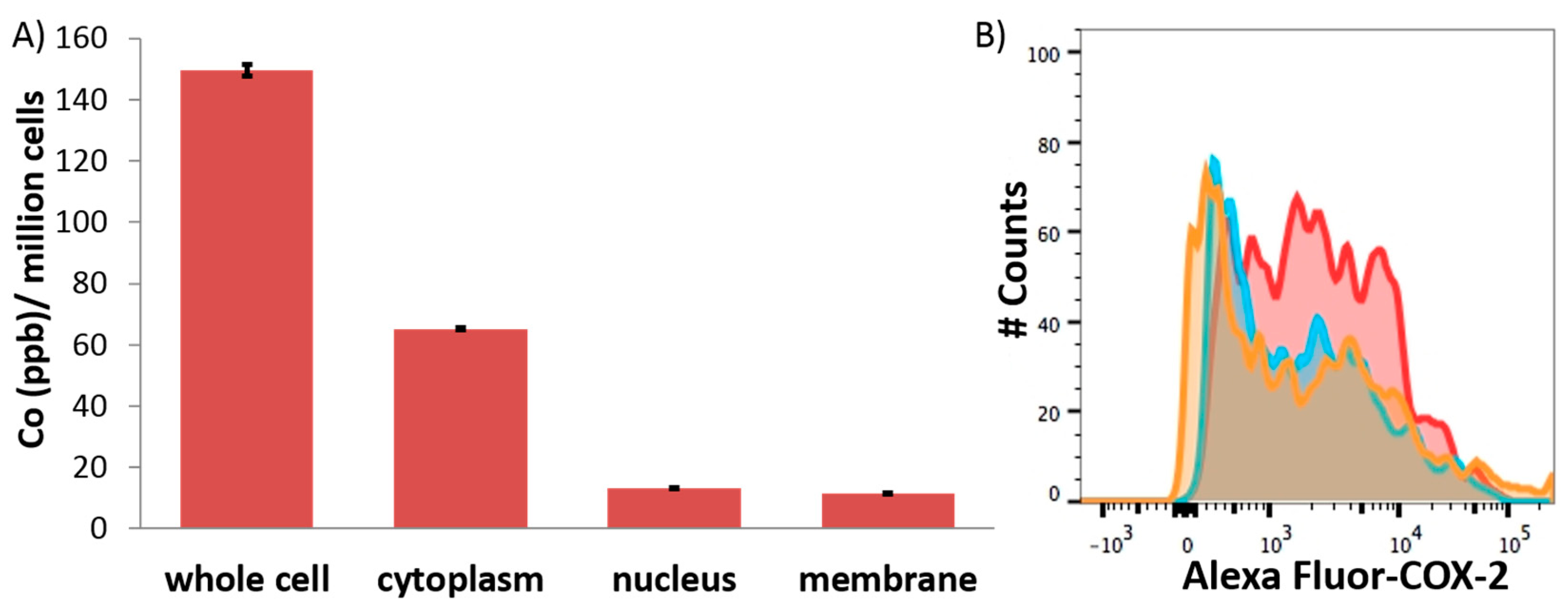
2. Results and Discussion
3. Materials and Methods
3.1. General Procedures
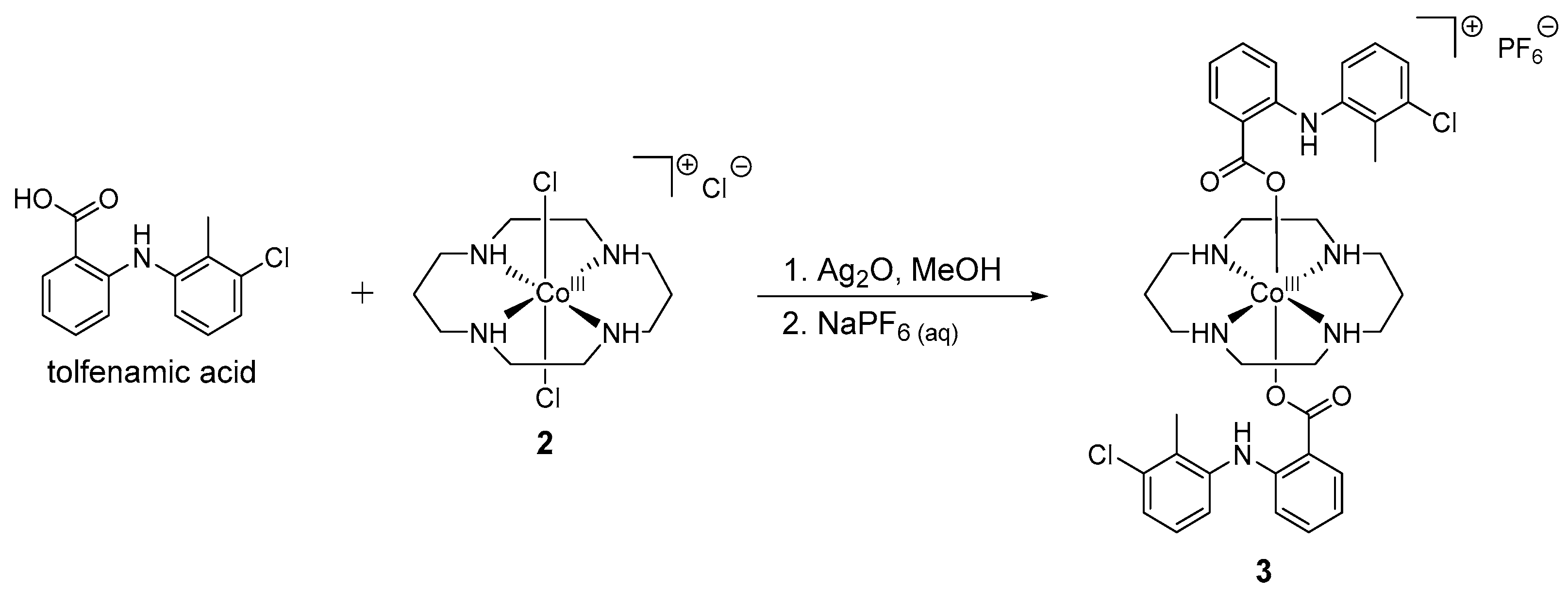
3.2. Synthesis of Co(1,4,8,11-Tetraazacyclotetradecane)(Tolfenamic Acid)2 (3)
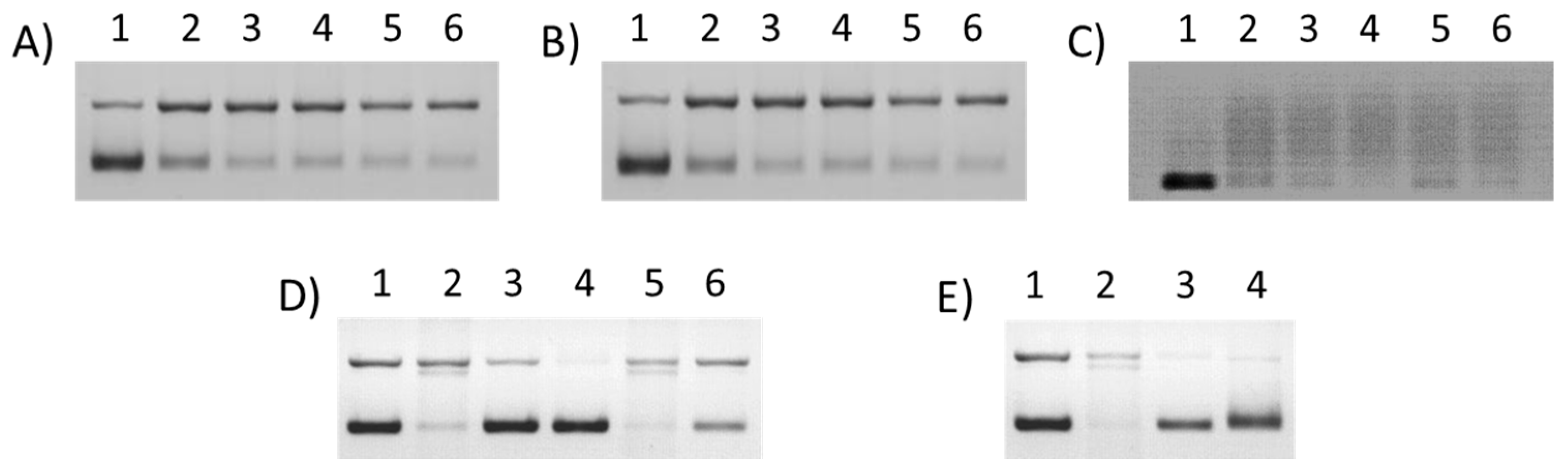
3.3. DNA Cleavage Studies
3.4. Measurement of Water–Octanol Partition Coefficient (LogP)
3.5. Cell Lines and Cell Culture Conditions
3.6. Cytotoxicity MTT Assay
3.7. Tumorsphere Formation and Viability Assay
3.8. Cellular Uptake
3.9. Immunoblotting Analysis
3.10. Flow Cytometry
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- World Health Organization. Fact Sheet; WHO Press: Geneva, Switzerland, 2015. [Google Scholar]
- Morrison, R.; Schleicher, S.M.; Sun, Y.; Niermann, K.J.; Kim, S.; Spratt, D.E.; Chung, C.H.; Lu, B. Targeting the mechanisms of resistance to chemotherapy and radiotherapy with the cancer stem cell hypothesis. J. Oncol. 2011, 2011, 941876. [Google Scholar] [CrossRef] [PubMed]
- Gupta, P.B.; Chaffer, C.L.; Weinberg, R.A. Cancer stem cells: Mirage or reality? Nat. Med. 2009, 15, 1010–1012. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, L.V.; Vanner, R.; Dirks, P.; Eaves, C.J. Cancer stem cells: An evolving concept. Nat. Rev. Cancer 2012, 12, 133–143. [Google Scholar] [CrossRef] [PubMed]
- Dean, M.; Fojo, T.; Bates, S. Tumour stem cells and drug resistance. Nat. Rev. Cancer 2005, 5, 275–284. [Google Scholar] [CrossRef] [PubMed]
- Tan, B.T.; Park, C.Y.; Ailles, L.E.; Weissman, I.L. The cancer stem cell hypothesis: A work in progress. Lab. Investig. 2006, 86, 1203–1207. [Google Scholar] [CrossRef] [PubMed]
- Kaiser, J. The cancer stem cell gamble. Science 2015, 347, 226–229. [Google Scholar] [CrossRef] [PubMed]
- Chen, K.; Huang, Y.-H.; Chen, J.-L. Understanding and targeting cancer stem cells: Therapeutic implications and challenges. Acta Pharmacol. Sin. 2013, 34, 732–740. [Google Scholar] [CrossRef] [PubMed]
- Marx, J. Cancer’s perpetual source? Science 2007, 317, 1029–1031. [Google Scholar] [CrossRef] [PubMed]
- Yu, Y.; Ramena, G.; Elble, R.C. The role of cancer stem cells in relapse of solid tumors. Front. Biosci. 2012, 4, 1528–1541. [Google Scholar] [CrossRef]
- Feng, Y.X.; Sokol, E.S.; Del Vecchio, C.A.; Sanduja, S.; Claessen, J.H.; Proia, T.A.; Jin, D.X.; Reinhardt, F.; Ploegh, H.L.; Wang, Q.; et al. Epithelial-to-mesenchymal transition activates PERK-eIF2α and sensitizes cells to endoplasmic reticulum stress. Cancer Discov. 2014, 4, 702–715. [Google Scholar] [CrossRef] [PubMed]
- Lamb, R.; Harrison, H.; Hulit, J.; Smith, D.L.; Lisanti, M.P.; Sotgia, F. Mitochondria as new therapeutic targets for eradicating cancer stem cells: Quantitative proteomics and functional validation via MCT1/2 inhibition. Oncotarget 2014, 5, 11029–11037. [Google Scholar] [CrossRef] [PubMed]
- Al-Hajj, M.; Wicha, M.S.; Benito-Hernandez, A.; Morrison, S.J.; Clarke, M.F. Prospective identification of tumorigenic breast cancer cells. Proc. Natl. Acad. Sci. USA 2003, 100, 3983–3988. [Google Scholar] [CrossRef] [PubMed]
- Eramo, A.; Lotti, F.; Sette, G.; Pilozzi, E.; Biffoni, M.; Di Virgilio, A.; Conticello, C.; Ruco, L.; Peschle, C.; De Maria, R. Identification and expansion of the tumorigenic lung cancer stem cell population. Cell Death Differ. 2008, 15, 504–514. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Heidt, D.G.; Dalerba, P.; Burant, C.F.; Zhang, L.; Adsay, V.; Wicha, M.; Clarke, M.F.; Simeone, D.M. Identification of pancreatic cancer stem cells. Cancer Res. 2007, 67, 1030–1037. [Google Scholar] [CrossRef] [PubMed]
- Prince, M.E.; Sivanandan, R.; Kaczorowski, A.; Wolf, G.T.; Kaplan, M.J.; Dalerba, P.; Weissman, I.L.; Clarke, M.F.; Ailles, L.E. Identification of a subpopulation of cells with cancer stem cell properties in head and neck squamous cell carcinoma. Proc. Natl. Acad. Sci. USA 2007, 104, 973–978. [Google Scholar] [CrossRef] [PubMed]
- Singh, S.K.; Hawkins, C.; Clarke, I.D.; Squire, J.A.; Bayani, J.; Hide, T.; Henkelman, R.M.; Cusimano, M.D.; Dirks, P.B. Identification of human brain tumour initiating cells. Nature 2004, 432, 396–401. [Google Scholar] [CrossRef] [PubMed]
- Janikova, M.; Skarda, J. Differentiation pathways in carcinogenesis and in chemo- and radioresistance. Neoplasma 2012, 59, 6–17. [Google Scholar] [CrossRef] [PubMed]
- Korkaya, H.; Paulson, A.; Charafe-Jauffret, E.; Ginestier, C.; Brown, M.; Dutcher, J.; Clouthier, S.G.; Wicha, M.S. Regulation of mammary stem/progenitor cells by PTEN/Akt/β-catenin signaling. PLoS Biol. 2009, 7, e1000121. [Google Scholar] [CrossRef] [PubMed]
- Takebe, N.; Harris, P.J.; Warren, R.Q.; Ivy, S.P. Targeting cancer stem cells by inhibiting Wnt, Notch, and Hedgehog pathways. Nat. Rev. Clin. Oncol. 2011, 8, 97–106. [Google Scholar] [CrossRef] [PubMed]
- Albini, A.; Bruno, A.; Gallo, C.; Pajardi, G.; Noonan, D.M.; Dallaglio, K. Cancer stem cells and the tumor microenvironment: Interplay in tumor heterogeneity. Connect. Tissue Res. 2015, 56, 414–425. [Google Scholar] [CrossRef] [PubMed]
- Kise, K.; Kinugasa-Katayama, Y.; Takakura, N. Tumor microenvironment for cancer stem cells. Adv. Drug Deliv. Rev. 2015, 99, 197–205. [Google Scholar] [CrossRef] [PubMed]
- Boodram, J.N.; McGregor, I.J.; Bruno, P.M.; Cressey, P.B.; Hemann, M.T.; Suntharalingam, K. Breast cancer stem cell potent copper(II)-non-steroidal anti-inflammatory drug complexes. Angew. Chem. Int. Ed. 2016, 55, 2845–2850. [Google Scholar] [CrossRef] [PubMed]
- Cressey, P.B.; Eskandari, A.; Bruno, P.M.; Lu, C.; Hemann, M.T.; Suntharalingam, K. The potent inhibitory effect of a naproxen-appended cobalt(III)–cyclam complex on cancer stem cells. ChemBioChem 2016, 17, 1713–1718. [Google Scholar] [CrossRef] [PubMed]
- Eskandari, A.; Boodram, J.N.; Cressey, P.B.; Lu, C.; Bruno, P.M.; Hemann, M.T.; Suntharalingam, K. The breast cancer stem cell potency of copper(II) complexes bearing nonsteroidal anti-inflammatory drugs and their encapsulation using polymeric nanoparticles. Dalton Trans. 2016, 45, 17867–17873. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez-Bartulos, M.; Aceves-Luquero, C.; Qualai, J.; Cusso, O.; Martinez, M.A.; Fernandez de Mattos, S.; Menendez, J.A.; Villalonga, P.; Costas, M.; Ribas, X.; et al. Pro-oxidant activity of amine-pyridine-based iron complexes efficiently kills cancer and cancer stem-like cells. PLoS ONE 2015, 10, e0137800. [Google Scholar] [CrossRef] [PubMed]
- Lum, C.T.; Wong, A.S.; Lin, M.C.; Che, C.M.; Sun, R.W. A gold(III) porphyrin complex as an anti-cancer candidate to inhibit growth of cancer-stem cells. Chem. Commun. 2013, 49, 4364–4366. [Google Scholar] [CrossRef] [PubMed]
- Suntharalingam, K.; Lin, W.; Johnstone, T.C.; Bruno, P.M.; Zheng, Y.R.; Hemann, M.T.; Lippard, S.J. A breast cancer stem cell-selective, mammospheres-potent osmium(VI) nitrido complex. J. Am. Chem. Soc. 2014, 136, 14413–14416. [Google Scholar] [CrossRef] [PubMed]
- Heddleston, J.M.; Li, Z.; Lathia, J.D.; Bao, S.; Hjelmeland, A.B.; Rich, J.N. Hypoxia inducible factors in cancer stem cells. Br. J. Cancer 2010, 102, 789–795. [Google Scholar] [CrossRef] [PubMed]
- Heddleston, J.M.; Li, Z.; McLendon, R.E.; Hjelmeland, A.B.; Rich, J.N. The hypoxic microenvironment maintains glioblastoma stem cells and promotes reprogramming towards a cancer stem cell phenotype. Cell Cycle 2009, 8, 3274–3284. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Bao, S.; Wu, Q.; Wang, H.; Eyler, C.; Sathornsumetee, S.; Shi, Q.; Cao, Y.; Lathia, J.; McLendon, R.E.; et al. Hypoxia-inducible factors regulate tumorigenic capacity of glioma stem cells. Cancer Cell 2009, 15, 501–513. [Google Scholar] [CrossRef] [PubMed]
- Soeda, A.; Park, M.; Lee, D.; Mintz, A.; Androutsellis-Theotokis, A.; McKay, R.D.; Engh, J.; Iwama, T.; Kunisada, T.; Kassam, A.B.; et al. Hypoxia promotes expansion of the CD133-positive glioma stem cells through activation of HIF-1α. Oncogene 2009, 28, 3949–3959. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Li, L. Stem cell niche: Microenvironment and beyond. J. Biol. Chem. 2008, 283, 9499–9503. [Google Scholar] [CrossRef] [PubMed]
- Abu-Surrah, A.S.; Kettunen, M. Platinum group antitumor chemistry: Design and development of new anticancer drugs complementary to cisplatin. Curr. Med. Chem. 2006, 13, 1337–1357. [Google Scholar] [CrossRef] [PubMed]
- Hall, M.D.; Failes, T.W.; Yamamoto, N.; Hambley, T.W. Bioreductive activation and drug chaperoning in cobalt pharmaceuticals. Dalton Trans. 2007, 3983–3990. [Google Scholar] [CrossRef] [PubMed]
- Heffern, M.C.; Yamamoto, N.; Holbrook, R.J.; Eckermann, A.L.; Meade, T.J. Cobalt derivatives as promising therapeutic agents. Curr. Opin. Chem. Biol. 2013, 17, 189–196. [Google Scholar] [CrossRef] [PubMed]
- Munteanu, C.R.; Suntharalingam, K. Advances in cobalt complexes as anticancer agents. Dalton Trans. 2015, 44, 13796–13808. [Google Scholar] [CrossRef] [PubMed]
- Chang, J.Y.; Lu, G.L.; Stevenson, R.J.; Brothers, P.J.; Clark, G.R.; Botting, K.J.; Ferry, D.M.; Tercel, M.; Wilson, W.R.; Denny, W.A.; et al. Cross-bridged cyclen or cyclam Co(III) complexes containing cytotoxic ligands as hypoxia-activated prodrugs. Inorg. Chem. 2013, 52, 7688–7698. [Google Scholar] [CrossRef] [PubMed]
- Chang, J.Y.-C.; Stevenson, R.J.; Lu, G.-L.; Brothers, P.J.; Clark, G.R.; Denny, W.A.; Ware, D.C. Syntheses of 8-quinolinolatocobalt(III) complexes containing cyclen based auxiliary ligands as models for hypoxia-activated prodrugs. Dalton Trans. 2010, 39, 11535–11550. [Google Scholar] [CrossRef] [PubMed]
- Renfrew, A.K.; Bryce, N.S.; Hambley, T. Cobalt(III) chaperone complexes of curcumin: Photoreduction, cellular accumulation and light-selective toxicity towards tumour cells. Chemistry 2015, 21, 15224–15234. [Google Scholar] [CrossRef] [PubMed]
- Yamamoto, N.; Danos, S.; Bonnitcha, P.D.; Failes, T.W.; New, E.J.; Hambley, T.W. Cellular uptake and distribution of cobalt complexes of fluorescent ligands. J. Biol. Inorg. Chem. 2008, 13, 861–871. [Google Scholar] [CrossRef] [PubMed]
- Yamamoto, N.; Renfrew, A.K.; Kim, B.J.; Bryce, N.S.; Hambley, T.W. Dual targeting of hypoxic and acidic tumor environments with a cobalt(III) chaperone complex. J. Med. Chem. 2012, 55, 11013–11021. [Google Scholar] [CrossRef] [PubMed]
- Abramson, S.B.; Weissmann, G. The mechanisms of action of nonsteroidal antiinflammatory drugs. Arthritis Rheum. 1989, 32, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Moon, C.M.; Kwon, J.H.; Kim, J.S.; Oh, S.H.; Jin Lee, K.; Park, J.J.; Pil Hong, S.; Cheon, J.H.; Kim, T.I.; Kim, W.H. Nonsteroidal anti-inflammatory drugs suppress cancer stem cells via inhibiting PTGS2 (cyclooxygenase 2) and NOTCH/HES1 and activating PPARG in colorectal cancer. Int. J. Cancer 2014, 134, 519–529. [Google Scholar] [CrossRef] [PubMed]
- Singh, B.; Berry, J.A.; Shoher, A.; Ramakrishnan, V.; Lucci, A. Cox-2 overexpression increases motility and invasion of breast cancer cells. Int. J. Oncol. 2005, 26, 1393–1399. [Google Scholar] [CrossRef] [PubMed]
- Sharma, V.; Dixit, D.; Ghosh, S.; Sen, E. Cox-2 regulates the proliferation of glioma stem like cells. Neurochem. Int. 2011, 59, 567–571. [Google Scholar] [CrossRef] [PubMed]
- Pentikainen, P.J.; Neuvonen, P.J.; Backman, C. Human pharmacokinetics of tolfenamic acid, a new anti-inflammatory agent. Eur. J. Clin. Pharmacol. 1981, 19, 359–365. [Google Scholar] [CrossRef] [PubMed]
- Meieranz, S.; Stefanopoulou, M.; Rubner, G.; Bensdorf, K.; Kubutat, D.; Sheldrick, W.S.; Gust, R. The biological activity of zeise’s salt and its derivatives. Angew. Chem. Int. Ed. 2015, 54, 2834–2837. [Google Scholar] [CrossRef] [PubMed]
- Ott, I.; Kircher, B.; Bagowski, C.P.; Vlecken, D.H.; Ott, E.B.; Will, J.; Bensdorf, K.; Sheldrick, W.S.; Gust, R. Modulation of the biological properties of aspirin by formation of a bioorganometallic derivative. Angew. Chem. Int. Ed. 2009, 48, 1160–1163. [Google Scholar] [CrossRef] [PubMed]
- Pathak, R.K.; Marrache, S.; Choi, J.H.; Berding, T.B.; Dhar, S. The prodrug platin-A: Simultaneous release of cisplatin and aspirin. Angew. Chem. Int. Ed. 2014, 53, 1963–1967. [Google Scholar] [CrossRef] [PubMed]
- Psomas, G.; Kessissoglou, D.P. Quinolones and non-steroidal anti-inflammatory drugs interacting with copper(II), nickel(II), cobalt(II) and zinc(II): Structural features, biological evaluation and perspectives. Dalton Trans. 2013, 42, 6252–6276. [Google Scholar] [CrossRef] [PubMed]
- Krstic, N.S.; Nikolic, R.S.; Stankovic, M.N.; Nikolic, N.G.; Dordevic, D.M. Coordination compounds of M(II) biometal ions with acid-type anti-inflammatory drugs as ligands—A review. Trop. J. Pharm. Res. 2015, 14, 337–349. [Google Scholar] [CrossRef]
- Banti, C.N.; Hadjikakou, S.K. Non-steroidal anti-inflammatory drugs (NSAIDS) in metal complexes and their effect at the cellular level. Eur. J. Inorg. Chem. 2016, 2016, 3048–3071. [Google Scholar] [CrossRef]
- Dontu, G.; Abdallah, W.M.; Foley, J.M.; Jackson, K.W.; Clarke, M.F.; Kawamura, M.J.; Wicha, M.S. In vitro propagation and transcriptional profiling of human mammary stem/progenitor cells. Genes Dev. 2003, 17, 1253–1270. [Google Scholar] [CrossRef] [PubMed]
- Roos, W.P.; Thomas, A.D.; Kaina, B. DNA damage and the balance between survival and death in cancer biology. Nat. Rev. Cancer 2016, 16, 20–33. [Google Scholar] [CrossRef] [PubMed]
- Singh, B.; Cook, K.R.; Vincent, L.; Hall, C.S.; Martin, C.; Lucci, A. Role of Cox-2 in tumorospheres derived from a breast cancer cell line. J. Surg. Res. 2011, 168, e39–e49. [Google Scholar] [CrossRef] [PubMed]
- Kanojia, D.; Zhou, W.; Zhang, J.; Jie, C.; Lo, P.K.; Wang, Q.; Chen, H. Proteomic profiling of cancer stem cells derived from primary tumors of HER2/Neu transgenic mice. Proteomics 2012, 12, 3407–3415. [Google Scholar] [CrossRef] [PubMed]
- Bosnich, B.; Poon, C.K.; Tobe, M.L. Complexes of cobalt(III) with a cyclic tetradentate secondary amine. Inorg. Chem. 1965, 4, 1102–1108. [Google Scholar] [CrossRef]
- Gupta, P.B.; Onder, T.T.; Jiang, G.; Tao, K.; Kuperwasser, C.; Weinberg, R.A.; Lander, E.S. Identification of selective inhibitors of cancer stem cells by high-throughput screening. Cell 2009, 138, 645–659. [Google Scholar] [CrossRef] [PubMed]

| Compound | HMLER IC50 (μM) | HMLER-shEcad IC50 (μM) | Mammosphere IC50 (μM) |
|---|---|---|---|
| 1 1 | 0.43 ± 0.05 | 0.11 ± 0.03 | 0.98 ± 0.02 |
| 3 | 0.22 ± 0.01 | 0.21 ± 0.01 | 1.83 ± 0.28 |
| salinomycin 1 | 12.17 ± 3.16 | 5.06 ± 1.45 | 14.05 ± 1.58 |
| tolfenamic acid 2 | 30.77 ± 4.69 | 57.95 ± 1.62 | >66 |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license ( http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cressey, P.B.; Eskandari, A.; Suntharalingam, K. A Cancer Stem Cell Potent Cobalt(III)–Cyclam Complex Bearing Two Tolfenamic Acid Moieties. Inorganics 2017, 5, 12. https://doi.org/10.3390/inorganics5010012
Cressey PB, Eskandari A, Suntharalingam K. A Cancer Stem Cell Potent Cobalt(III)–Cyclam Complex Bearing Two Tolfenamic Acid Moieties. Inorganics. 2017; 5(1):12. https://doi.org/10.3390/inorganics5010012
Chicago/Turabian StyleCressey, Paul B., Arvin Eskandari, and Kogularamanan Suntharalingam. 2017. "A Cancer Stem Cell Potent Cobalt(III)–Cyclam Complex Bearing Two Tolfenamic Acid Moieties" Inorganics 5, no. 1: 12. https://doi.org/10.3390/inorganics5010012
APA StyleCressey, P. B., Eskandari, A., & Suntharalingam, K. (2017). A Cancer Stem Cell Potent Cobalt(III)–Cyclam Complex Bearing Two Tolfenamic Acid Moieties. Inorganics, 5(1), 12. https://doi.org/10.3390/inorganics5010012