Abstract
Variations in the thickness of retinal layers serve as early diagnostic indicators for various fundus diseases, and precise segmentation of these layers is essential for accurately measuring their thickness. Optical Coherence Tomography (OCT) is an important non-invasive tool for diagnosing various eye diseases through the acquisition and layering of retinal images. However, noise and artifacts in images present significant challenges in accurately segmenting retinal layers. We propose a novel method for retinal layer segmentation that addresses these issues. This method utilizes ConvNeXt as the backbone network to enhance multi-scale feature extraction and incorporates a Transformer–CNN module to improve global processing capabilities. This method has achieved the highest segmentation accuracy on the Retina500 dataset, with a mean Intersection over Union (mIoU) of 81.26% and an accuracy (Acc) of 91.38%, and has shown excellent results on the public NR206 dataset.
1. Introduction
In recent years, the widespread use of electronic devices in daily work and leisure activities has placed great strain on our visual system. Coupled with environmental influences, genetic factors, and the natural aging process, these changes have collectively contributed to an increase in the prevalence of eye diseases [1]. The effective prevention and treatment of these conditions have become increasingly crucial due to their profound impact on individuals’ quality of life. The predominant eye diseases today include glaucoma [2], diabetic retinopathy [3], and age-related macular degeneration [4]. These conditions often induce alterations in retinal thickness, and in severe cases may lead to the disappearance of retinal cell layers [5]. For instance, glaucoma is associated with degeneration of the nerve fiber layer [6], while age-related macular degeneration can cause the thinning or disappearance of the ganglion cell layer [7]. Additionally, diabetes often leads to the development of macular edema [8]. Early detection of these subtle retinal changes through screening can significantly enhance disease prevention efforts and minimize the risk of vision impairment. Therefore, the precise quantitative analysis of the thickness of each retinal cell layer is essential for assessing the severity of these diseases and monitoring their progression [9].
OCT represents a significant advancement in the field of in vivo biological tissue imaging and has rapidly evolved in recent years [10]. Widely adopted in ophthalmological clinical diagnosis, OCT is prized for its non-contact, high-resolution imaging, and non-invasive properties [11]. Typically, researchers utilize retinal layer boundary segmentation to measure thickness changes, which is vital for the effective detection and prevention of retinal diseases [12]. However, manual segmentation of these layers in OCT images is both time-consuming and subjective, thereby impacting the efficiency and accuracy of clinical diagnoses [13]. The advent of deep learning has significantly advanced retinal layer segmentation technology. In contrast to traditional methods that require the construction of mathematical models, deep learning approaches offer substantial generalizability with minimal need for prior knowledge, as exemplified by ReLayNet. This model enhances clinical diagnostics by automating the segmentation process, significantly reducing the time and subjectivity associated with manual methods while maintaining high accuracy in the delineation of retinal layers [14]. Moreover, OctNet serves as a notable example, utilizing a deep learning-based three-dimensional convolutional network capable of effectively handling the spatial complexities of retinal OCT images and achieving efficient segmentation of multiple retinal layers [15]. By leveraging a deeper network architecture and optimized feature learning strategies, OctNet significantly enhances the ability to extract precise layer boundaries from noisy data, thereby providing a more accurate and reliable tool for clinical diagnostics. The development of these technologies marks substantial progress in the application of deep learning in medical image processing.
In contemporary deep learning frameworks, Convolutional Neural Networks (CNN) and Transformer are cornerstone technologies. CNN excels in extracting detailed features from images, making it ideal for identifying complex structures and patterns. However, it often falters in synthesizing global contextual information, affecting the coherence and overall understanding of the image content [16]. Conversely, Transformer utilizes its global attention mechanisms to effectively process extensive contextual information, yet sometimes struggles with detail precision in regions with intricate local characteristics [17]. Furthermore, in the domain of OCT imaging, challenges such as blood flow noise and other artifacts complicate the segmentation process, highlighting the need for advanced methodologies. This scenario underscores the imperative for developing novel retinal layer segmentation strategies that synergistically harness the strengths of both CNN and Transformer, thereby mitigating individual limitations and enhancing diagnostic accuracy.
In this paper, we introduce a novel retinal layer segmentation method, employing a multi-scale structure. Our method utilizes the advanced ConvNeXt architecture as the backbone for robust feature extraction. Uniquely, we design a Transformer–CNN feature decoder module that employs the Transformer block not as a traditional encoding module but as an optimization tool to fully leverage the global attention mechanism inherent in the Transformer architecture. We validate the effectiveness of our approach through rigorous testing on two distinct datasets: one proprietary dataset, as illustrated in Figure 1, and another dataset, which is publicly available. Our results, including detailed ablation studies, demonstrate the superiority of our enhanced structural design.
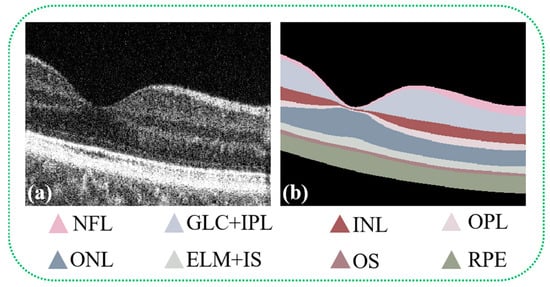
Figure 1.
OCT B-scan images display the retinal layers of healthy human eyes alongside annotations of each specific retinal tissue layer. Figure (a) presents the original B-scan image of the retinal layers, while Figure (b) illustrates the annotated ground truth, identifying the eight distinct layers of the retina: the Nerve Fiber Layer (NFL), Ganglion Cell Layer + Inner Plexiform Layer (GCL + IPL), Inner Nuclear Layer (INL), Outer Plexiform Layer (OPL), Outer Nuclear Layer (ONL), External Limiting Membrane + Inner Segments (ELM + IS), Outer Segments (OS), and Retinal Pigment Epithelium (RPE). Annotations for regions classified as background are also included.
2. Related Work
Recently, a variety of methods have been employed extensively in the study of retinal layer boundary segmentation. These include thresholding [18], active contour models [19], Markov random field models [20], level set models, and graph theory [21]. For instance, Chiu et al. developed a method combining graph theory with dynamic programming to automatically segment seven retinal layers. In their approach, each pixel in an OCT image is mapped to a node in a graph, with edge weights between neighboring nodes calculated based on the vertical luminance gradient values of the pixels [22]. Similarly, Chen et al. introduced a graph-cutting algorithm incorporated into a 3D graph search method, which segments retinal layers and extracts fluid regions within the retina [23]. Naz et al. employed a combination of structural tensor and kernel regression models to segment retinal layer boundaries while preserving the complex structural information inherent in OCT images [24]. Furthermore, Hussain et al. proposed a novel graph model construction method that initially extracts groups of candidate boundary pixels using the Canny edge detection operator, and subsequently using their endpoints as graph nodes. The connections between these nodes are weighted based on three attributes: Euclidean distance, slope similarity, and disjunction [25]. Although these traditional methods rely on a priori knowledge derived from extensive projections and are supported by strong theoretical foundations, they often lack generalizability. Specifically designed for certain types of retinal images, their performance can be suboptimal in cases involving retinal layer deformation, interlayer fluid accumulation, or the presence of scattering noise.
Deep learning has delivered impressive outcomes across various applications [26,27], particularly in the rapid advancement of semantic segmentation within medical imaging [28,29,30]. Notably, the U-net model, introduced in 2015, has set a high standard in medical semantic segmentation by combining efficiency with high performance [31]. Building on this, the TransUnet model has been established as a benchmark for semantic segmentation tasks. In the specific area of retinal layer segmentation, significant advancements have been made. Roy et al. first introduced a fully convolutional network, ReLayNet, which segments multiple retinal layers and delineates fluid regions. Following this, Iqbal et al. developed the G-Net, enhancing it to reduce the complexity of vascular segmentation architecture by optimizing the number of filters per layer and minimizing feature overlap, thereby achieving superior performance in this field [32]. Furthermore, Gao et al. improved the TransUnet network model for the automatic semantic segmentation of inner retinal layers in OCT images, significantly enhancing the performance of the retinal segmentation architecture. This method not only improves the overall semantic accuracy but also reduces computational demands, leading to improved outcomes in the segmentation of inner retinal layers, thereby aiding ophthalmologists in clinical diagnostics [33]. Additionally, Yao et al. introduced a network that utilizes global information fusion and dual decoder collaboration for the joint segmentation of hard exudates and microglia in OCT images, achieving notable success [34]. Finally, He et al. proposed an innovative end-to-end retinal layer segmentation network based on ConvNeXt. This network employs a novel depth-deficit-attention module and a multiscale structure to measure retinal layer thickness with greater accuracy and stability [35].
3. Methodology
3.1. Overall Framework
Our proposed MT_Net employs a U-shaped architecture inspired by the U-Net framework, with ConvNeXt serving as its core component. ConvNeXt, a Transformer-based visual model, is designed for the efficient extraction of multi-scale deep features, which are crucial for image processing tasks. Specifically, our model processes retinal images with dimensions (2, 1, H, W) as input, where the input image undergoes multi-scale feature extraction through the ConvNeXt network, and the features are designated as F_1, F_2, F_3, and F_4. Initially, feature F_4 is upsampled and merged with feature F_3. To maintain a balance between computational efficiency and effective feature transmission, a Transformer module is integrated at this stage. After integration, the output from the Transformer module undergoes depthwise separable convolution and further upsampling before fusing with feature F_2. This procedure is repeated in subsequent layers. As the network approaches its final layer, the processed feature maps are sequentially fused using Concat modules and further refined through depthwise separable convolution. Finally, these feature maps are processed by the Output Completion (OC) module, resulting in the generation of the final output image. This structured approach not only ensures efficient handling of features across multiple scales but also enhances the model’s capability to process complex image data, making it highly suitable for sophisticated image processing tasks. The overall network framework is depicted in Figure 2.
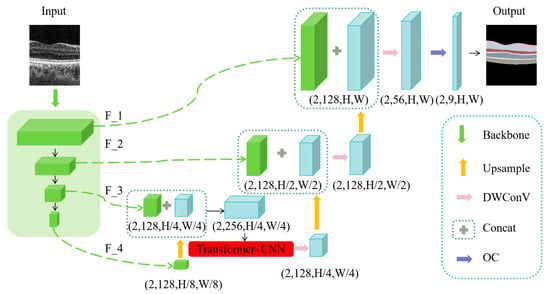
Figure 2.
Overall Network Architecture of MT_Net.
3.2. ConvNeXt
ConvNeXt serves as the foundational backbone for feature extraction within our proposed framework. Leveraging the inherent strengths of CNN, ConvNeXt adeptly captures local feature information from input data. Its multi-layered architecture enables hierarchical feature learning, allowing for the extraction of increasingly abstract and discriminative features as data progress through the network. Furthermore, its parameter-efficient design ensures computational efficiency without compromising the quality of extracted features.
Figure 3 illustrates the overall structure of the ConvNeXt network, which comprises several normalization layers (LayerNorm), ConvNeXt Block modules, and Downsample modules. These components systematically process the pre-processed input image to sequentially extract features labeled as F1, F2, F3, and F4. Additionally, to meet specific application requirements and enhance processing speed, we have refined the traditional ConvNeXt by standardizing the dimensions across all stages to (128,128,128,128). This modification optimizes the network’s computational efficiency while ensuring that it remains highly adaptable to various image-processing tasks.
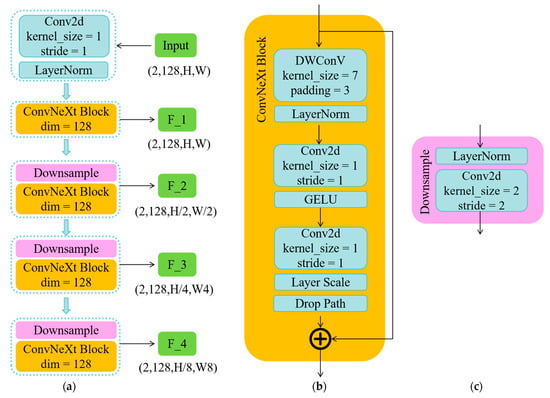
Figure 3.
The proposed method framework. Figure (a) depicts the overall framework of the ConvNeXt network, Figure (b) details the components of the ConvNeXt Block module, and Figure (c) illustrates the composition of the Downsample module.
3.3. Transformer–CNN Feature Decoder Module
For feature extraction, CNN boasts remarkable efficacy in discerning local features, while exhibiting limitations in comprehensively capturing global feature contexts. Due to the exceptional ability of the Transformer block to capture long-range dependencies, it inherently excels at processing global information, thereby demonstrating superior performance in processing global information. In MT_Net, we introduce a novel Transformer–CNN feature decoder block tailored to simultaneous global and local feature acquisition. Specifically, we opted to integrate the original Transformer block into our framework to enhance model compatibility and minimize computational complexity.
Before the feature is processed by the Transformer block, the feature map F is first converted into a 2D patch sequence as
We use a trainable linear project to map the vector patch to a latent D-dimensional embedding space. To encode the spatial information of the patches, we learn a specific position embedding that is added to preserve the positional information in the patch embedding as
where represents the patch embedding projection, e is an additional learnable vector concatenated with the remaining vectors to integrate the information of all remaining vectors, is the concatenation operator, and represents the position embedding. Then, is input into the Transformer layer
As shown in Figure 4, first through the first Layer Norm (LN) layer and then through the Multi-head Self-attention (MSA) layer, forms the residual structure and obtains . Then, after passing through the second LN layer, it enters the Multi-Layer Perceptron (MLP) layer; here . once again forms the residual structure and is obtained (Equations (2) and (3)). At this point, the first Transformer layer is finished. We have set three Transformer layers in our approach, so we have to cycle three times.
When the outputs from the Transformer block, we have to remove the vector from the , because only then can we reshape it to the same size of
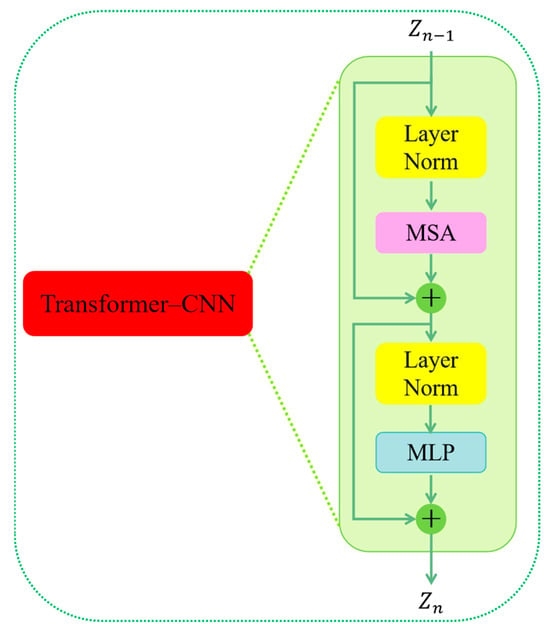
Figure 4.
The flowchart of the Transformer–CNN block.
4. Results and Discussion
4.1. Datasets
We conducted experimental studies utilizing the Retina500 and NR206 datasets, with their settings detailed in Table 1.

Table 1.
Setup of datasets Retina500 and NR206.
4.1.1. Retina500 Dataset
We independently design and develop the Retina500 dataset employing a specially engineered visible light OCT system. Figure 5 displays the arrangement of the optical setup used in our experimentation. A broad-spectrum laser output is produced by the SuperK supercontinuum laser manufactured by NKT Photonics (Birkerød, Denmark). This laser output is split into visible and near-infrared (NIR) light by a dichroic mirror (DM1) with a cut-off wavelength set at 650 nm. The visible light is polarized utilizing a polarization beam splitter (PBS) and subsequently expanded by a pair of prisms. The polarization is optimized for interference efficiency using polarization controllers (PCs). A specific range within the visible spectrum is identified using a slit aperture and redirected by a mirror (M). The NIR light is separated by another dichroic mirror (DM2) at a cut-off wavelength of 900 nm and filtered to a bandwidth of 800 nm to 875 nm by edge filters. These spectral segments are then combined using a custom wavelength division multiplexer (WDM) and fed into an optical fiber coupler (TW670R2A2, Thorlabs, Newton, NJ, USA). In the sample arm, the beam is collimated with a 6 mm lens (CL), corrected with an achromatizing lens (AL), controlled by galvanometer mirrors, and focused onto the pupil through a 2:1 telescope, attaining a 2 mm beam diameter on the cornea. The reference arm consists of collimated light that is reflected back. Dispersion compensation in the sample arm is accomplished using BK7 glass plates (DC), and light intensity is regulated using a variable ND filter (ND). Additionally, dispersion matching can be carried out using a water cuvette. Light from both arms is combined in the fiber coupler, and then divided into two spectrometers through another WDM. These spectrometers are equipped with line scanning cameras (spl2048-140km, Basler, Ahrensburg, Germany) capturing spectral ranges from 535 to 600 nm and 780 to 880 nm, respectively. The spectrometer converts the return beam into an electrical signal, which is processed by a computer to produce a final OCT image.
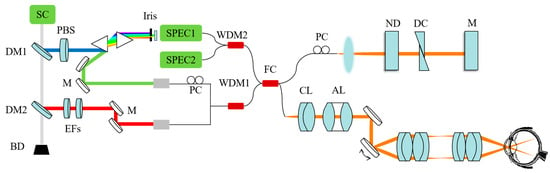
Figure 5.
System setup for vnOCT for human retinal imaging. BD: beam dump; PC: polarization controller. EFs: two edge filters.
For annotation purposes, we selected the advanced graphics software, Inkscape (version number: 1.1.2), under the guidance of our ophthalmology experts, to precisely label the eight retinal layers: NFL, GCL + IPL, INL, OPL, ONL, ELM + IS, OS, and RPE. Each layer is distinguished by unique colors, with non-layer areas marked in black as the background. Before finalizing and exporting each image as a 480 × 400 pixel PNG file, it undergoes meticulous examination and validation by our professional ophthalmologists to ensure data accuracy and reliability. Additionally, to facilitate the effective training and evaluation of machine learning models, we segment the dataset into a training set with 400 images, a validation set with 50 images, and a test set with 50 images.
4.1.2. NR206 Dataset
The NR206 dataset is acquired from the Sankara Nethralaya Eye Hospital in Chennai, India, using a Cirrus HD-OCT machine (Carl Zeiss Meditec, Inc., Dublin, CA, USA) [36]. Each image in the dataset, selected by an experienced clinical optometrist, is centered on the fovea of the volumetric scan. The OCT machine employs a superluminescent diode as a light source, emitting at a wavelength of 840 nanometers, and achieves an axial resolution of 5 microns and a lateral resolution of 15 microns. The dataset is systematically organized into three subsets: training, validation, and testing, containing 126, 40, and 40 images, respectively.
4.2. Implementation Details
Our experiments are conducted on a machine equipped with an NVIDIA A100 GPU, utilizing the PyTorch framework. Considering the memory constraints of the GPU, we standardize the training batch size across all comparative methods to ensure fairness. We employ the Adam optimizer coupled with the StepLR learning rate scheduler to dynamically adjust the learning rate throughout the training process. The cross-entropy loss function is used for model training. To augment the training dataset, we apply various data augmentation techniques, including horizontal flipping and random image rotation. The training is set to automatically terminate after 200 epochs, with the selection of the best-performing model weights from the validation set for further testing. Specifically, each input image was subjected to horizontal flipping with a probability of 0.5, and to random rotations with the same probability, where the angle of rotation varied between −10 and 10 degrees. For the Retina500 dataset, the input images are resized to 400 × 480 pixels, while for the NR206 dataset, the images are cropped to 480 × 400 pixels. An initial learning rate of 0.002 is set for both datasets.
4.3. Performance Metrics
To consider the class imbalance problem in the dataset, we quantitatively evaluate the segmentation performance using the Dice score, mIoU, Acc, and mPA. They are computed using the following formulas:
where TP stands for true positive, i.e., the predicted result is consistent with the ground truth label; TN stands for true negative, i.e., both the predicted result and the ground truth label are negative; FP stands for false positive, i.e., the predicted result is positive but the ground truth label is negative; FN stands for false negative, i.e., the predicted result is negative but the ground truth label is positive; and k stands for the number of categories.
4.4. Results and Discussion
4.4.1. Experimentation and Analysis on the Retina500 Dataset
In this study, we benchmark our method against several key competitors in the field of retinal layer segmentation. These include ReLayNet, a well-established method, as well as OS_MGUNet and EMV_Net, which are recent advancements introduced over the past two years. Additionally, we compare our approach with DeepLab_v3+, UNETR_2D, and SegFormer, which are noted for their efficacy in semantic segmentation. Noteworthy is the fact that both SegFormer and our method utilize a hybrid architecture combining transformers with CNNs. Despite these architectural similarities, the results in Table 2 show that our method outperforms these established models on the Retina500 dataset. It achieves the highest mIoU, Acc, and mPA, surpassing the second-ranked method by margins of 0.39%, 0.61%, and 0.10%, respectively.

Table 2.
Quantitative comparison of evaluation indicators of various methods on the Retina500 dataset.
Table 3 presents a comparison of Dice scores across various layers of the Retina500 dataset. Our method generally outperforms the six aforementioned methods, particularly in terms of the segmentation accuracy of the NFL, which is critical due to its association with a higher disease prevalence. Although OS_MGUNet demonstrates a slight advantage in the ELM and RPE layers, it falls behind our method in the overall metrics—mIoU, Acc, and mPA—by 0.39%, 0.61%, and 0.10%, respectively. Compared to EMV_Net, which utilizes a multi-scale feature segmentation approach, our method exhibits superior performance on most layers. This superiority is likely attributable to the integration of the Transformer module, which enhances our framework’s effectiveness. In contrast, DeepLab_v3+, despite its standing as a classic in semantic segmentation, significantly underperforms relative to our method. This underperformance is partly because DeepLab_v3+’s backbone network is limited to extracting only two levels of features, primarily from the RPE layer. Furthermore, in comparison with SegFormer, our methodology exhibits consistently superior segmentation performance.

Table 3.
Dice score (%) of the segmentation results on the Retina500 dataset obtained by different methods.
Furthermore, we conduct a statistical analysis of the pixel proportions across each retinal layer within the entire Retina500 dataset, with the statistical outcomes displayed in Figure 6. Our findings align with those of He et al. [41], demonstrating that the segmentation accuracy for specific retinal layers such as GCL + IPL and ONL is significantly higher compared to other layers. This discrepancy is attributed to class imbalance within the dataset. Specifically, in the Retina500 dataset, the average pixel proportions for GCL + IPL and ONL classes are the highest, accounting for 23.28% and 21.81% of the total retinal layer pixels, respectively, which is significantly higher than in other classes. This imbalance leads to model overfitting on these more-frequently occurring classes during training, resulting in higher segmentation accuracy. Conversely, the classes that appear less frequently in the dataset do not provide sufficient examples for the model to train with comparable precision, resulting in lower accuracy for these categories. This underscores the importance of balancing class distribution in datasets for retinal layer segmentation research in the field of deep learning.
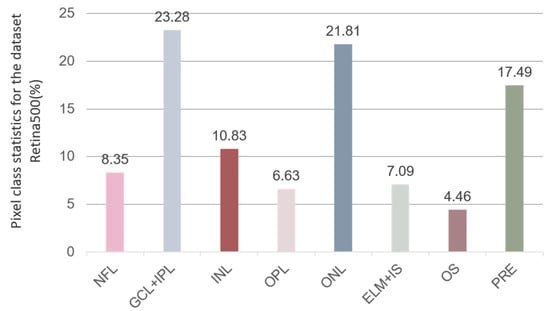
Figure 6.
The average percentage of pixels in each retinal layer in the Retina500 dataset among all retinal layers (excluding background).
In addition to quantitative assessments, our study incorporates a detailed qualitative analysis, as depicted in Figure 7. Our method exhibits superior segmentation performance, which is particularly evident in Figure 7i. When comparing our results to other methods, segmentation errors typically fall into two main categories: intra-class and inter-class errors. Intra-class errors, which pertain primarily to misclassifications within the same class, including the background, are marked by white dashed lines in Figure 7d,f,g. These errors are typically due to similarities in texture or color within the same class, leading to segmentation challenges. Inter-class errors, depicted by red dashed lines in Figure 7c–h, occur when the boundaries between different classes are inaccurately identified. In contrast, Figure 7i shows that our proposed method achieves accurate, continuous, and complete segmentation across each layer without interruptions, yielding superior results. These visual comparisons further substantiate the effectiveness of our segmentation approach.
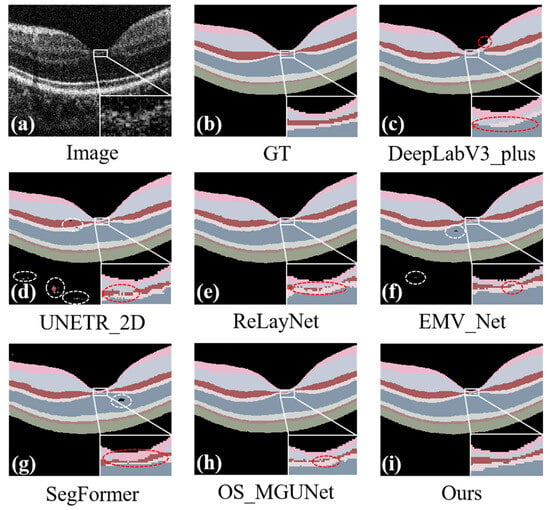
Figure 7.
Predicted maps of retinal layer segmentation for randomized test images from the Retina500 dataset. Panel (a) displays the original image, and panel (b) shows the ground truth. The prediction maps are generated via various segmentation methods: DeepLab_v3+ in panel (c), UNETR_2D in panel (d), ReLayNet in panel (e), EMV_Net in panel (f), SegFormer in panel (g), OS_MGUNet in panel (h), and our proposed method in panel (i). Each panel demonstrates the effectiveness of the respective methods in segmenting the complex structures of the retinal layers.
Noise and artifacts in OCT images are caused by various factors, among which motion artifacts are notably significant. These artifacts typically result from movements of the patient’s eye or head and are manifested as thin vertical white or black lines on the retinal layer images. This phenomenon is exemplified in the area highlighted by the red arrow in Figure 8a. As shown in Figure 8d–h, compared to our method, other methods display various degrees of intra-class errors in the presence of artifacts. Although there are no evident intra-class errors in Figure 8c, artifacts still lead to varying extents of under-segmentation in the GLC + IPL, INL, and OPL, which severely impacts the accuracy of disease diagnosis. Our method remains unaffected by noise and artifacts, achieving precise segmentation.
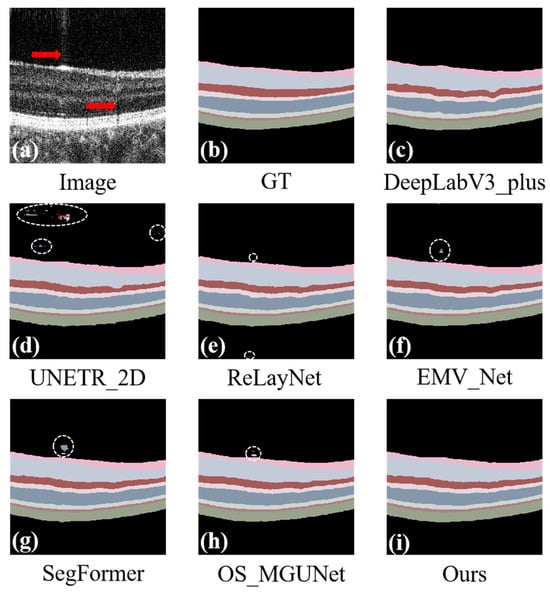
Figure 8.
Segmentation effects of the model under the influence of noise and artifacts. Panel (a) displays the original image, and panel (b) shows the ground truth. The prediction maps are generated via various segmentation methods: DeepLab_v3+ in panel (c), UNETR_2D in panel (d), ReLayNet in panel (e), EMV_Net in panel (f), SegFormer in panel (g), OS_MGUNet in panel (h), and our proposed method in panel (i). Each panel demonstrates the effectiveness of the respective methods in segmenting the complex structures of the retinal layers.
4.4.2. Experiments and Analysis on the NR206 Dataset
To further substantiate the effectiveness of our approach, we extended our comparisons to include experiments on the third-party public dataset, NR206. Table 4 presents a quantitative comparison of our method against other established methods, where our approach demonstrates superior performance. Specifically, it outperforms the second-ranked method by 0.51% in mIoU and by 0.03% in mPA.

Table 4.
Quantitative comparison of evaluation indicators of various methods on the NR206 dataset.
Additionally, Table 5 presents the Dice score performance of various methods across different layers of the NR206 dataset. The results indicate that our methods achieve the highest Dice scores in most of the layers, notably excelling in the segmentation accuracy of the NFL, where they outperform the second-best method by 0.22%. This enhancement underscores the improved capability of our method in capturing and processing complex patterns more efficiently than traditional models.

Table 5.
Dice score (%) of the segmentation results on the NR206 dataset obtained by different methods.
We also analyze the average pixel proportions within the NR206 dataset, with the statistical outcomes displayed in Figure 9. Specific retinal layers such as GCL + IPL and ONL consistently show higher segmentation accuracies compared to other layers. Notably, GCL + IPL and ONL account for 22.89% and 21.77% of the total retinal layer pixels, respectively, significantly exceeding other layers. Layers with lower pixel proportions also exhibit lower accuracies. These results further confirm that differences in accuracy primarily stem from class imbalance in the dataset. Therefore, to enhance overall segmentation precision, it is necessary to implement specific measures to address the recognition and segmentation challenges of different categories.
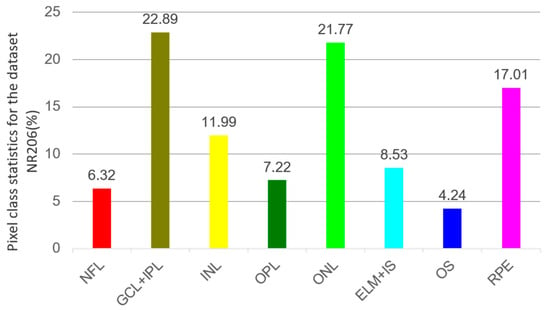
Figure 9.
The average percentage of pixels in each retinal layer in the NR206 dataset among all retinal layers (excluding background).
Figure 10 displays a qualitative assessment of our method using the NR206 dataset. As previously mentioned, segmentation errors are divided into intra-class errors, represented by white dashed lines as shown in Figure 10d, and inter-class errors, indicated by red dashed lines in Figure 10c,e–h. When comparing our results with other methods, it is evident that other approaches exhibit noticeable intra-class and inter-class errors, whereas our method, as illustrated in Figure 10i, shows neither type of error. This demonstrates that our approach achieves the accurate, continuous, and complete segmentation of each retinal layer. These visual comparisons further validate the effectiveness and precision of our segmentation method.
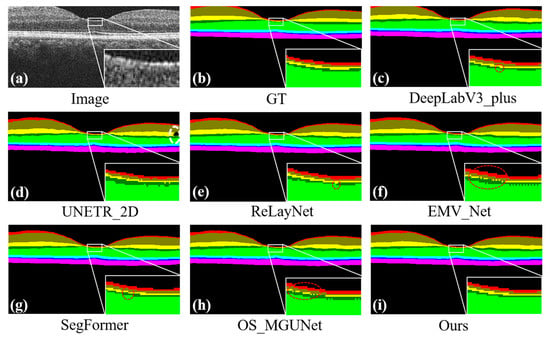
Figure 10.
Predicted maps of retinal layer segmentation for randomized test images from the NR206 dataset. Panel (a) displays the original image, while panel (b) shows the ground truth. The subsequent panels illustrate the prediction maps generated by various segmentation methods: DeepLab_v3+ in panel (c), UNETR_2D in panel (d), ReLayNet in panel (e), EMV_Net in panel (f), SegFormer in panel (g), OS_MGUNet in panel (h), and our method in panel (i). Additionally, each panel includes a zoomed-in view to highlight the local details of the predictions, providing a closer look at the segmentation accuracy of each method.
4.4.3. Inference Time Statistics
In Table 6, we present the inference times recorded for each method when applied to the Retina500 dataset. While our approach exhibits superior segmentation accuracy, it also incurs a slightly longer inference time compared to other methods. This observation underscores the need for optimization in future developments, aiming to balance computational efficiency with performance accuracy.

Table 6.
Inference time (s) statistics for various methods.
4.5. Ablation Study without Transformer
Our ablation study conducted on the Retina500 dataset evaluates the effectiveness of the Transformer module within our framework. In particular, Figure 11c,d demonstrate that the inclusion of the Transformer module significantly enhances the model’s capability in segmenting different categories, effectively reducing intra-class and inter-class errors. This improvement not only strengthens the model’s generalization ability but also enhances its capacity to capture details, enabling the model to more accurately distinguish between closely related or similar categories.
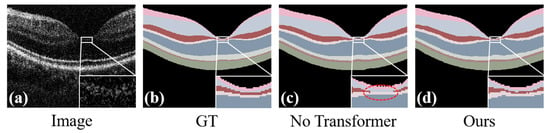
Figure 11.
Ablation experiments performed on the Retina500 dataset, where (a) is the original image, (b) is the ground-truth, (c) is the prediction map without the Transformer module in our method, and (d) is the prediction map with the Transformer module in our method.
The results presented in Table 7 demonstrate that the implementation of the Transformer module significantly enhances the performance of the model. Specifically, following the incorporation of this module, the Dice score shows an average increase of 2.16%, while the mIoU, Acc, and mPA improve by 3.61%, 0.30%, and 1.90%, respectively. These improvements highlight the capability of the Transformer module to boost segmentation performance by effectively extracting global features. This aggregation of global information not only improves the model’s performance in handling local details but also enhances its accuracy in terms of its overall structure.

Table 7.
Comparison of quantitative analysis on the Retina500 dataset through Transformer ablation experiments performed in our framework.
5. Conclusions
In this study, we have developed an advanced technique for the segmentation of retinal layers, specifically designed to tackle the challenges posed by noise and artifacts in OCT images, which hinder precise layer segmentation. Our innovative method utilizes a multi-scale framework that leverages the strengths of the ConvNeXt backbone network along with the dynamic capabilities of the Transformer module, significantly enhancing the segmentation of retinal images. The ConvNeXt architecture ensures consistent and efficient feature extraction from retinal images, while the inclusion of the Transformer module employs its global attention mechanism to manage complex information across the images more effectively. The strategic architecture of our model’s multi-scale structure allows it to adapt to the varying scales of retinal layers, thereby enhancing both the accuracy and robustness of segmentation. Rigorous evaluations conducted on the Retina500 and NR206 datasets have shown the superior performance of our method, achieving benchmark metrics of mIoU, Acc, and mPA at 81.26%, 91.38%, and 96.77%, respectively, on the Retina500 dataset. These results not only confirm the effectiveness of our approach in segmenting fundus images but also underscore its significant potential for advancing the early diagnosis of fundus diseases.
In future work, our model will be rigorously tested across a broader spectrum of disease datasets to further substantiate its performance. Additionally, to bolster the model’s ability to generalize across different clinical scenarios, we will evaluate it with images obtained from various diagnostic devices. Efforts will also be made to refine the model for lightweight deployment, with the dual objectives of enhancing diagnostic accuracy and reducing the incidence of false positives in medical imaging.
Author Contributions
Conceptualization, E.L. and X.H.; methodology, E.L.; software, J.Y.; validation, L.Z., A.W. and J.L.; formal analysis, E.L. and S.Y.; resources, W.S.; data curation, E.L. and Y.G.; writing—original draft preparation, E.L.; writing—review and editing, E.L. All authors have read and agreed to the published version of the manuscript.
Funding
This work was funded by the National Natural Science Foundation of China, Youth Science Foundation Program grant number 62205181, the Shandong University Education and Teaching Reform Research Program grant number 2022Y126, the National Foundation of China, Youth Science Foundation Program grant number 62205181, the Shandong Provincial Science and Technology Department, Excellent Youth Fund (Overseas) grant number 2023HWYQ-023, the Natural Science Foundation of Shandong Province, Youth Fund grant number ZR2022QF017, the Organization Department of Shandong Provincial Committee, Taishan Scholars grant number tsqn202211038, and Shandong University Education and the Teaching Reform Research Program grant number XYJG2023058.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The data presented in this study are available on request from the corresponding author. The data are not publicly available due to privacy.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Wang, J.; Song, W.; Sadlak, N.; Fiorello, M.G.; Manishi, D.; Yi, J. Oxygen Saturation of Macular Vessels in Glaucoma Subjects Using Visible Light Optical Coherence Tomography. Investig. Ophthalmol. Vis. Sci. 2023, 64, 1357. [Google Scholar]
- Song, W.; Zhang, S.; Mun Kim, Y.; Sadlak, N.; Fiorello, M.G.; Desai, M.; Yi, J. Visible Light Optical Coherence Tomography of Peripapillary Retinal Nerve Fiber Layer Reflectivity in Glaucoma. Trans. Vis. Sci. Technol. 2022, 11, 28. [Google Scholar] [CrossRef]
- Solano, A.; Dietrich, K.N.; Martínez-Sober, M.; Barranquero-Cardeñosa, R.; Vila-Tomás, J.; Hernández-Cámara, P. Deep Learning Architectures for Diagnosis of Diabetic Retinopathy. Appl. Sci. 2023, 13, 4445. [Google Scholar] [CrossRef]
- He, Y.; Carass, A.; Liu, Y.; Calabresi, P.A.; Saidha, S.; Prince, J.L. Longitudinal deep network for consistent OCT layer segmentation. Biomed. Opt. Express 2023, 14, 1874. [Google Scholar] [CrossRef]
- Hsia, W.P.; Tse, S.L.; Chang, C.J.; Huang, Y.L. Automatic Segmentation of Choroid Layer Using Deep Learning on Spectral Domain Optical Coherence Tomography. Appl. Sci. 2021, 11, 5488. [Google Scholar] [CrossRef]
- Bowd, C.; Weinreb, R.N.; Williams, J.M.; Zangwill, L.M. The Retinal Nerve Fiber Layer Thickness in Ocular Hypertensive, Normal, and Glaucomatous Eyes with Optical Coherence Tomography. Arch. Ophthalmol. 2000, 118, 22–26. [Google Scholar] [CrossRef] [PubMed]
- Yenice, E.; Şengün, A.; Soyugelen Demirok, G.; Turaçlı, E. Ganglion cell complex thickness in nonexudative age-related macular degeneration. Eye 2015, 29, 1076–1080. [Google Scholar] [CrossRef]
- Tatsumi, T. Current Treatments for Diabetic Macular Edema. Int. J. Mol. Sci. 2023, 24, 9591. [Google Scholar] [CrossRef]
- Abramoff, M.D.; Garvin, M.K.; Sonka, M. Retinal Imaging and Image Analysis. IEEE Rev. Biomed. Eng. 2010, 3, 169–208. [Google Scholar] [CrossRef] [PubMed]
- Song, W.; Shao, W.; Yi, J. Wide-Field and Micron-Resolution Visible Light Optical Coherence Tomography in Human Retina by a Linear-K Spectrometer. In Biophotonics Congress 2021; Boudoux, C.M.K.H., Ed.; Optica Publishing Group: Washington, DC, USA, 2021; pp. DM2A–DM4A. [Google Scholar]
- Fujimoto, J.G.; Drexler, W.; Schuman, J.S.; Hitzenberger, C.K. Optical Coherence Tomography (OCT) in Ophthalmology: Introduction. Opt. Express 2009, 17, 3978–3979. [Google Scholar] [CrossRef]
- Frohman, E.M.; Fujimoto, J.G.; Frohman, T.C.; Calabresi, P.A.; Cutter, G.; Balcer, L.J. Optical coherence tomography: A window into the mechanisms of multiple sclerosis. Nat. Rev. Neurol. 2008, 4, 664–675. [Google Scholar] [CrossRef]
- Liu, W.; Sun, Y.; Ji, Q. MDAN-UNet: Multi-Scale and Dual Attention Enhanced Nested U-Net Architecture for Segmentation of Optical Coherence Tomography Images. Algorithms 2020, 13, 60. [Google Scholar] [CrossRef]
- Roy, A.G.; Conjeti, S.; Karri, S.P.K.; Sheet, D.; Katouzian, A.; Wachinger, C.; Navab, N. ReLayNet: Retinal layer and fluid segmentation of macular optical coherence tomography using fully convolutional networks. Biomed. Opt. Express 2017, 8, 3627–3642. [Google Scholar] [CrossRef]
- Sunija, A.P.; Kar, S.; Gayathri, S.; Gopi, V.P.; Palanisamy, P. OctNET: A Lightweight CNN for Retinal Disease Classification from Optical Coherence Tomography Images. Comput. Meth. Prog. Biomed. 2021, 200, 105877. [Google Scholar]
- Lam, C.; Yu, C.; Huang, L.; Rubin, D. Retinal Lesion Detection with Deep Learning Using Image Patches. Investig. Ophthalmol. Vis. Sci. 2018, 59, 590–596. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Y.; Liang, J.; Cheng, T.; Lin, X.; Zhang, Y.; Dong, J. MTPA_Unet: Multi-Scale Transformer-Position Attention Retinal Vessel Segmentation Network Joint Transformer and CNN. Sensors 2022, 22, 4592. [Google Scholar] [CrossRef] [PubMed]
- Lang, A.; Carass, A.; Hauser, M.; Sotirchos, E.S.; Calabresi, P.A.; Ying, H.S.; Prince, J.L. Retinal layer segmentation of macular OCT images using boundary classification. Biomed. Opt. Express 2013, 4, 1133–1152. [Google Scholar] [CrossRef]
- Yazdanpanah, A.; Hamarneh, G.; Smith, B.R.; Sarunic, M.V. Segmentation of Intra-Retinal Layers from Optical Coherence Tomography Images Using an Active Contour Approach. IEEE Trans. Med. Imaging 2011, 30, 484–496. [Google Scholar] [CrossRef] [PubMed]
- Koozekanani, D.; Boyer, K.; Roberts, C. Retinal thickness measurements from optical coherence tomography using a Markov boundary model. IEEE Trans. Med. Imaging 2001, 20, 900–916. [Google Scholar] [CrossRef]
- Xiang, D.; Tian, H.; Yang, X.; Shi, F.; Zhu, W.; Chen, H.; Chen, X. Automatic Segmentation of Retinal Layer in OCT Images with Choroidal Neovascularization. IEEE Trans. Image Process. 2018, 27, 5880–5891. [Google Scholar] [CrossRef]
- Chiu, S.J.; Li, X.T.; Nicholas, P.; Toth, C.A.; Izatt, J.A.; Farsiu, S. Automatic segmentation of seven retinal layers in SDOCT images congruent with expert manual segmentation. Opt. Express 2010, 18, 19413–19428. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Niemeijer, M.; Zhang, L.; Lee, K.; Abramoff, M.D.; Sonka, M. Three-dimensional segmentation of fluid-associated abnormalities in retinal OCT: Probability constrained graph-search-graph-cut. IEEE Trans. Med. Imaging 2012, 31, 1521–1531. [Google Scholar] [CrossRef] [PubMed]
- Naz, S.; Akram, M.U.; Khan, S.A. Automated segmentation of retinal layers from OCT images using structure tensor and kernel regression + GTDP approach. In Proceedings of the 2017 1st International Conference on Next Generation Computing Applications (NextComp), Pointe aux Piments, Mauritius, 19–21 July 2017; pp. 98–102. [Google Scholar]
- Hussain, M.A.; Bhuiyan, A.; Turpin, A.; Luu, C.D.; Smith, R.T.; Guymer, R.H.; Kotagiri, R. Automatic Identification of Pathology-Distorted Retinal Layer Boundaries Using SD-OCT Imaging. IEEE Trans. Biomed. Eng. 2017, 64, 1638–1649. [Google Scholar] [CrossRef] [PubMed]
- Tao, H.; Duan, Q.; Lu, M.; Hu, Z. Learning discriminative feature representation with pixel-level supervision for forest smoke recognition. Pattern Recognit. 2023, 143, 109761. [Google Scholar] [CrossRef]
- Tao, H. Smoke Recognition in Satellite Imagery via an Attention Pyramid Network with Bidirectional Multilevel Multigranularity Feature Aggregation and Gated Fusion. IEEE Internet Things J. 2024, 11, 14047–14057. [Google Scholar] [CrossRef]
- Hu, K.; Zhang, Z.; Niu, X.; Zhang, Y.; Cao, C.; Xiao, F.; Gao, X. Retinal vessel segmentation of color fundus images using multiscale convolutional neural network with an improved cross-entropy loss function. Neurocomputing 2018, 309, 179–191. [Google Scholar] [CrossRef]
- van Grinsven, M.J.J.P.; van Ginneken, B.; Hoyng, C.B.; Theelen, T.; Sanchez, C.I. Fast Convolutional Neural Network Training Using Selective Data Sampling: Application to Hemorrhage Detection in Color Fundus Images. IEEE Trans. Med. Imaging 2016, 35, 1273–1284. [Google Scholar] [CrossRef] [PubMed]
- Xie, H.; Yang, D.; Sun, N.; Chen, Z.; Zhang, Y. Automated pulmonary nodule detection in CT images using deep convolutional neural networks. Pattern Recognit. 2019, 85, 109–119. [Google Scholar] [CrossRef]
- Ronneberger, O.; Fischer, P.; Brox, T. In U-Net: Convolutional Networks for Biomedical Image Segmentation. In Proceedings of the Medical Image Computing and Computer-Assisted Intervention–MICCAI 2015: 18th International Conference, Munich, Germany, 5–9 October 2015; Proceedings, Part III 18. Springer International Publishing: Cham, Switzerland, 2015; pp. 234–241. [Google Scholar]
- Iqbal, S.; Naqvi, S.S.; Khan, H.A.; Saadat, A.; Khan, T.M. G-Net Light: A Lightweight Modified Google Net for Retinal Vessel Segmentation. Photonics 2022, 9, 923. [Google Scholar] [CrossRef]
- Gao, Z.; Wang, Z.; Li, Y. A Novel Intraretinal Layer Semantic Segmentation Method of Fundus OCT Images Based on the TransUNet Network Model. Photonics 2023, 10, 438. [Google Scholar] [CrossRef]
- Yao, C.; Wang, M.; Zhu, W.; Huang, H.; Shi, F.; Chen, Z.; Wang, L.; Wang, T.; Zhou, Y.; Peng, Y.; et al. Joint Segmentation of Multi-Class Hyper-Reflective Foci in Retinal Optical Coherence Tomography Images. IEEE Trans. Biomed. Eng. 2022, 69, 1349–1358. [Google Scholar] [CrossRef] [PubMed]
- He, X.; Wang, Y.; Poiesi, F.; Song, W.; Xu, Q.; Feng, Z.; Wan, Y. Exploiting multi-granularity visual features for retinal layer segmentation in human eyes. Front. Bioeng. Biotechnol. 2023, 11, 1191803. [Google Scholar] [CrossRef] [PubMed]
- Gholami, P.; Roy, P.; Parthasarathy, M.K.; Lakshminarayanan, V. OCTID: Optical coherence tomography image database. Comput. Electr. Eng. 2020, 81, 106532. [Google Scholar] [CrossRef]
- Chen, L.; Zhu, Y.; Papandreou, G.; Schroff, F.; Adam, H. Encoder-Decoder with Atrous Separable Convolution for Semantic Image Segmentation. In Proceedings of the European Conference on Computer Vision (ECCV), Munich, Germany, 8–14 September 2018; pp. 801–818. [Google Scholar]
- Hatamizadeh, A.; Tang, Y.; Nath, V.; Yang, D.; Myronenko, A.; Landman, B.; Roth, H.R.; Xu, D. UNETR: Transformers for 3D Medical Image Segmentation. In Proceedings of the IEEE/CVF Winter Conference on Applications of Computer Vision (WACV), Waikoloa, HI, USA, 3–8 January 2022; pp. 1748–1758. [Google Scholar]
- Xie, E.; Wang, W.; Yu, Z.; Anandkumar, A.; Alvarez, J.M.; Luo, P. SegFormer: Simple and Efficient Design for Semantic Segmentation with Transformers. Adv. Neural Inf. Process. Syst. 2021, 34, 12077–12090. [Google Scholar]
- Li, J.; Jin, P.; Zhu, J.; Zou, H.; Xu, X.; Tang, M.; Zhou, M.; Gan, Y.; He, J.; Ling, Y.; et al. Multi-scale GCN-assisted two-stage network for joint segmentation of retinal layers and discs in peripapillary OCT images. Biomed. Opt. Express 2021, 12, 2204–2220. [Google Scholar] [CrossRef]
- He, X.; Song, W.; Wang, Y.; Poiesi, F.; Yi, J.; Desai, M.; Xu, Q.; Yang, K.; Wan, Y. Lightweight Retinal Layer Segmentation with Global Reasoning. IEEE Trans. Instrum. Meas. 2024, 73, 2520214. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).