1. Introduction
In science, understanding the physical and chemical properties of materials is of great significance, as these properties depend primarily on their molecular structure. Predicting the atomic structure of a molecule using computational methods, starting from an arbitrary initial position of its atoms, is a complex task, since the system may include a large number of local minima on the potential energy surface (PES) [
1,
2]. In recent decades, several methods have been developed to optimize molecular structures, such as minimum jumping [
3,
4], simulated annealing [
5,
6], the random sampling method [
7,
8], basin-hopping (BH) [
9,
10], and metadynamics [
10,
11].
As the system to be studied increases in size with the number of atoms, the problem becomes more complex, increasing the computational cost. An important aspect of the research on structural configurations of clusters is the determination of their lowest potential energy. This step is essential for the study of the properties of the clusters since the structure with the lowest potential energy corresponds to the most stable configuration [
12]. This problem is known as global optimization (GO) since it involves the determination of the lowest minimum value of the energy based on the atomic coordinates of the clusters in the PES. Due to the inherent complexity of the problem, it is impossible to solve it using analytical calculation methods. In practice, the problem can only be addressed through numerical techniques. Furthermore, any algorithm that searches for a global minimum must broadly explore the energy landscape, going deep into its most relevant regions, and thus perform a more efficient exploration [
13].
GO is an arduous task due to the enormous number of local equilibrium configurations (local energy minima) of the cluster structures in the PES, which increase exponentially with the number of atoms
N in the system [
2,
14,
15,
16,
17,
18,
19]. These local minima correspond to different geometric structures, which may present different properties. Thus, the further the optimized structural parameters are separated from those reported experimentally by X-ray diffraction, the less accurate the properties of the modeled structure will be.
Advances in energy landscape theory have helped narrow the search space for the diversity of more stable cluster structures in the PES [
20,
21,
22]. Some studies reported in the literature implement approximations with harmonic or Hookeian potentials to estimate the structure of some clusters [
23,
24]. A simple harmonic oscillator is a model used to study a wide range of phenomena, such as pendulum oscillations [
25], sound waves [
26], and bonds between atoms [
27]. The latter considers the mass of the atom as a particle oscillating around an equilibrium point under the action of a restoring force, which is proportional to the distance from the equilibrium point, like the mass-spring system in Hooke’s Law, from which the oscillations or vibrations of the bonds are obtained, showing the existence of a vibrational spectrum in the infrared region [
18].
One of the methods that is receiving the most attention in the optimization of molecular structures is the Particle Swarm Optimization (PSO) [
28]. The algorithm is based on circumstances similar to the movement of a school of fish, a swarm of bees, or a flock of birds in motion by adjusting their positions and velocities. In general, a colony of animals, apparently without a leader, will change their speeds randomly in search of food, following the group member closest to the food source (possible solution). The group of animals optimizes its positions by following the members that have already reached a better orientation. This exploration process is repeated until the best position within the group is obtained, which is considered to be the position vector in the search space, and the velocity vector regulates the subsequent distribution or movement of the particles. The PSO method adjusts or updates the position and velocity of each particle at each step until achieving the best position experienced by the collective or swarm [
29,
30].
Another alternative is the basin-hopping (BH) method, which is a metaheuristic global optimization method combining local searches with stochastic jumps between different regions of the [
9] space. This method involves two-step loops, a perturbation with good candidate solutions, and the application of the local search to the perturbed solution, transforming the complex energy landscape into a collection of basins that are explored by jumps. Monte Carlo random moves and the Metropolis criterion’s acceptance or rejection of the solutions perform these jumps. Alternatively, this method is beneficial for solving problems with multiple local minima, such as particle cluster optimization [
9,
31].
In the present work, we are interested in optimizing two types of clusters: carbon and those formed by oxygen atoms and transition metals. The first group is important for organic, inorganic, and physical chemistry [
32]. Furthermore, these molecules are also of great importance in astrophysics, especially regarding the chemistry of carbon stars [
33], comets [
34], and interstellar molecular clouds [
35]. On the other hand, carbon atom clusters are vital elements in hydrocarbon flames [
36], and play a crucial role in gas-phase carbon chemistry, acting as precursors in the production of fullerenes, carbon nanotubes, diamond films, and silicon carbides [
37,
38,
39]. Studying and synthesizing these molecules in the laboratory is challenging due to their high reactivity [
40], underscoring the interest in studying the structural information of carbon clusters in theoretical research [
41,
42,
43]. On the other hand, clusters consisting of oxygen atoms and transition metals with a high oxidation state (
,
, or
) have an electronic structure that allows them to act as oxidizing agents, making them good candidates for obtaining new materials. These clusters are precursors of polyoxometalate compounds (POM), which are formed from condensation reactions through the self-assembly of some oxometalates, such as
,
,
,
, or
[
44,
45].
The optimization was carried out using a PSO algorithm written in Fortran 90 to locate global minimum energy structures. We have used the algorithm on clusters consisting of carbon atoms
(
n = 3–5) and tungsten–oxygen atoms
(
n = 4–6,
), where
n is the number of mol of atoms of the element in the cluster, and
m is the charge of the anion. In this model, the atoms have been considered rigid spheres joined by a spring (link) with a harmonic potential, where the restoring force (Hooke’s Law) is proportional to the displacement from the equilibrium length. The algorithm was based on the search for the global minimum of the potential energy function of the aforementioned clusters in a multidimensional hyperspace
(where
N is the number of atoms in the system). The motions of the conformers of the clusters are guided by their own most favorable known atomic positions as well as by the best-known position of the entire swarm in the search space
. We have implemented a basin-hopping (BH) method and performed the DFT calculations through the Gaussian 09 software to validate our results. Furthermore, for the case of carbon clusters, we have compared with the results obtained by Jana and collaborators [
46] who used a PSO algorithm written in Python, which combines an evolutionary subroutine with a variational optimization technique through an interface of the PSO algorithm with the Gaussian 09 software.
The article is divided into the following sections:
Section 2, Discussion and Results, presents the results and compares them with the literature;
Section 3, Methodology, presents the details of the algorithm; and
Section 4, Conclusions, outlines the main results.
2. Discussion and Results
Tables S1–S4 of the supplementary material present the initial structures of the conformers of 10 clusters (swarms) of three-, four-, and five-carbon atoms, from which Jana et al. initiated their study of the PSO algorithm [
46]. Using structures optimized with the commercial software Gaussian 09 as a reference, we observed that our results with PSO and BH showed good agreement with the conformations of the optimized structures reported by Jana et al. [
46], for carbon clusters of 3–5 atoms (
–
; see
Table 1).
The Python-based PSO algorithm by Jana et al. [
46], implemented with a swarm of 10 clusters, operates in synergy with the Gaussian software (version 09). This commercial software optimizes cluster structures at each iteration through the gradient method; however, the article by Jana [
46] does not specify whether these optimizations are sequential or parallel. At every step:
Gaussian software provides optimized atomic coordinates for each cluster to the PSO algorithm.
The PSO modifies these coordinates through displacements in the hyperspace
.
The new coordinates are fed back to Gaussian for the subsequent iteration.
Gaussian optimizes cluster structures in Hilbert space
, using the Schlegel algorithm [
47], which begins by minimizing the electronic energy density functional using the conjugate gradient method at each step. The optimized cluster structure is obtained by minimizing the gradient of the electronic energy concerning the nuclear coordinates, through a series of single-point calculations in the PES, applying the Hellmann–Feynman theorem [
48].
The new atomic positions found by the Gaussian software are used by the PSO algorithm to locate the optimal nuclear coordinates, for each cluster (both
and
), based on the electronic energy system.
The cycle repeats during each optimization step performed by the Gaussian software until the electronic energy of the clusters converges.
The critical dependence lies in the fact that the PSO algorithm by Jana [
46] lacks an intrinsic objective function. It exclusively utilizes the electronic energy computed by Gaussian software as a reference to guide atomic displacements in the hyperspace
. This dependence implies two fundamental limitations:
- •
If the electronic energy of the system converges to a local minimum on the PES through Gaussian software, the PSO algorithm is hopelessly trapped in this minimum.
- •
The absence of an intrinsic model to determine the energy of the system in the hyperspace
restricts the PSO to functioning as an atomic configuration generator, without independent evaluation capability during the structural optimization process.
In essence, the algorithm by Jana et al. [
46] acts as a conformational exploration mechanism whose efficacy is contingent upon external quantum chemical computations from Gaussian software.
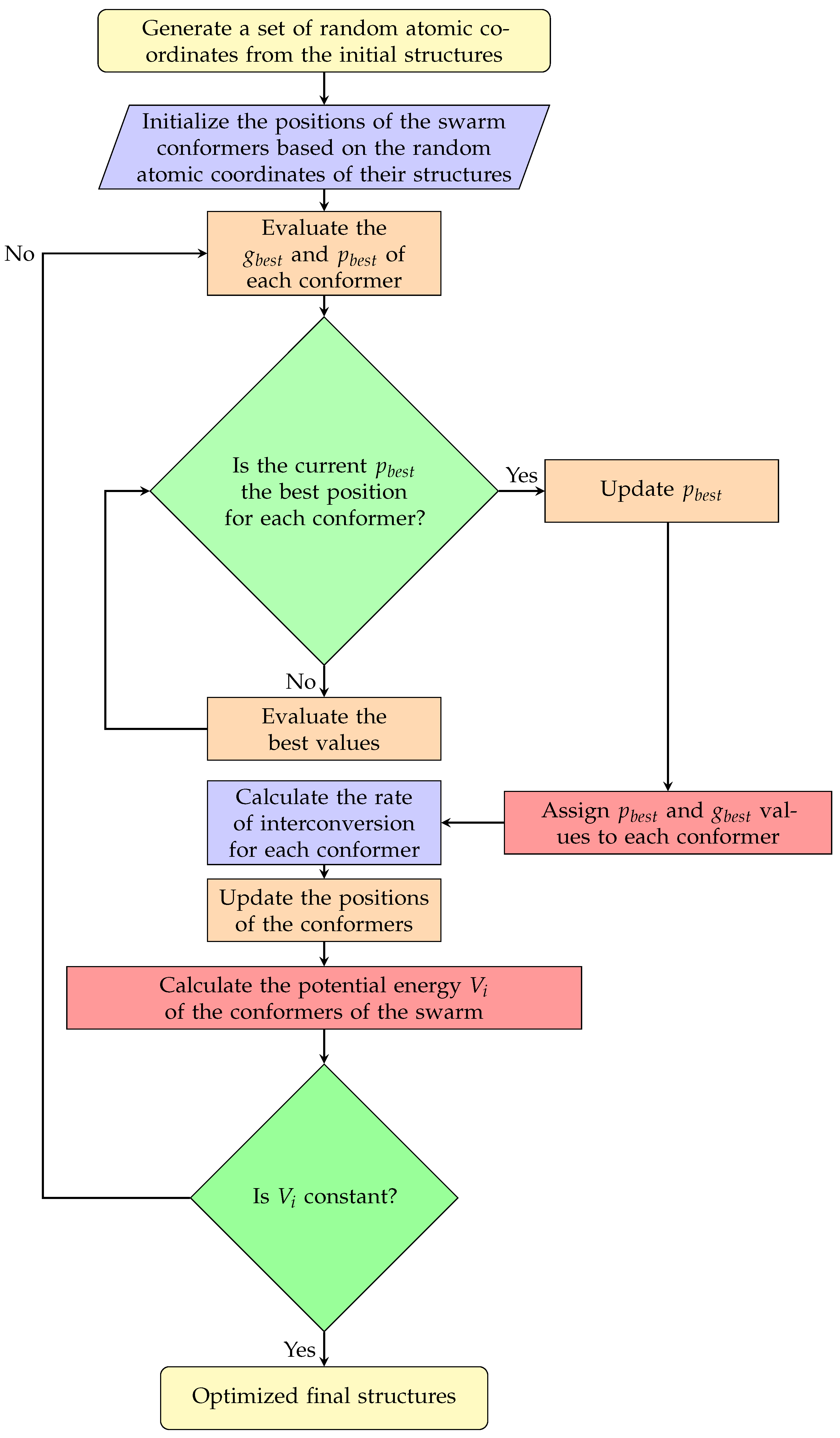
However, in this work, the PSO algorithm independently optimizes the structures of 10 cluster conformers simultaneously, without executing the Gaussian software. The algorithm models atoms as rigid spheres connected by springs and minimizes the potential energy (Equation (5)) for each conformer to determine its optimal positions in the search space
. At each step t, the algorithm updates the best positions
and
until the potential energy of each conformer converges. This convergence is achieved when the velocity and positions vectors satisfy
and
, respectively (Equations (3) and (4)). During PES exploration, each cluster individually stores its atomic positions from iteration
to
; this enables each cluster to retain its exploration history of the hyperspace
, providing a global perspective of the energy landscape. Consequently, the possibility of cluster swarm trapping in local minima during geometric optimization is significantly reduced.
The BH algorithm is a global optimization method designed to find the global minimum energy in systems with multiple local minima. This approach combines local optimization with random jumps to explore the configuration space of molecular structures associated with local minima. The algorithm starts by assigning trial positions to each atom, and the structure is defined using the Atomic Simulation Environment (ASE) library. An energy calculator, such as the effective medium theory (EMT) model, is then used to compute the total energy of the configuration. Restrictions based on Hooke’s Law were applied between specific pairs of atoms, imposing a restoring force proportional to the deviation from a target distance, to maintain physically reasonable structures during the optimization process. The algorithm initiates local optimization from the trial structure configuration using the Limited-Memory Broyden–Fletcher–Goldfarb–Shanno (L-BFGS) algorithm [
49]. This step searches for the local energy minimum near the current configuration. Once a local minimum is found, the algorithm makes a random jump in the configuration space. This jump allows the exploration of new configurations that may have lower energies. The energy of the new configuration is evaluated, and if it is lower than that of the previous configuration, it is accepted as the new current configuration. The new configuration remains acceptable under specific conditions, determined by the system temperature, which enables the algorithm to escape deep local minima when the energy is higher.
Tables S5–S7 in the supplementary material present the initial structures of the 10 conformers of the tungsten–oxygen clusters
,
, and
used for geometry optimization.
Table 2 displays the optimized structures obtained with the BH and PSO algorithms (this work), the commercial Gaussian 09 software, and the experimentally reported structures from single-crystal X-ray diffraction [
50,
51,
52]. The structural conformations of the tungsten–oxygen clusters
(
n = 4–6,
) indicate that the PSO and BH algorithms provide a close approximation to both the commercial Gaussian 09 software results and the experimentally reported structures from single-crystal X-ray diffraction (see
Table 2).
The system energy was computed using a Hookeian potential (Equation (5)), with explicit omission of the electrostatic interaction term. This approach is justified because electrostatic contributions are implicitly incorporated in the force parameters (bond constants
and
in Equation (5)), which were adjusted using experimental Differential Scanning Calorimetry (DSC) data [
53,
54,
55,
56]. Consequently, the force constants inherently account for electrostatic effects associated with interatomic bond energies.
The structural parameters obtained with the PSO algorithm, shown in
Table 3 for the
–
carbon clusters, are comparable to those derived from the BH algorithm and the Gaussian 09 software. The optimization of the three-carbon cluster showed the highest accuracy in its average bond lengths and angles, except for the C–C single bond length predicted by the BH algorithm, which was slightly elongated compared to those from the PSO algorithms (Jana et al. [
46] and the one in this work) and the commercial software Gaussian 09. The average bond lengths for both cyclic and acyclic four-carbon cluster structures showed good agreement, with deviations of 0.02–0.30 Å relative to the Gaussian 09 results. However, a discrepancy of
was observed in the angle of the acyclic
cluster structures optimized using the PSO algorithm from Jana et al. [
46]. For the
cluster, the average bond angles (∼
) obtained with our PSO algorithm demonstrated higher accuracy than those from the algorithm reported by Jana et al. [
46], aligning closely with the structural parameters derived from Gaussian 09 optimizations.
Table 4 presents the average bond lengths and angles of tungsten–oxygen clusters
(
n = 4–6,
), optimized using BH and our PSO algorithms, Gaussian 09 software, and the values reported from single-crystal X-ray diffraction studies [
50,
51,
52]. The results show that the structural parameters of the
(
n = 4–6,
) tungsten–oxygen clusters obtained via the BH and PSO methods are highly consistent with those obtained with Gaussian 09. Furthermore, the calculated average bond lengths and angles closely match the experimental XRD data [
50,
51,
52]. Notably, only the optimized angle
of the
cluster exhibited a deviation
compared to the XRD result, likely due to intermolecular interactions associated with crystal lattice packing [
51,
52].
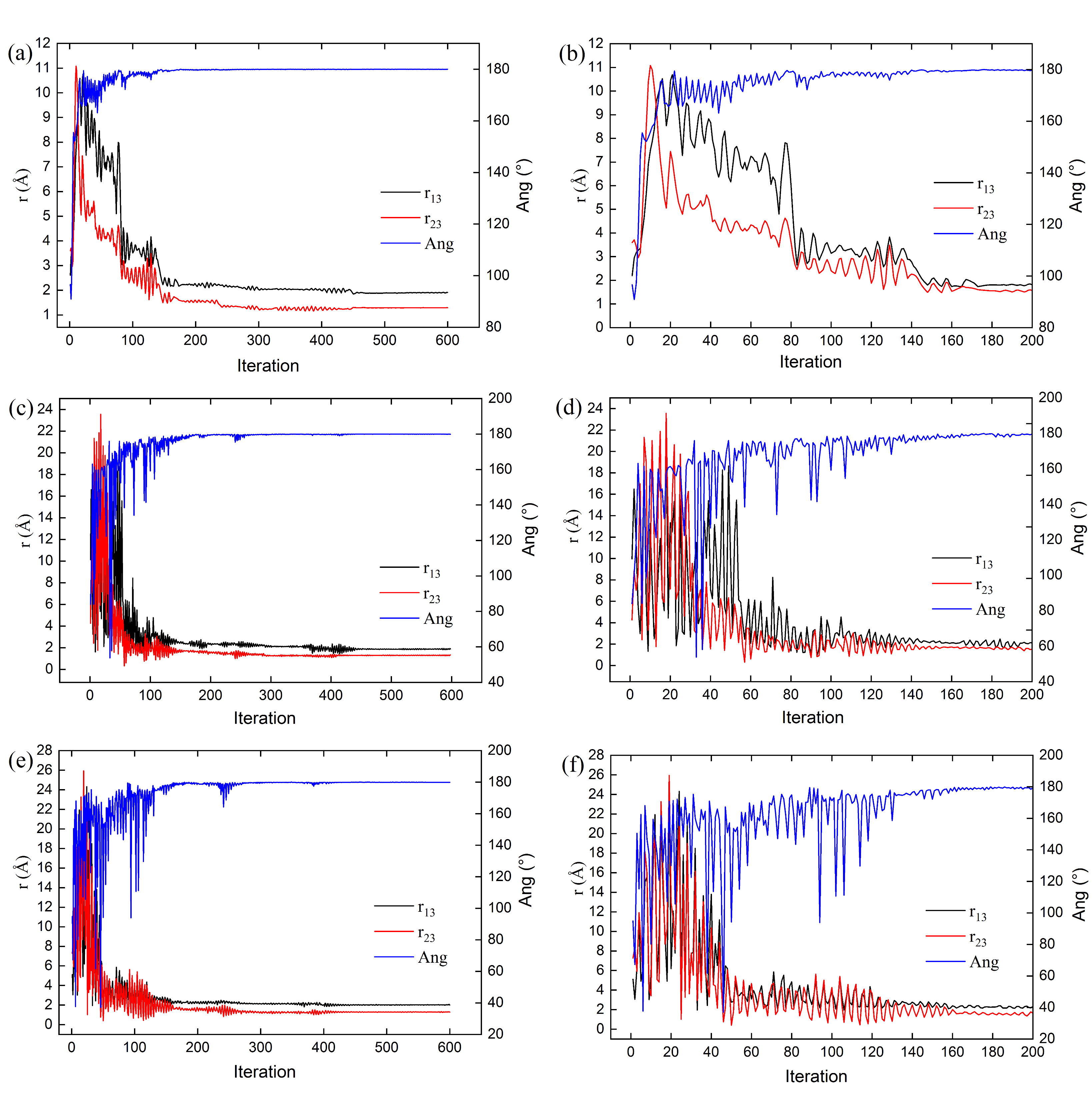
The evolution of the geometric structure optimization for cluster
1 (composed of three carbon atoms,
) is illustrated in
Figure 1, which displays snapshots of the process across iterations. The cluster bonds (
and
) are initially elongated during the first 20 iterations (
Figure 2b), after they gradually decrease to reach a minimum value of 1.29 Å (
Figure 2a and
Table 5). Concurrently, the angle formed by the three carbon atoms undergoes minor variations, increasing from
to
as the iterations progress. This angle stabilizes near
when the potential energy is minimized (
Figure 2a,b and
Table 5). Similar harmonic motion trends—though with higher frequency—are observed in the C–C bond lengths and the angle increase (denoted as Ang) for clusters
8 and 10 (
Figure 2c–f and
Table 5). Notably, within the swarm of 10 clusters, each conformation exhibits distinct dynamics, geometric configurations, and energy profiles during optimization via the PSO algorithm (see
Figure 2,
Figure 3,
Figure 4,
Figure 5 and
Figure 6 and
Table 5).
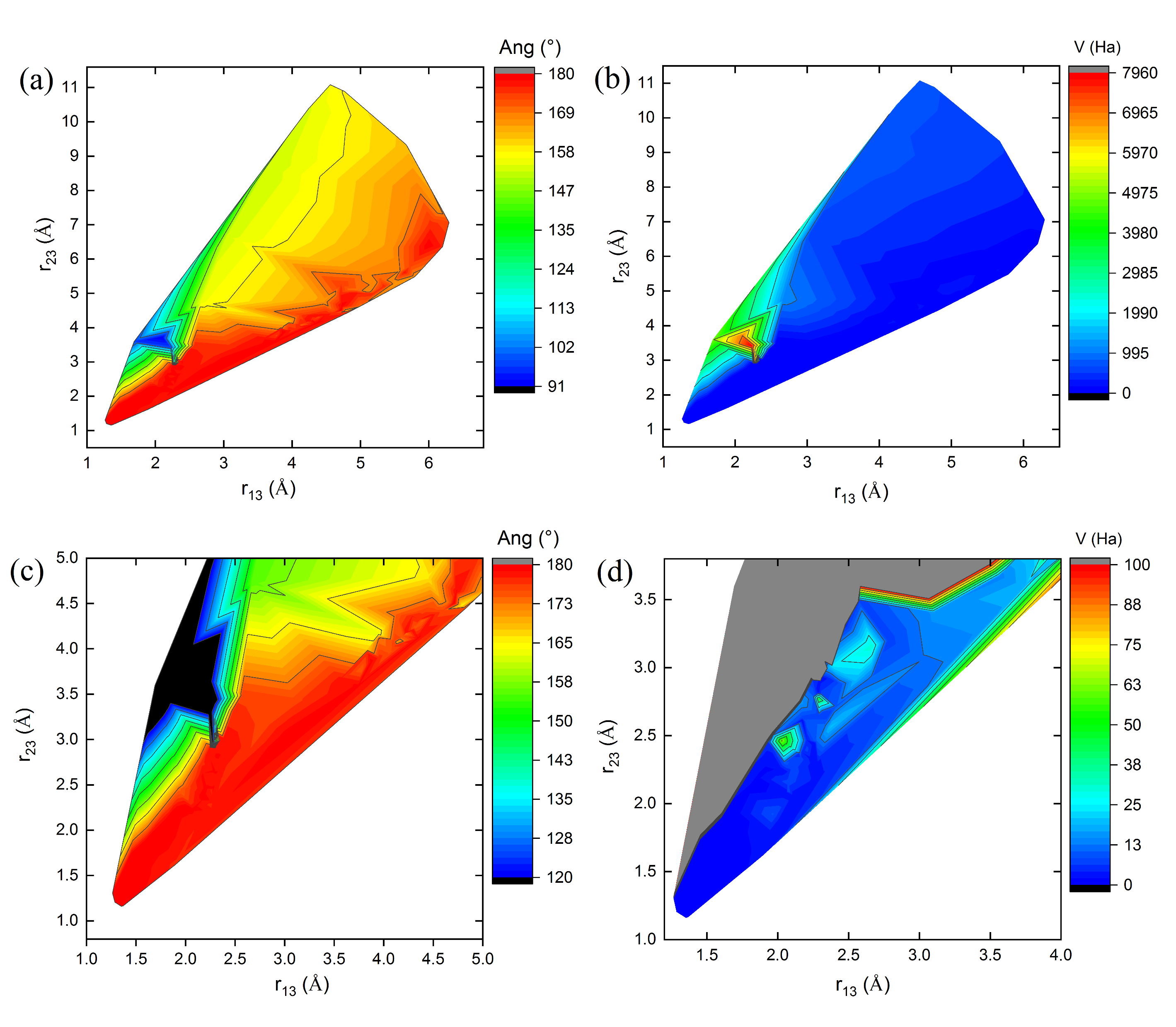
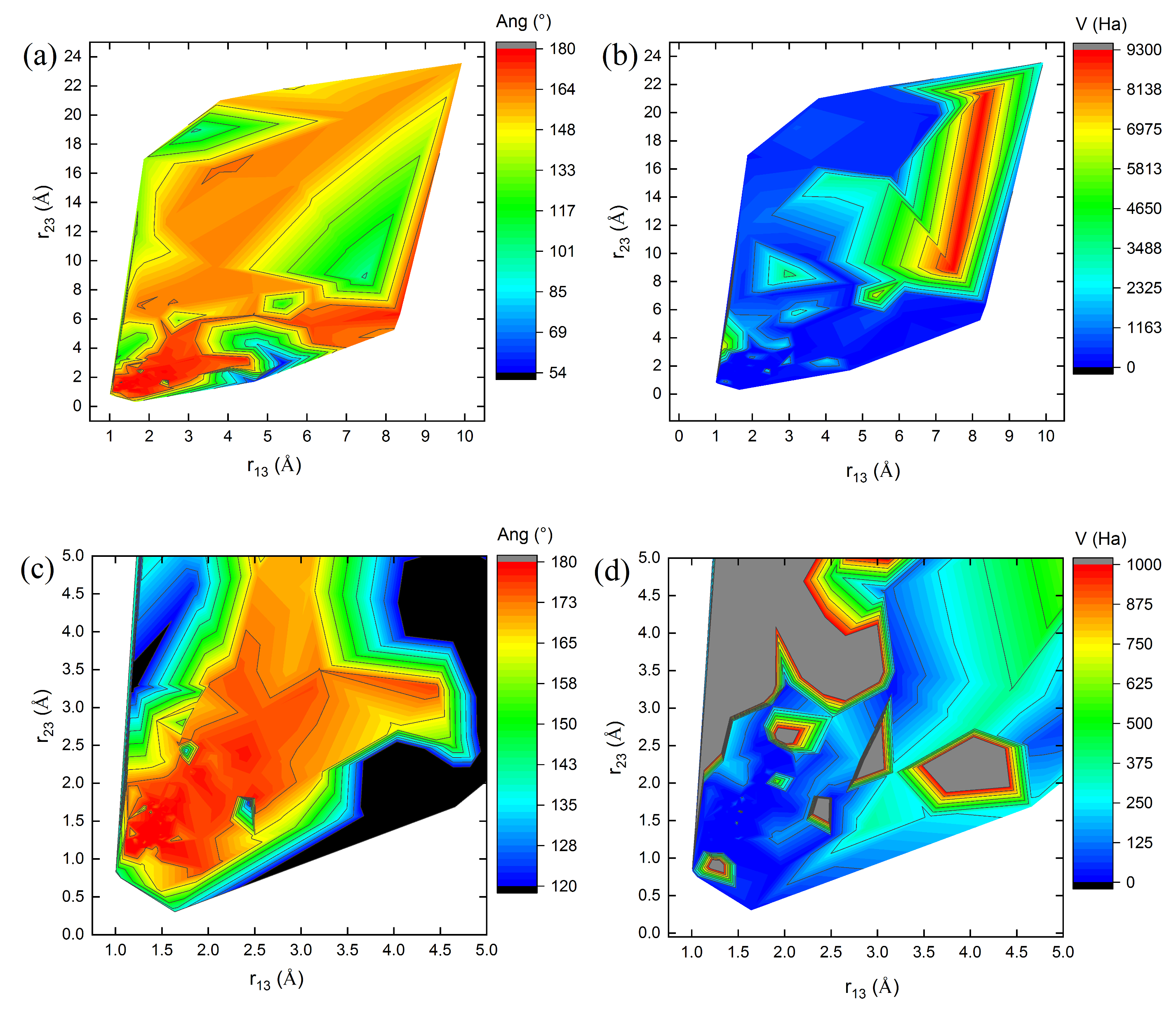
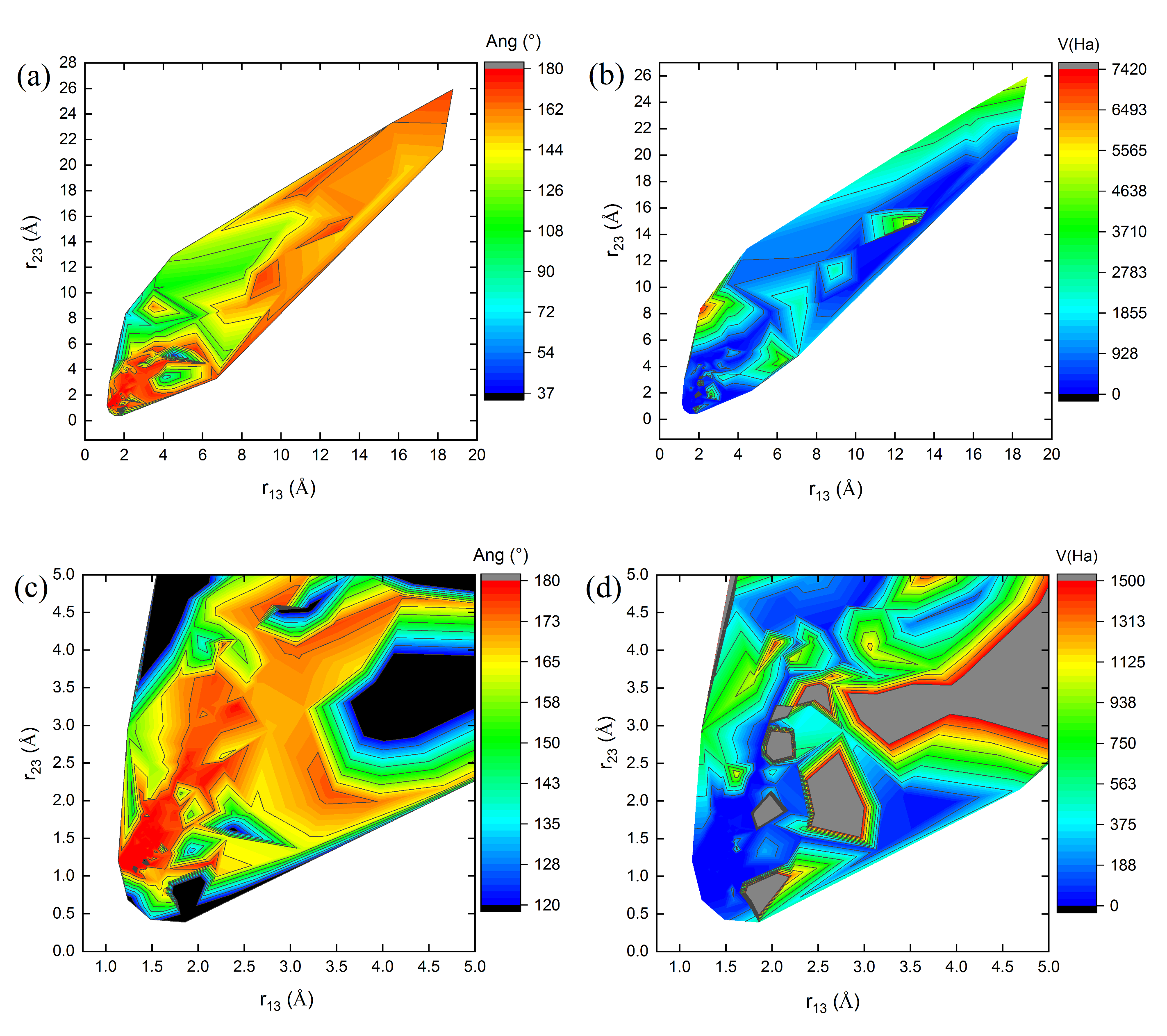
Figure 3,
Figure 4 and
Figure 5 show 3D relief maps of the angle Ang (a) and the potential energy
V (b), with their respective magnifications (c,d), as functions of the lengths
and
, for the
clusters
1, 8, and 10, over 600 iterations. Each
conformer within the swarm exhibits distinct energy landscapes. However, all converge to the same minimum potential energy value,
, as a function of the positional changes of the clusters during the dynamics of the swarm in the 3N-dimensional hyperspace
. The blank spaces correspond to regions where bond lengths, angles, and energies of the
cluster conformations are undefined in the energy landscape during geometry optimization. The highest Ang values are located at the peaks (red values near
in parts (a,c) of
Figure 3,
Figure 4 and
Figure 5), protruding from the paper plane. In contrast, the lowest values are found in the valleys (dark blue and black regions near
in the same figures), oriented towards the paper plane. These features occur when the C–C bond lengths,
and
, approach 1.29 Å (see
Table 5 and
Figure 3,
Figure 4 and
Figure 5).
Similarly, the highest potential energy values
V are observed at the peaks (red regions in parts (b) of
Figure 3,
Figure 4 and
Figure 5). Additional intermediate values lie on the slopes (gray regions in the magnified parts (d) of
Figure 3,
Figure 4 and
Figure 5), extending outward from the paper plane. The lowest energy values, however, are concentrated in the valleys of the energy landscape (dark blue regions in parts (b,d) of
Figure 3,
Figure 4 and
Figure 5), oriented toward the paper plane. These observations correspond to the geometry optimization process of the
cluster structures.
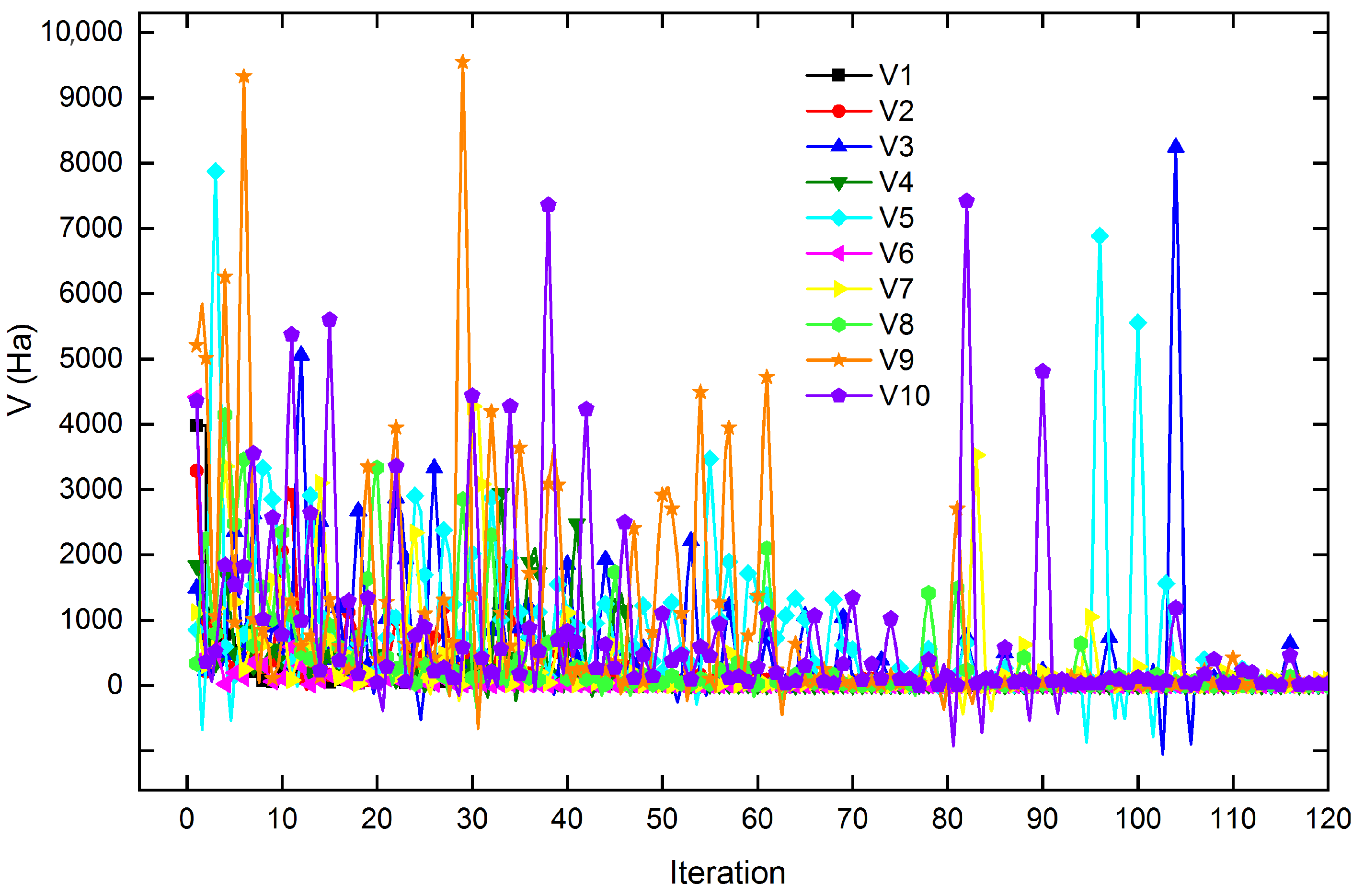
Figure 6 shows the potential energy behavior of the swarm of 10 three-carbon-atom clusters over the first 120 iterations. The cluster
8 exhibits an intermediate potential energy during the first 60 iterations, subsequently reaching one of the lowest potential energy values in the swarm as its trajectory progresses. However, clusters 4, 5, 9, and 10 exhibit conformations with alternating high potential energy values. At the same time, the remaining conformers maintain a potential energy close to the average for the swarm during the energy minimization of each conformer.
The potential energy behavior of the clusters, as their atoms undergo positional changes, illustrates the various conformations adopted during the search for the global potential energy minimum, a process often characterized by cooperative swarm dynamics. Conformers with higher potential energy require larger displacements within the
hyperspace (relative to the average position of the swarm) to contribute to the energy minimization potential [Equation (5)]; this accounts for the bond elongation observed in the three-carbon-atom cluster
during early iterations. Only by iteration 22 (
Figure 1 and
Figure 2) do these bond lengths begin to shorten, demonstrating the integration of the cluster
conformer into the swarm collective, where it assumes positions similar to the other swarm conformers, thereby reducing its potential energy as the system progresses toward convergence.
Table 6 presents the calculated relative electronic energy values for carbon, tungsten, and oxygen cluster structures studied in this work. To evaluate the precision of the electronic energies for the structures optimized with the PSO and BH methods, we used the absolute electronic energy obtained from Gaussian as a reference (
Table 6).
Table 6 shows the difference between the electronic energy calculated by Gaussian in a single-point calculation for a optimized structure obtained with PSO/BH, and the corresponding electronic energy obtained directly from Gaussian. In this way, values closest to zero indicate higher precision in the respective optimization method. The small relative electronic energy differences (∼
–
Ha) between BH/PSO-optimized and DFT-optimized structures suggest that both BH and PSO exhibit high accuracy in predicting the electronic energies of these molecular structures, carbon, tungsten and oxygen clusters systems. However, PSO demonstrated slightly higher accuracy compared to BH, particularly for carbon clusters
(
n = 3–5) and the tungsten–oxygen clusters
(
n = 4–6,
), where lower relative energy electronic values were observed.
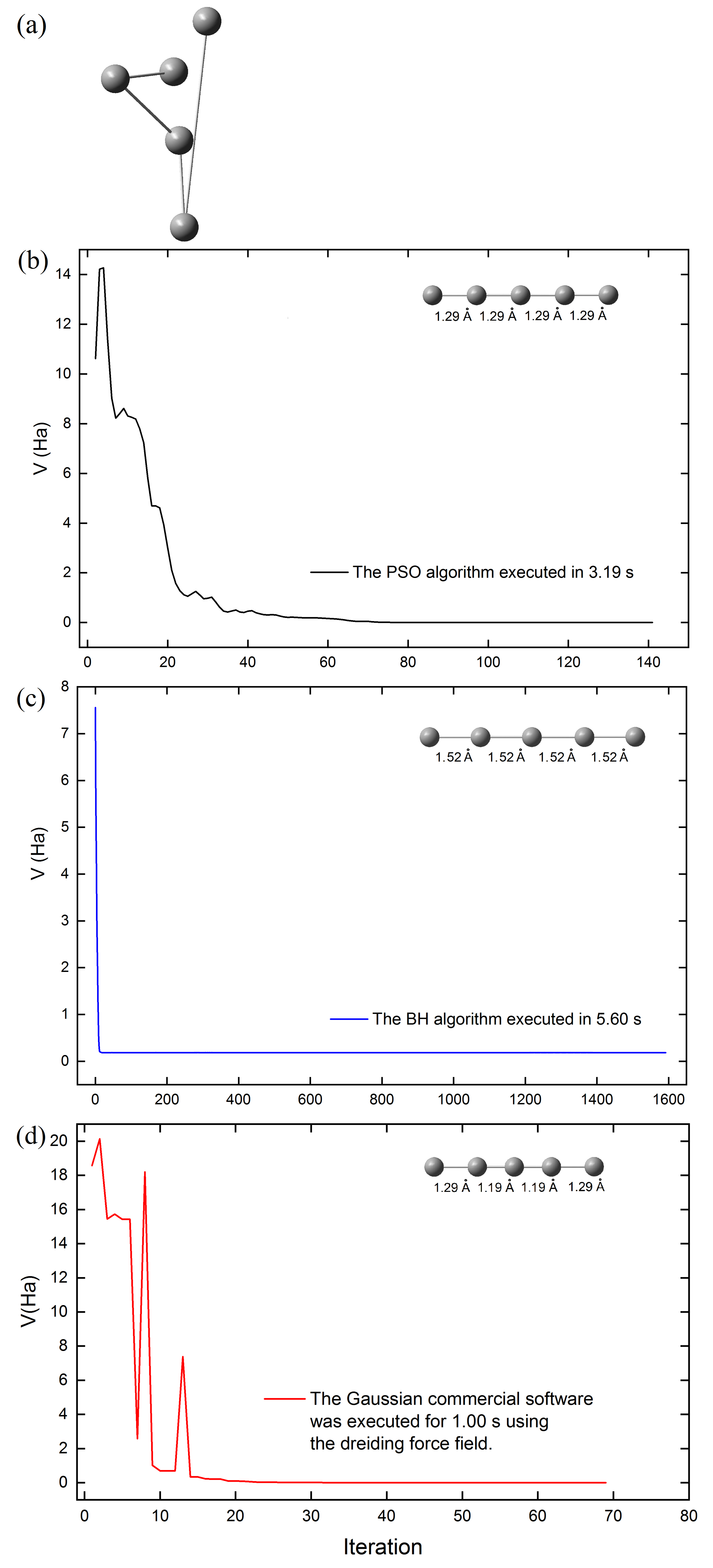
The computational cost of the PSO, BH, and molecular mechanics (using the Dreiding force field in Gaussian), was assessed by tracking the energy minimization of cluster
during geometric optimization as a function of optimization steps (
Figure 7b,d), starting from the initial structure of cluster
(
Figure 7a).
Figure 7 highlights that the structural optimization of the
cluster required the least time with the Gaussian software (69 steps), followed by the PSO algorithm (141 steps), and the BH algorithm (1592 steps). All calculations employed the same initial structure of the
cluster. Although Gaussian software optimizes the
cluster structure more quickly, it predicts triple bond lengths (1.19 Å) for the carbon atoms inside the cluster. This value differs slightly from the results obtained (1.29 Å in
Table 3) when the same structure is optimized at the DFT level of theory using the same software. However, our PSO algorithm predicts double bonds (1.29 Å) in the optimized structure of the
cluster (see
Figure 7b). Additionally, the structure of the
cluster was optimized using the UFF (Unified Force Field) in Gaussian software, but an unexpected cyclic structure was obtained.
Contrarily, it is important to note that the PSO algorithm implemented by Jana et al. [
46] uses Gaussian 09 software to calculate the atomic positions at each step of the geometric optimization of cluster structures via DFT. In contrast, our PSO algorithm does not depend on any external software to determine atomic positions during the simultaneous optimization of ten conformer structures. Instead, it relies solely on Equations (
1)–(4) to locate each atom of each cluster within the
search hyperspace at each iteration and uses the potential energy of each cluster [Equation (5)] as the objective function to be minimized.
A precise understanding of the structures of these clusters is crucial, particularly for compounds of tungsten and oxygen atoms, as they serve as fundamental building blocks for the growth of larger metal oxide arrangements. Through self-assembly, larger compounds, such as polyoxometalates, are formed, which have numerous applications due to their electronic and magnetic properties, which in turn depend on their structural arrangement [
57,
58].