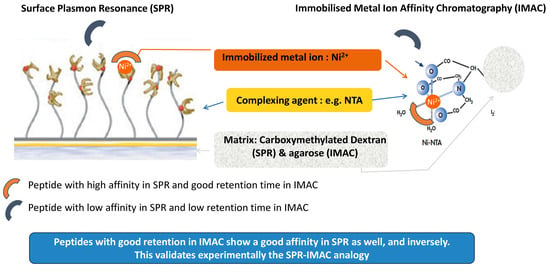
Deciphering Interactions Involved in Immobilized Metal Ion Affinity Chromatography and Surface Plasmon Resonance for Validating the Analogy between Both Technologies
Abstract
1. Introduction
2. Materials and Methods
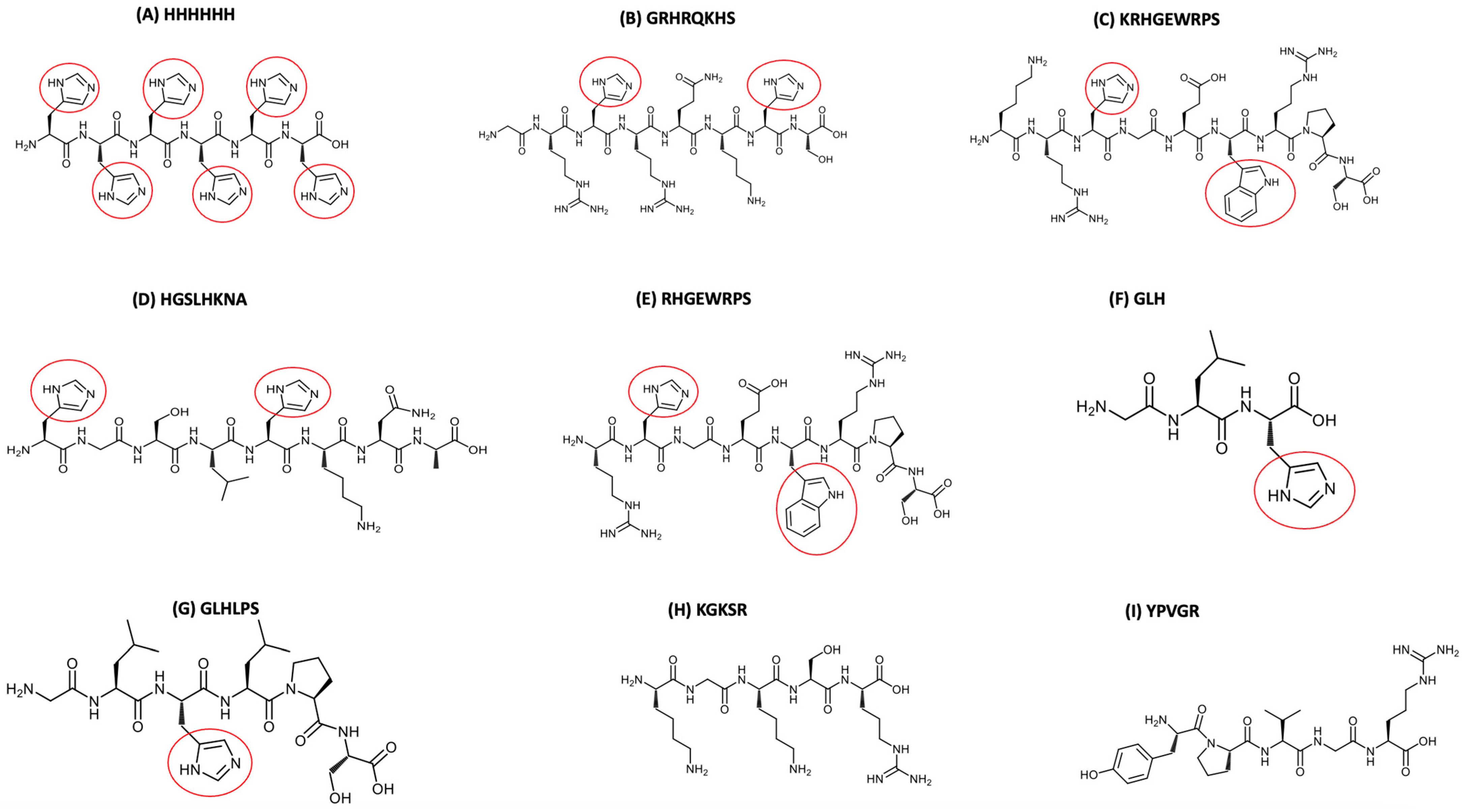
2.1. Synthetic Peptides Investigated and Other Chemicals
2.2. Immobilized Metal Ion Affinity Chromatography
2.2.1. HPLC Equipment and IMAC Columns Used
2.2.2. IMAC Cycle and Elution Program
2.2.3. Treatment of IMAC Data
2.3. LC-MS
2.4. Surface Plasmon Resonance
2.5. Statistical Analysis
3. Results and Discussion
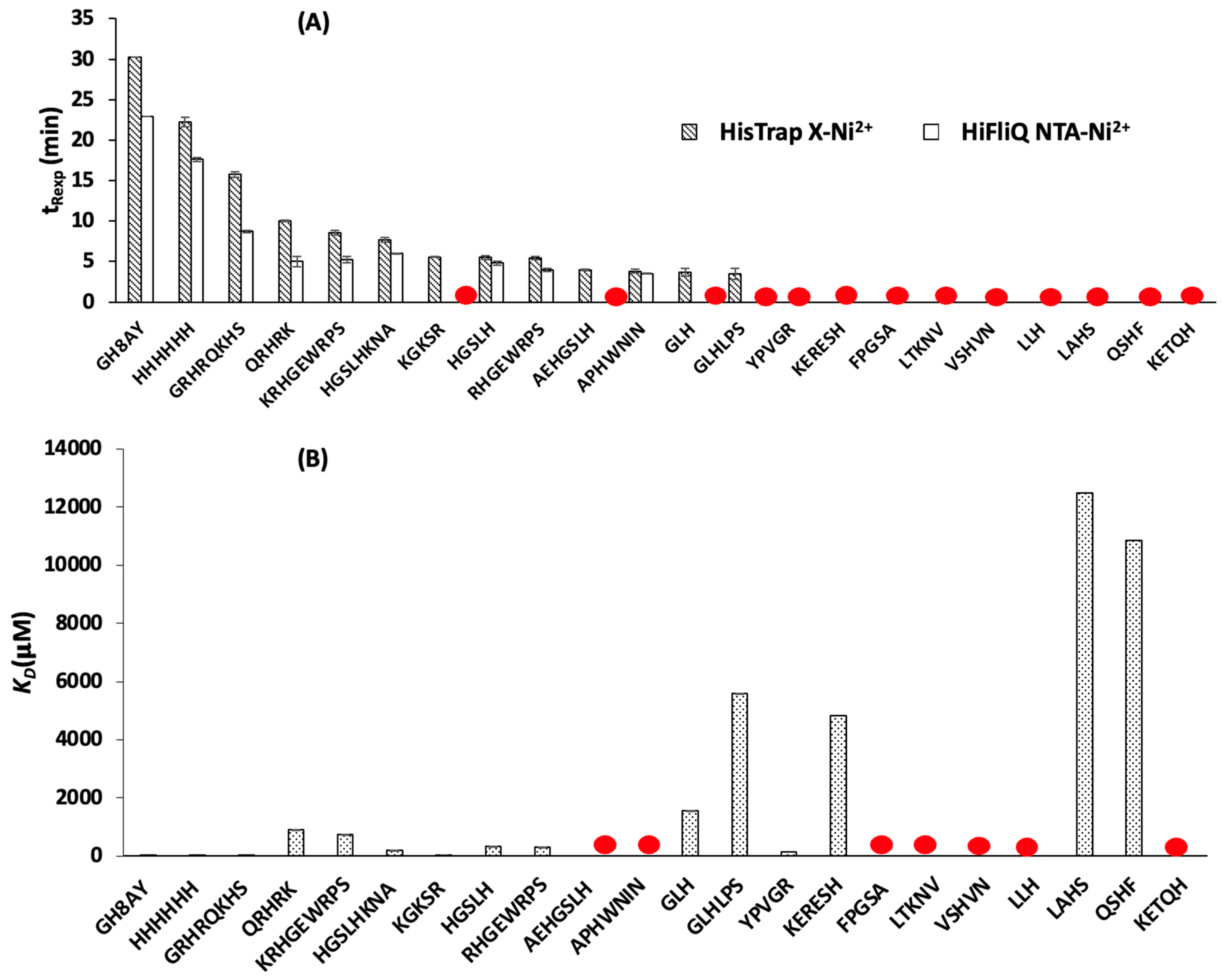
3.1. Determination of Peptide Retention Time Using IMAC
3.1.1. Peptide Retention Time Using HisTrap X-Ni2+ Column
3.1.2. Peptide Retention Time Using HiFliQ NTA-Ni2+ Column
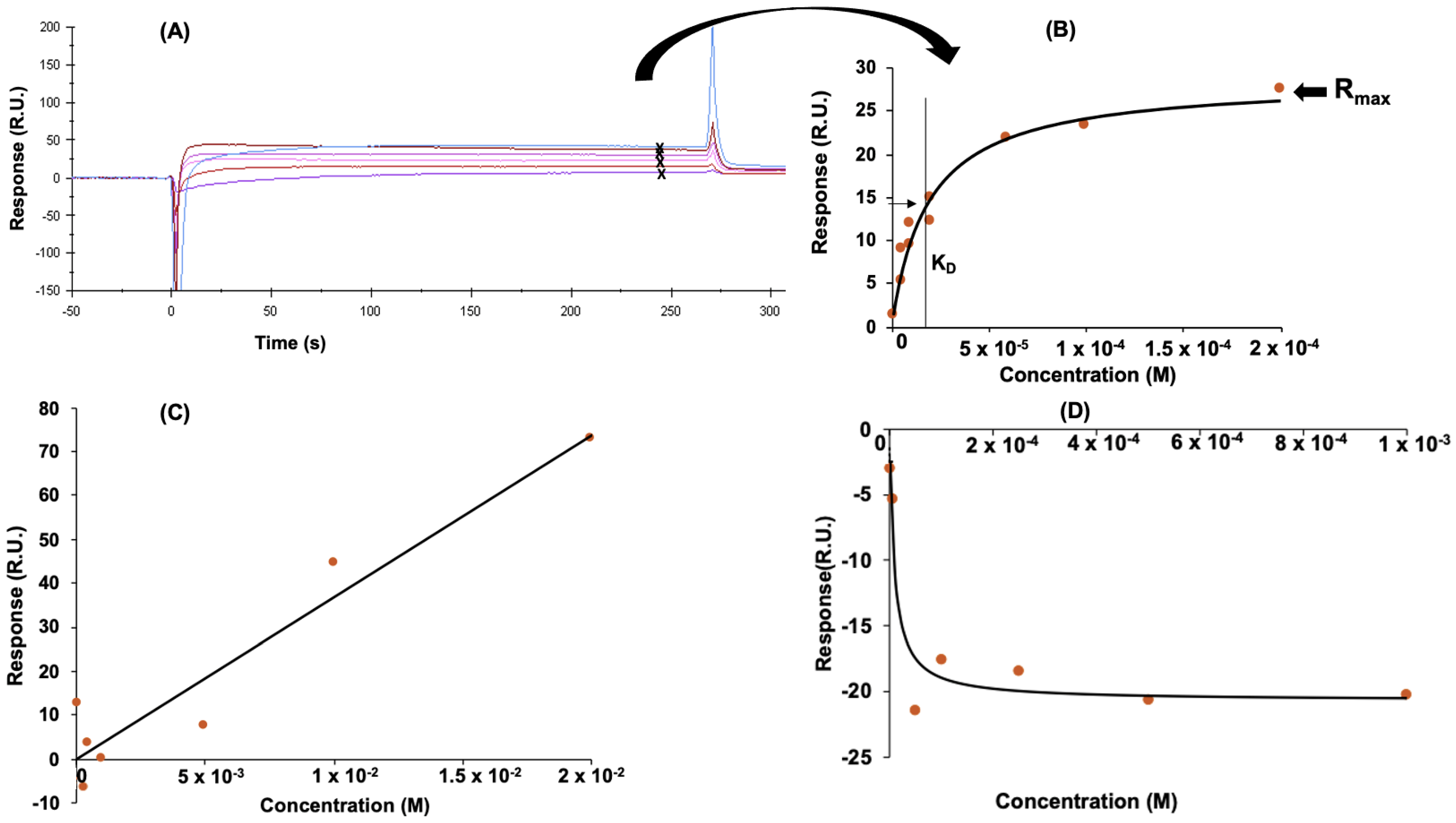
3.2. Determination of Peptide Affinity Parameters Using SPR
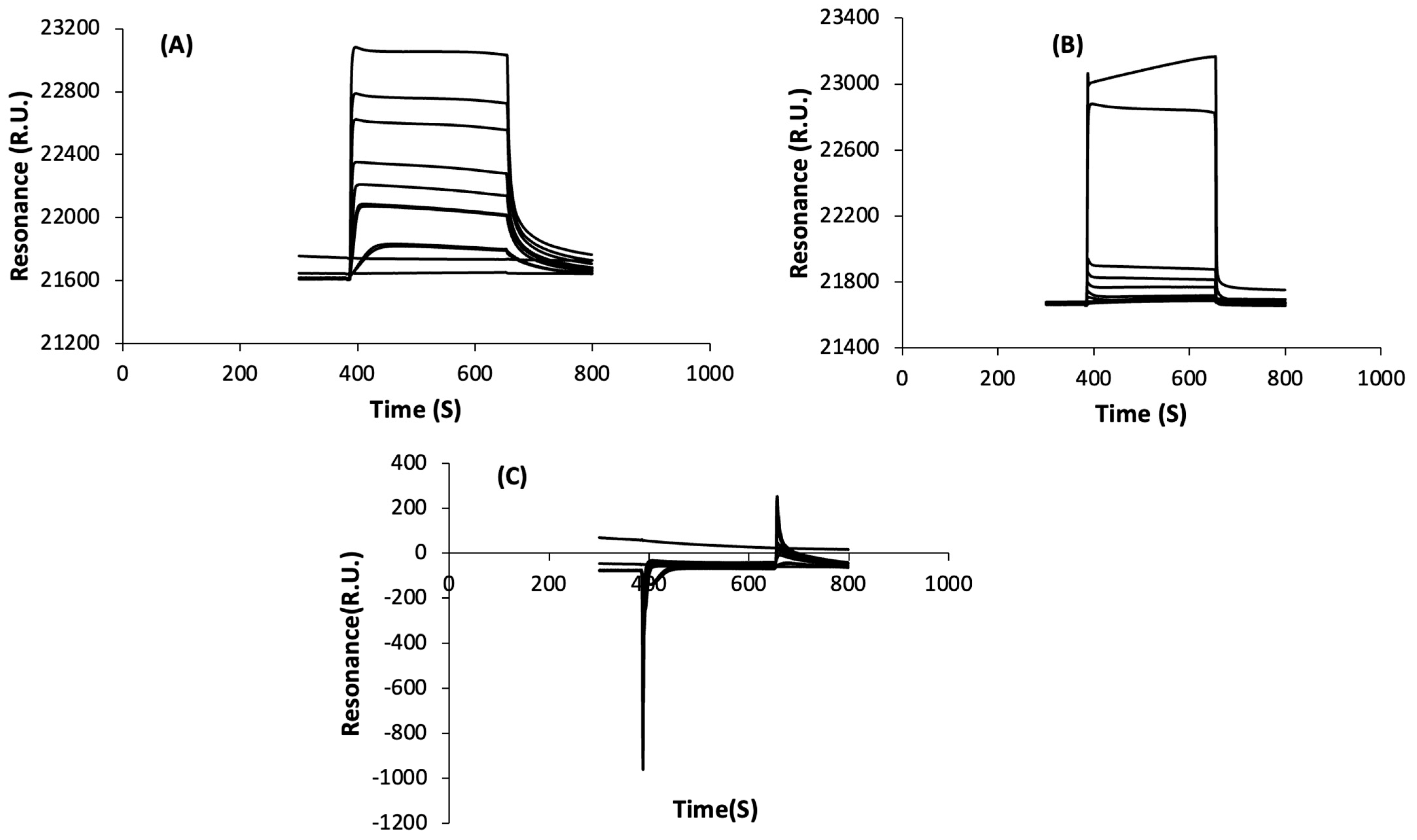
3.2.1. Non-Specific Interactions
3.2.2. Specific Interactions
3.3. Understanding the Peptide-Ni2+ Interaction Based on the Analogy between SPR and IMAC
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Guo, L.; Harnedy, P.A.; Li, B.; Hou, H.; Zhang, Z.; Zhao, X.; FitzGerald, R.J. Food protein-derived chelating peptides: Biofunctional ingredients for dietary mineral bioavailability enhancement. Trends Food Sci. Technol. 2014, 37, 92–105. [Google Scholar] [CrossRef]
- Yesiltas, B.; García-Moreno, P.J.; Gregersen, S.; Olsen, T.H.; Jones, N.C.; Hoffmann, S.V.; Marcatili, P.; Overgaard, M.T.; Hansen, E.B.; Jacobsen, C. Antioxidant peptides derived from potato, seaweed, microbial and spinach proteins: Oxidative stability of 5% fish oil-in-water emulsions. Food Chem. 2022, 385, 132699. [Google Scholar] [CrossRef]
- Lv, Y.; Liu, Q.; Bao, X.; Tang, W.; Yang, B.; Guo, S. Identification and Characteristics of Iron-Chelating Peptides from Soybean Protein Hydrolysates Using IMAC-Fe3+. J. Agric. Food Chem. 2009, 57, 4593–4597. [Google Scholar] [CrossRef] [PubMed]
- Lv, Y.; Bao, X.; Liu, H.; Ren, J.; Guo, S. Purification and characterization of caclium-binding soybean protein hydrolysates by Ca2+/Fe3+ immobilized metal affinity chromatography (IMAC). Food Chem. 2013, 141, 1645–1650. [Google Scholar] [CrossRef]
- Wang, C.; Li, B.; Ao, J. Separation and identification of zinc-chelating peptides from sesame protein hydrolysate using IMAC-Zn2+ and LC–MS/MS. Food Chem. 2012, 134, 1231–1238. [Google Scholar] [CrossRef] [PubMed]
- Caetano-Silva, M.E.; Simabuco, F.M.; Bezerra, R.M.N.; da Silva, D.C.; Barbosa, E.A.; Moreira, D.C.; Brand, G.D.; de Almeida Leite, J.R.d.S.; Pacheco, M.T.B. Isolation and Sequencing of Cu-, Fe-, and Zn-Binding Whey Peptides for Potential Neuroprotective Applications as Multitargeted Compounds. J. Agric. Food Chem. 2020, 68, 12433–12443. [Google Scholar] [CrossRef] [PubMed]
- Kangsanant, S.; Thongraung, C.; Jansakul, C.; Murkovic, M.; Seechamnanturakit, V. Purification and characterisation of antioxidant and nitric oxide inhibitory peptides from Tilapia (Oreochromis niloticus) protein hydrolysate. Int. J. Food Sci. Technol. 2015, 50, 660–665. [Google Scholar] [CrossRef]
- Sulkowski, E. Purification of proteins by IMAC. Trends Biotechnol. 1985, 3, 1–7. [Google Scholar] [CrossRef]
- Ueda, E.K.M.; Gout, P.W.; Morganti, L. Current and prospective applications of metal ion–protein binding. J. Chromatogr. A 2003, 988, 1–23. [Google Scholar] [CrossRef]
- Caetano-Silva, M.E.; Bertoldo-Pacheco, M.T.; Paes-Leme, A.F.; Netto, F.M. Iron-binding peptides from whey protein hydrolysates: Evaluation, isolation and sequencing by LC–MS/MS. Food Res. Int. 2015, 71, 132–139. [Google Scholar] [CrossRef]
- Yamauchi, O.; Odani, A.; Takani, M. Metal–amino acid chemistry. Weak interactions and related functions of side chain groups. J. Chem. Soc. Dalton Trans. 2002, 18, 3411–3421. [Google Scholar] [CrossRef]
- Braun, R.; Bachmann, S.; Schönberger, N.; Matys, S.; Lederer, F.; Pollmann, K. Peptides as biosorbents—Promising tools for resource recovery. Res. Microbiol. 2018, 169, 649–658. [Google Scholar] [CrossRef] [PubMed]
- El Hajj, S.; Sepúlveda Rincón, C.T.; Girardet, J.-M.; Cakir-Kiefer, C.; Stefan, L.; Zapata Montoya, J.E.; Boschi-Muller, S.; Gaucher, C.; Canabady-Rochelle, L. Electrically switchable nanolever technology for the screening of metal-chelating peptides in hydrolysates. J. Agric. Food Chem. 2021, 69, 8819–8827. [Google Scholar] [CrossRef]
- Canabady-Rochelle, L.L.S.; Selmeczi, K.; Collin, S.; Pasc, A.; Muhr, L.; Boschi-Muller, S. SPR screening of metal chelating peptides in a hydrolysate for their antioxidant properties. Food Chem. 2018, 239, 478–485. [Google Scholar] [CrossRef]
- Irankunda, R.; Camaño Echavarría, J.A.; Paris, C.; Stefan, L.; Desobry, S.; Selmeczi, K.; Muhr, L.; Canabady-Rochelle, L. Metal-Chelating Peptides Separation Using Immobilized Metal Ion Affinity Chromatography: Experimental Methodology and Simulation. Separations 2022, 9, 370. [Google Scholar] [CrossRef]
- El Hajj, S.; Irankunda, R.; Andrés Camaño Echavarría, J.; Arnoux, P.; Paris, C.; Stefan, L.; Gaucher, C.; Boschi-Muller, S.; Canabady-Rochelle, L. Metal-chelating activity of soy and pea protein hydrolysates obtained after different enzymatic treatments from protein isolates. Food Chem. 2023, 405, 134788. [Google Scholar] [CrossRef] [PubMed]
- Bernaudat, F.; Bülow, L. Rapid evaluation of nickel binding properties of His-tagged lactate dehydrogenases using surface plasmon resonance. J. Chromatogr. A 2005, 1066, 219–224. [Google Scholar] [CrossRef] [PubMed]
- Muhr, L.; Pontvianne, S.; Selmeczi, K.; Paris, C.; Boschi-Muller, S.; Canabady-Rochelle, L. Chromatographic separation simulation of metal-chelating peptides from surface plasmon resonance binding parameters. J. Sep. Sci. 2020, 43, 2031–2041. [Google Scholar] [CrossRef] [PubMed]
- García Arteaga, V.; Apéstegui Guardia, M.; Muranyi, I.; Eisner, P.; Schweiggert-Weisz, U. Effect of enzymatic hydrolysis on molecular weight distribution, techno-functional properties and sensory perception of pea protein isolates. Innov. Food Sci. Emerg. Technol. 2020, 65, 102449. [Google Scholar] [CrossRef]
- Boye, J.; Zare, F.; Pletch, A. Pulse proteins: Processing, characterization, functional properties and applications in food and feed. Food Res. Int. 2010, 43, 414–431. [Google Scholar] [CrossRef]
- Tamm, F.; Herbst, S.; Brodkorb, A.; Drusch, S. Functional properties of pea protein hydrolysates in emulsions and spray-dried microcapsules. Food Hydrocoll. 2016, 58, 204–214. [Google Scholar] [CrossRef]
- Lam, A.C.Y.; Can Karaca, A.; Tyler, R.T.; Nickerson, M.T. Pea protein isolates: Structure, extraction, and functionality. Food Rev. Int. 2018, 34, 126–147. [Google Scholar] [CrossRef]
- Hou, Y.; Wu, Z.; Dai, Z.; Wang, G.; Wu, G. Protein hydrolysates in animal nutrition: Industrial production, bioactive peptides, and functional significance. J. Anim. Sci. Biotechnol. 2017, 8, 24. [Google Scholar] [CrossRef]
- Knecht, S.; Ricklin, D.; Eberle, A.N.; Ernst, B. Oligohis-tags: Mechanisms of binding to Ni2+-NTA surfaces. J. Mol. Recognit. 2009, 22, 270–279. [Google Scholar] [CrossRef] [PubMed]
- Kermani, B.G.; Kozlov, I.; Melnyk, P.; Zhao, C.; Hachmann, J.; Barker, D.; Lebl, M. Using support vector machine regression to model the retention of peptides in immobilized metal-affinity chromatography. Sens. Actuators B Chem. 2007, 125, 149–157. [Google Scholar] [CrossRef][Green Version]
- Sofer, G.K.; Hagel, L. Handbook of Process Chromatography: A Guide to Optimization, Scale up, and Validation; Academic Press: San Diego, CA, USA, 1997. [Google Scholar]
- Block, H.; Maertens, B.; Spriestersbach, A.; Brinker, N.; Kubicek, J.; Fabis, R.; Labahn, J.; Schäfer, F. Methods in Enzymology; Elsevier: Amsterdam, The Netherlands, 2009; pp. 439–473. [Google Scholar]
- Ren, D.; Penner, N.A.; Slentz, B.E.; Regnier, F.E. Histidine-Rich Peptide Selection and Quantification in Targeted Proteomics. J. Proteome Res. 2004, 3, 37–45. [Google Scholar] [CrossRef]
- Sun, X.; Chiu, J.-F.; He, Q.-Y. Application of immobilized metal affinity chromatography in proteomics. Expert Rev. Proteom. 2005, 2, 649–657. [Google Scholar] [CrossRef] [PubMed]
- Karavelas, T.; Mylonas, M.; Malandrinos, G.; Plakatouras, J.C.; Hadjiliadis, N.; Mlynarz, P.; Kozlowski, H. Coordination properties of Cu(II) and Ni(II) ions towards the C-terminal peptide fragment –ELAKHA– of histone H2B. J. Inorg. Biochem. 2005, 99, 606–615. [Google Scholar] [CrossRef] [PubMed]
- Sóvágó, I.; Ősz, K. Metal ion selectivity of oligopeptides. Dalton Trans. 2006, 32, 3841–3854. [Google Scholar] [CrossRef] [PubMed]
- Mesmin, C.; Domon, B. Improvement of the Performance of Targeted LC–MS Assays through Enrichment of Histidine-Containing Peptides. J. Proteome Res. 2014, 13, 6160–6168. [Google Scholar] [CrossRef] [PubMed]
- Ren, D.; Penner, N.A.; Slentz, B.E.; Mirzaei, H.; Regnier, F. Evaluating Immobilized Metal Affinity Chromatography for the Selection of Histidine-Containing Peptides in Comparative Proteomics. J. Proteome Res. 2003, 2, 321–329. [Google Scholar] [CrossRef] [PubMed]
- Cheung, R.C.F.; Wong, J.H.; Ng, T.B. Immobilized metal ion affinity chromatography: A review on its applications. Appl. Microbiol. Biotechnol. 2012, 96, 1411–1420. [Google Scholar] [CrossRef] [PubMed]
- GE Healthcare. Biacore X100 Getting Started 28-9615-81 Edition AA. 2009. Available online: http://www.molekulske-interakcije.si/data/equipment/GettingStarted.pdf (accessed on 17 March 2023).
- GE Healthcare. Biacore X100 Handbook BR-1008-10 Edition AB. 2007. Available online: https://www.imb.sinica.edu.tw/core/biophysicals/form/Biacore_X100_2_0_Handbook.pdf (accessed on 17 March 2023).
- Nieba, L.; Nieba-Axmann, S.E.; Persson, A.; Hämäläinen, M.; Edebratt, F.; Hansson, A.; Lidholm, J.; Magnusson, K.; Karlsson, Å.F.; Plückthun, A. BIACORE Analysis of Histidine-Tagged Proteins Using a Chelating NTA Sensor Chip. Anal. Biochem. 1997, 252, 217–228. [Google Scholar] [CrossRef]
- Fischer, M.; Leech, A.P.; Hubbard, R.E. Comparative Assessment of Different Histidine-Tags for Immobilization of Protein onto Surface Plasmon Resonance Sensorchips. Anal. Chem. 2011, 83, 1800–1807. [Google Scholar] [CrossRef]
- Bal, W.; Dyba, M.; Kozlowski, H. ChemInform Abstract: The Impact of the Amino Acid Sequence on the Specificity of Copper(II) Interactions with Peptides Having Non-Coordinating Side-Chains. ChemInform 1997, 44, 467–476. [Google Scholar] [CrossRef]
| UniprotKB | PepCalc | Percepta | ||||
|---|---|---|---|---|---|---|
| Protein Code | Protein Name | Peptide Sequence | n Residues | MW (g/mol) | Iso-Electric Point (PI) | % of Global Charge at pH 7.4 |
| n.c | n.c | GHHHHHHHHAY | 11 | 1406.43 | 8.21 | 80% +, 3% - |
| n.c | n.c | HHHHHH | 6 | 840.87 | 7.97 | 70% +, 6% - |
| P05692 | Leguminin J | GRHRQKHS | 8 | 1005.12 | 12.13 | 100% + |
| O24294_PEA | Legumin (Minor small) | QRHRK | 5 | 723.9 | 12.13 | 100% + |
| P13915 | Convicilin | KRHGEWRPS | 9 | 1152.27 | 11.22 | 100% + |
| P02857 | Leguminin A | HGSLHKNA | 8 | 862.95 | 9.88 | 65% +, 0% - |
| P02857 | Leguminin A | KGKSR | 5 | 574.67 | 11.57 | 100% + |
| P02857 | Leguminin A | HGSLH | 5 | 549.59 | 7.72 | 26% +, 25% - |
| P13915 | Convicilin | RHGEWRPS | 8 | 1024.09 | 10.39 | 44% +, 0% - |
| P02857 | Leguminin A | AEHGSLH | 7 | 749.77 | 6.05 | 11% +, 48% - |
| P05692 | Leguminin J | APHWNIN | 7 | 850.92 | 7.88 | 20% +, 21% - |
| P05692 | Leguminin J | GLH | 3 | 325.36 | 7.81 | 34% +, 13% - |
| P05692 | Leguminin J | GLHLPS | 6 | 622.71 | 7.80 | 19% +, 21% - |
| P05692 | legumin J | YPVGR | 5 | 590.67 | 9.57 | 52% +, 0% - |
| O24294_PEA | Legumin (Minor small) | KERESH | 6 | 784.82 | 7.60 | 24% +, 27% - |
| P13918 | vicilin | FPGSA | 5 | 477.51 | 3.38 | 0%-, 56% - |
| P62926 | Albumin-1 A | LTKNV | 5 | 573.68 | 10.12 | 70% +, 0% - |
| O24294_PEA | Legumin (Minor small) | VSHVN | 5 | 554.60 | 7.78 | 18% +, 25% - |
| Q7M1N3_PEA | Legumin L1 beta chain | LLH | 3 | 381.47 | 7.81 | 34% +, 13% - |
| O24294_PEA | Legumin (Minor small) | LAHS | 4 | 426.47 | 7.81 | 22% +, 19% - |
| P13918 | Vicilin | QSHF | 4 | 517.53 | 7.52 | 15%+, 34% - |
| P13918 | vicilin | KETQH | 5 | 641.67 | 7.60 | 24% +, 26% - |
| Peptides | HisTrap X-Ni2+ Mean tR (min) | Group | HiFliQ NTA-Ni2+ Mean tR (min) | Group |
|---|---|---|---|---|
| GH8AY | 30.25 ± 0 g | 1 | 22.93 ± 0 h | I |
| HHHHHH | 22.20 ± 0.58 f | 1 | 17.62 ± 0.23 g | I |
| GRHRQKHS | 15.79 ± 0.34 e | 1 | 8.72 ± 0.18 e | I |
| QRHRK | 10.01 ± 0.06 d | 2 | 5.01 ± 0.62 c,d | II |
| KRHGEWRPS | 8.58 ± 0.29 c | 2 | 5.24 ± 0.36 c,d | II |
| HGSLHKNA | 7.71 ± 0.29 c | 2 | 6.02 ± 0.05 d | II |
| KGKSR | 5.57 ± 0.02 b | 2 | not retained | III |
| HGSLH | 5.48 ± 0.20 b | 2 | 4.82 ± 0.24 b,c | II |
| RHGEWRPS | 5.41 ± 0.19 b | 2 | 3.98 ± 0.22 a,b | II |
| AEHGSLH | 3.93 ± 0.09 a | 3 | not retained | III |
| APHWNIN | 3.81 ± 0.22 a | 3 | 3.50 ± 0.04 a | II |
| GLH | 3.68 ± 0.46 a | 3 | not retained | III |
| GLHLPS | 3.44 ± 0.67 a | 3 | not retained | III |
| YPVGR | not retained | 4 | not retained | III |
| KERESH | 4 | III | ||
| FPGSA | 4 | III | ||
| LTKNV | 4 | III | ||
| VSHVN | 4 | III | ||
| LLH | 4 | III | ||
| LAHS | 4 | III | ||
| QSHF | 4 | III | ||
| KETQH | 4 | III |
| Group | Peptide | KD (μM) | SE (KD) (μM) | Rmax (R.U.) | SE (Rmax) (R.U.) | Concentration Range (µM) |
|---|---|---|---|---|---|---|
| A | HHHHHH | 0.7 | 27.0 | 197.90 | 27.00 | 0.4–50 |
| A | GH8AY | 1.1 | 0.5 | 164.8 | 11 | 0.01–100 |
| A | GRHRQKHS | 18.7 | 4.9 | 28.70 | 1.90 | 1–200 |
| A | KGKSR | 45.6 | 37.0 | 24.9 | 7.6 | 20–5000 |
| B | YPVGR | 156.6 | 420.0 | 15 | 9 | 5–5000 |
| B | HGSLHKNA | 195.5 | 64.0 | 72.38 | 5.70 | 5–1000 |
| B | RHGEWRPS | 303.9 | 210.0 | 81.3 | 14 | 20–1000 |
| B | HGSLH | 341.5 | 74.0 | 38.40 | 2.20 | 5–5000 |
| B | QRHRK | 910.0 | 230.0 | 64.30 | 5.30 | 20–5000 |
| B | KRHGEWRPS | 737.9 | 250.0 | 366.3 | 30 | 20–5000 |
| C | GLH | 1567.0 | 760.0 | 19.30 | 2.9 | 20–5000 |
| C | KERESH | 4835.0 | 5400.0 | 76.9 | 25 | 500–10,000 |
| C | GLHLPS | 5586.0 | 5700.0 | 54.5 | 22 | 100–10,000 |
| C | QSHF | 10,840.0 | 13,000.0 | 86.9 | 25 | 1000–30,000 |
| C | LAHS | 12,480.0 | 5400.0 | 105.8 | 23 | 50–15,000 |
| D | VSHVN | n.d | n.d | n.d | n.d | 100–10,000 |
| D | APHWNIN | n.d | n.d | n.d | n.d | 5–5000 |
| D | AEHGSLH | n.d | n.d | n.d | n.d | 5–10,000 |
| D | KETQH | n.d | n.d | n.d | n.d | 50–20,000 |
| D | LLH | n.d | n.d | n.d | n.d | 100–30,000 |
| D | FPGSA | n.d | n.d | n.d | n.d | 1–10,000 |
| D | LTKNV | n.d | n.d | n.d | n.d | 1–10,000 |
| Peptide | HisTrap X-Ni2+ | HiFliQ NTA-Ni2+ |
|---|---|---|
| GH8AY | 1A | IA |
| HHHHHH | 1A | IA |
| GRHRQKHS | 1A | IA |
| QRHRK | 2B | IIB |
| KRHGEWRPS | 2B | IIB |
| HGSLHKNA | 2B | IIB |
| KGKSR | 2A | IIIA |
| HGSLH | 2B | IIB |
| RHGEWRPS | 2B | IIB |
| AEHGSLH | 3D | IIID |
| APHWNIN | 3D | IID |
| GLH | 3C | IIIC |
| GLHLPS | 3C | IIIC |
| YPVGR | 4B | IIIB |
| KERESH | 4C | IIIC |
| FPGSA | 4D | IIID |
| LTKNV | 4D | IIID |
| VSHVN | 4D | IIID |
| LLH | 4D | IIID |
| LAHS | 4C | IIIC |
| QSHF | 4C | IIIC |
| KETQH | 4D | IIID |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Irankunda, R.; Camaño Echavarría, J.A.; Paris, C.; Selmeczi, K.; Stefan, L.; Boschi-Muller, S.; Muhr, L.; Canabady-Rochelle, L. Deciphering Interactions Involved in Immobilized Metal Ion Affinity Chromatography and Surface Plasmon Resonance for Validating the Analogy between Both Technologies. Inorganics 2024, 12, 31. https://doi.org/10.3390/inorganics12010031
Irankunda R, Camaño Echavarría JA, Paris C, Selmeczi K, Stefan L, Boschi-Muller S, Muhr L, Canabady-Rochelle L. Deciphering Interactions Involved in Immobilized Metal Ion Affinity Chromatography and Surface Plasmon Resonance for Validating the Analogy between Both Technologies. Inorganics. 2024; 12(1):31. https://doi.org/10.3390/inorganics12010031
Chicago/Turabian StyleIrankunda, Rachel, Jairo Andrés Camaño Echavarría, Cédric Paris, Katalin Selmeczi, Loïc Stefan, Sandrine Boschi-Muller, Laurence Muhr, and Laetitia Canabady-Rochelle. 2024. "Deciphering Interactions Involved in Immobilized Metal Ion Affinity Chromatography and Surface Plasmon Resonance for Validating the Analogy between Both Technologies" Inorganics 12, no. 1: 31. https://doi.org/10.3390/inorganics12010031
APA StyleIrankunda, R., Camaño Echavarría, J. A., Paris, C., Selmeczi, K., Stefan, L., Boschi-Muller, S., Muhr, L., & Canabady-Rochelle, L. (2024). Deciphering Interactions Involved in Immobilized Metal Ion Affinity Chromatography and Surface Plasmon Resonance for Validating the Analogy between Both Technologies. Inorganics, 12(1), 31. https://doi.org/10.3390/inorganics12010031