1. Introduction
Holography is a technique by which a wave front reflected from an object can be recorded and later reconstructed in the absence of the original object. In conventional Gabor holography [
1], the holograms are recorded photographically and reconstructed optically. Holographic microscopy was first proposed by Gabor [
1]. It offers high-contrast 3D images of microscopic samples in depth. Dennis Gabor invented holography in an attempt to improve the resolution of an electron microscope. Gabor realized that the interference pattern of a wave going through an object or reflected by it (object wave) with the initial undisturbed wave (reference wave) holds the whole information of the sample in it. He named the new imaging principle holography, because of its ability to record the whole optical field.
The electronic recording of a hologram and its digital reconstruction opened a wide range of new imaging possibilities. When the amplitude and phase of the light wave are digitally recorded, it is possible afterwards to process these data via a number of numerical manipulations. Digital holography refers to the acquisition and processing of holograms with the help of a digital sensor array [
2] such as a charge-coupled device (CCD) or CMOS sensors. The digital reconstruction of the electronically recorded holograms is carried out using appropriate software. As a result, quantitative information about the intensity and phase distribution of the electronically reconstructed wave field is obtained. Fast imaging is possible, limited only by video acquisition rate.
Digital holographic microscopy (DHM) is an advanced imaging technology, because a single digital hologram contains the information necessary for the microscopic volume reconstruction of the object [
3]. Numerical reconstruction of the digital holograms gives an efficient methodology of virtual focusing throughout the depth of a microscopic sample, which is a unique feature of DHM.
McLeod et al. [
4,
5,
6] reviewed lens-free holographic imaging, giving detailed information of how images are reconstructed, and the phase recovery techniques used.
Digital holographic microscopy is a suitable technique for 3D visualization and investigation of transparent and semi-transparent objects. DHM can be applied for fast imaging and quantitative marker-free analysis of living cells in usual laboratory conditions. One of many interesting and now-routine applications of DHM is studying cells without staining or labelling them.
We review the main applications of DHM for imaging of live cells, microscopic organisms and drug-induced morphological changes in biological tissues. The novel microscopic technique is non-destructive and label-free and offers unmatched imaging capabilities for biological and bio-medical applications. Live-cell imaging by DHM allows investigation of the dynamic processes underlying the function and morphology of cells.
2. Digital Holographic Microscopy Technique
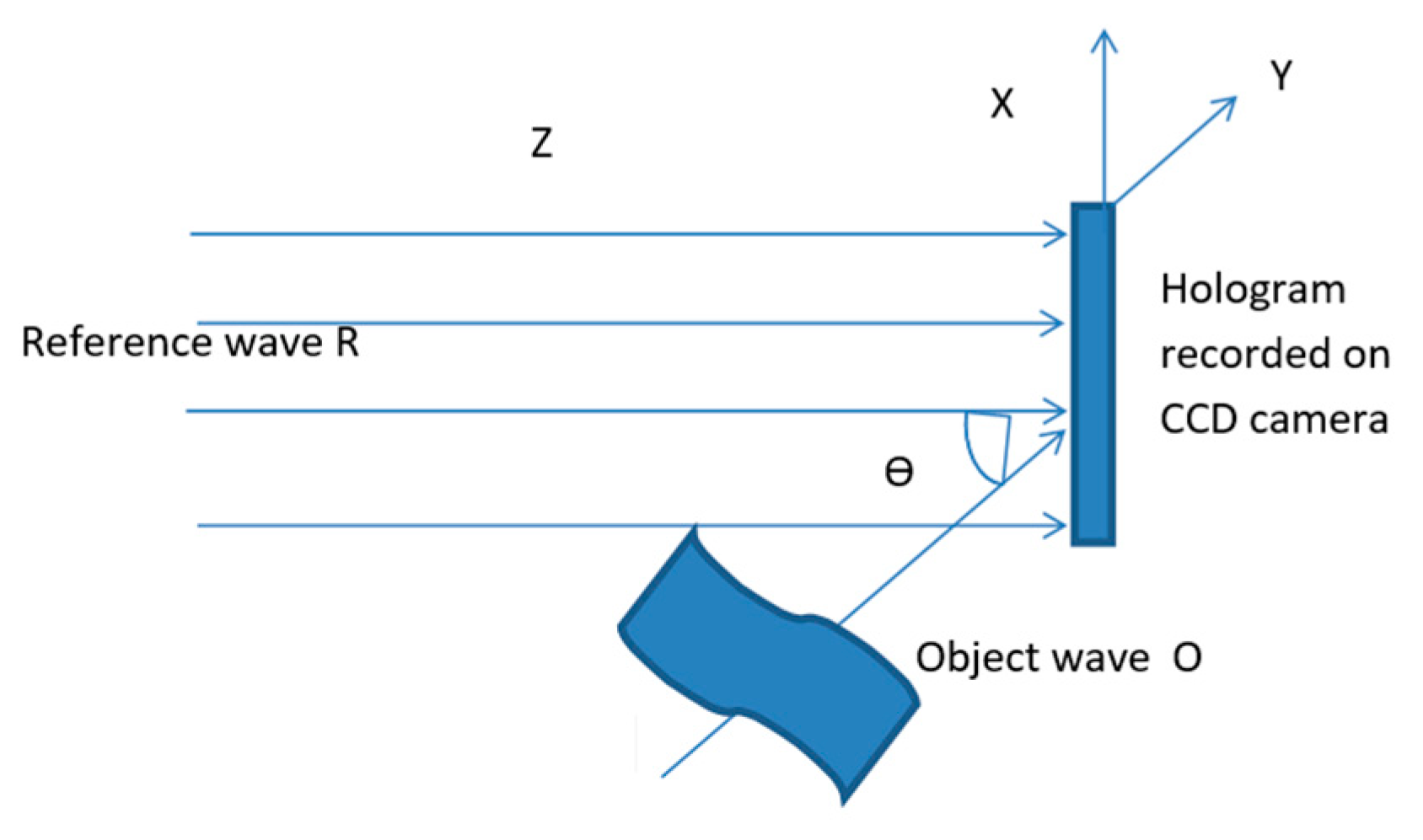
The basic principle of holography is the recording of the interference pattern of an electromagnetic wave that is reflected by or passes through an object (the so-called object wave) and a reference wave which comes from the same laser source. The recorded interference pattern is called a hologram. In classical holography, the image of the object is restored from the hologram, recorded in holographic material, after irradiation with the reference wave. In digital holography, the restoration is done by mathematically transforming the hologram, recorded in a CCD camera (
Figure 1) or using a CMOS sensor.
If R(x,y) denotes the reference wave (which for simplicity is assumed to be flat and collimated), and O(x,y) denotes the wave scattered by the object, then
The interference of the two waves gives the following result on the screen (CCD):
The reconstruction of the wave field is performed by numerical methods. The numerical reconstruction process is based on the Fresnel–Kirchhoff integral, which describes the diffraction of the reconstructing wave at the micro-structure of the hologram.
In the numerical reconstruction process, not only the intensity, but also the phase distribution of the stored wave field can be computed from the digital hologram. This offers new possibilities for a variety of applications.
The mathematical principles of digital reconstruction and digital reconstruction algorithms are well described in the comprehensive review by Kim M. K. [
7]. Kim [
7] also presents several techniques of digital holographic microscopy. We will present here two classic interferometric configurations, which are most commonly used in practice.
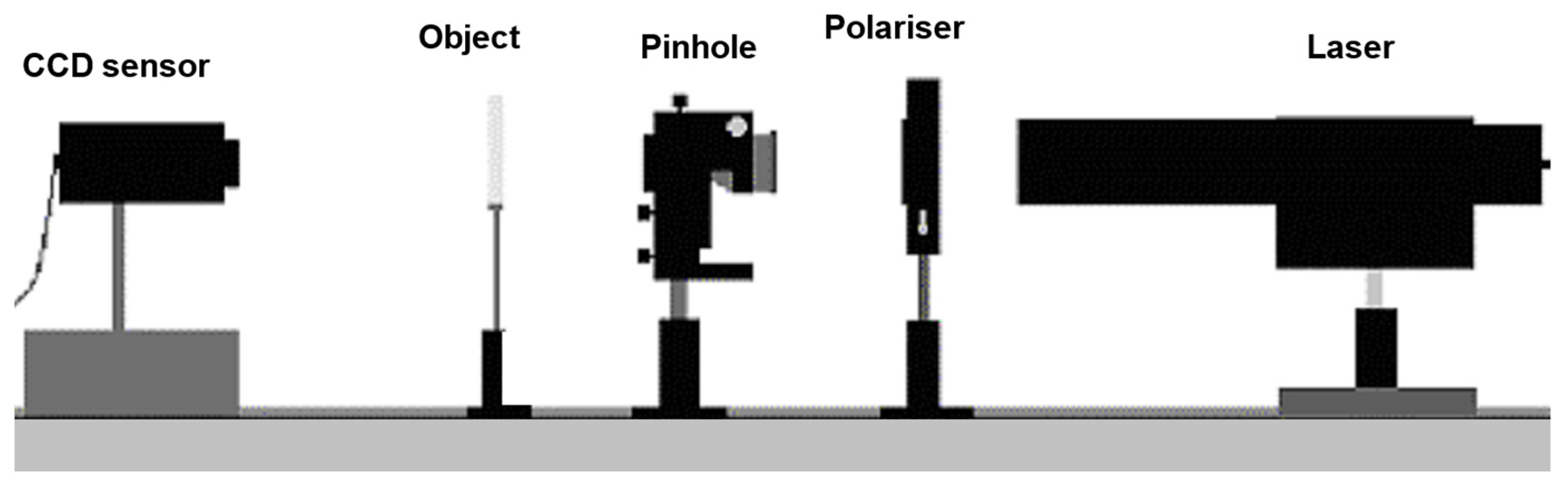
One basic digital holographic microscopy setup consists of an illumination source, a polarizer, a digitizing camera, and a computer with the necessary program for reconstruction of digital holograms. Often, laser is used for illumination, having the necessary coherence to produce interference. This optical setup is called a digital in-line holographic microscope, since all optical components are aligned in a line (
Figure 2). The name ‘in-line’ derives from the fact that a laser beam passing through a semitransparent sample is directly imaged on an imaging sensor (e.g., CCD or CMOS). Thus, the image is made of the overlap of the unscattered laser light (reference beam) and light scattered by the sample (object beam). This overlap produces a holographic interference pattern. Typically, a CCD or CMOS cameras are used to record and digitize a holographic interference pattern. The recorded hologram pattern is digitized and input to the computer as a 2D array of integers with high grayscale resolution.
The second classical interferometric configuration used in a DHM layout is presented in
Figure 3. An incoming laser beam is split into two by a beam splitter (BS). The transmitted beam illuminates the object under investigation. The diffracted wave front is magnified by a microscope lens onto the CCD sensor. The reflected beam at the BS closest to the CCD sensor provides the reference beam for the off-axis holographic recording when interfering with the object beam.
A digital in-line holographic microscope (DIHM) was developed at the Agricultural University of Plovdiv [
8]. The light source is a diode laser (Lasiris) with wavelength of 673.2 nm and output of 7 mW. The laser radiation is focused onto a pinhole, after which the intensity is controlled by a polarizer (
Figure 2). After the pinhole, the spherical wave passes through the object, which is positioned on a microscope slide: the wave diffracted by the object and the non-diffracted wave interfere and are recorded as a hologram on a CCD sensor. The intensity and the phase are reconstructed numerically.
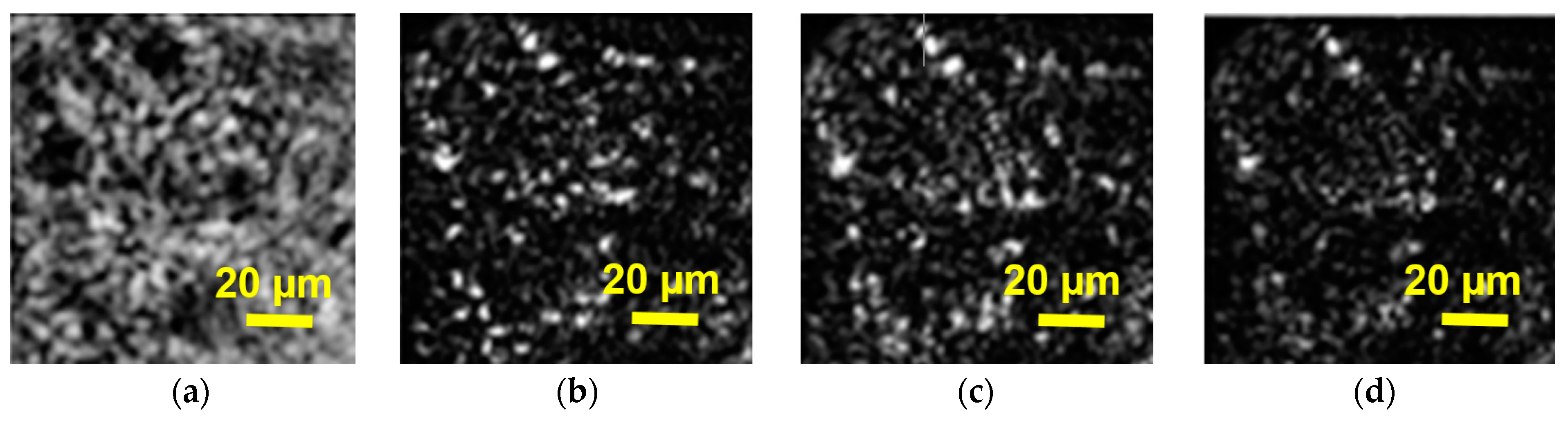
An example of DHM application for imaging of algae is shown in
Figure 4. A hologram (4a) and three reconstructed wave front intensities of it (4b–d) to represent the object are shown. The reconstructions are performed in three consecutive planes with the distance between them changing by 2 μm. A well-known distinctive feature of holography is the 3D content of the image information. In DHM, a single hologram is used to reconstruct the optical field at any distance from the hologram, within the limitation of the approximation method used. From a single hologram, the image is calculated at various distances, which are then compared and the image with the best focus is found (Kim M. K.) [
7]. The reconstructed intensities illustrate the unique ability of DHM to allow observation of different sections in a live cell, obtained from one digital hologram only. This makes it possible to obtain many cuts of one live object from one digital hologram of the object.
A single digital hologram contains the information necessary for the microscopic volume reconstruction of the object. Quantitative information about the intensity and the phase distribution of the numerically reconstructed wave field can be obtained from the digital holograms.
Figure 4 shows that numerical reconstruction of the digital holograms provides an efficient method of virtual focusing throughout the depth of a microscopic sample, which is a unique feature of DHM.
3. Digital Holographic Microscopy for Life Science Applications
Quantitative phase microscopy by digital holography has been applied to observe various cell types, including SKOV-3 ovarian cancer cells [
9,
10], fibroblast cells [
11], amoeba cells [
12], diatom skeleton [
13] and red blood cells [
14]. It has also been used to monitor cell dynamics upon drug delivery into pancreatic tumor cells [
15].
3.1. Imaging of Microbes, Marine Plankton and Algae
Digital holography provides an excellent mechanism for imaging the 3D distribution of point-like microbes [
16,
17]. By observing the difference between successive frames of a time series of holographic photographs, the movement of microbes can be tracked. At the same time, this differential technique effectively suppresses side effects in the image. In another application of the holographic technique, underwater instruments have been constructed to observe marine plankton and other particles [
18,
19]. Digital Gabor holography has also been used to study the flow velocity field generated by crustacean organelles [
20].
Microscopy of cell adhesion is important for a deeper understanding of cell movement and morphogenesis. The main tool in studying these surface processes is total internal reflection fluorescence microscopy and reflection interference microscopy. The evanescent boundary field is used to selectively illuminate the surface layer of the cell. In the holographic variant of this method, the interaction of the damping part of the field with the contact layer leads to modulation of the field phase of the light reflected in total internal reflection [
21,
22]. The phase modulation is then detected and imaged by quantitative phase microscopy. The method is non-invasive, does not require fluorophores and uses exclusively the incoming radiation.
In [
23], digital holographic microscopy was applied for the first time to optical tomographic spectroscopy of pollen grains. Phase images with nanometer axial accuracy were digitally reconstructed from holograms captured at different sample orientations; then, the three-dimensional refractive index was calculated by inverse Radon transformation. An accuracy of 0.01 for the refractive index and a resolution in the micrometer range have been demonstrated.
Mihaylova et al. [
24] have shown that visualization of live cells without any preliminary preparation is possible.
Figure 5 presents three digital holograms of algae cells
Tetraselmis suecica and the wave front intensities of the corresponding images. The morphology of the cells is clearly visible on the images shown in
Figure 4 in terms of reconstructed intensities.
These experiments clearly show that DHM can be used for the following:
label-free morphology imaging of live cells
label-free investigation of cell division and movement
label-free analysis of subcellular motion in living cells, etc.
Applying digital holographic microscopy for visualization of biological samples is extremely appealing because it is only necessary for the investigated objects to be transparent or semi-transparent in order to scatter the laser light and thus to form the object beam. This means that there is no need for any preliminary preparation, labeling or use of fluorescent dyes. It also means that live imaging of moving objects can be done.
Swimming patterns of zoospores of the green alga
Ulva linza near the surface were investigated by Digital Line Holography in [
25]. Complete 3D movement patterns were derived from the measurements and the resulting tracks were compared to known swimming patterns of spores from the brown alga
Hincksia irregularis and the green alga
Ulva linza observed with a conventional optical microscope. Quantitative information about swimming speed can also be obtained from the 3D traces.
The results demonstrate the potential of digital linear holography to image and quantitatively study the behavior patterns of motile spores near the surface. This technique may enable penetration of surface colonization mechanisms by spores and larvae in response to changes in surface properties.
In an article by Garcia-Sucerquia J. et al. [
19], important features of DIHM are listed: 1. Simple hardware of the microscope: DIHM is microscopy without objective lenses. The necessary hardware for it is a laser, pinhole and CCD camera. For the underwater version of DIHM, the same elements are used, contained in a submersible hermetic shell. 2. One hologram contains all the information about the object, including its 3D structure. 3. Optimal resolution on the order of the laser wavelength can be easily achieved. 4. No staining and no separation of biological samples: the living cells and samples can be observed in depth. For the underwater digital holographic microscope, there is no sample preparation required and real-time information on living organisms can be extracted. The resolution achieved is of the order of the wavelength of light. Thus, it is possible to track small organisms such as bacteria and plankton in water.
3.2. Imaging of Living Cells and Cellular Samples
Digital holographic microscopy (DHM) enables quantitative, multifocal, phase-contrast imaging [
26], which is suitable for viewing and quantitative imaging of living cells. The combination of DHM with a fast and stable autofocus algorithm allows subsequent automation of focusing by numerical propagation of the reconstructed holographic object wave. Combined with a calibrated optical imaging system, the acquired data quantify the axial positions of the object under study. Evaluation of quantitative phase contrast images allows efficient determination of lateral cell displacements. Results from studies on sedimentation of red blood cells and HT-1080 fibrosarcoma cells in collagen tissue demonstrate that DHM enables automated quantitative, marker-free, and dynamic 3D cell tracking without mechanical focus adjustments.
First results are presented on a method [
27] that, by means of digital holographic microscopy at multiple wavelengths, enables non-mechanical scanning tomography with sub-micrometer axial resolution. By sequentially acquiring and summing 20 wave fronts of reflection holograms evenly spaced in the wave range 485–670 nm, slice-by-slice tomographic reconstruction with 0.6–1 μm axial resolution is achieved in a biological environment. The method is applied to the study of erythrocytes to extract a cell profile of their membrane in three dimensions.
Optofluidic devices offer flexibility for a variety of tasks that involve biological samples. A system [
28] for three-dimensional (3D) observation and identification of biological microorganisms has been proposed. This system consists of a microfluidic device together with a digital holographic microscope and the corresponding statistical recognition algorithms. A microfluidic channel is used to house the microorganisms, and a holographic microscope and CCD camera record digital holograms. Holograms can be digitally reconstructed in 3D using various algorithms, such as the Fresnel transform. Statistical analysis algorithms are used to recognize and identify the microorganisms from the reconstructed wavefront.
Many biological objects are mostly transparent and weakly scattering, so the way to observe their images is to look at variations in the refractive index. A method was proposed [
29] that allows obtaining 3D images of the refractive index distribution by a tomographic approach. The great possibilities of obtaining 3D images using diffraction tomography in biology are shown.
In [
30], multi-wavelength digital holography in combination with epi-fluorescence microscopy was applied to generate quantitative phase and fluorescence information from cellular samples. While digital holography provides high-precision morphological information, the addition of fluorescence information provides the specificity needed to identify cellular constituents. By applying digital holography at several wavelengths, it is possible to obtain complete wave front data in real time. The potential of this dual mode for imaging in live cell studies is demonstrated.
To study the morphology and dynamics of live red blood cells from individuals who suffer from sickle cell anemia (SCA) (a genetic disease that affects the structure and mechanical properties of red blood cells), wide-field digital interferometry (WFDI) is applied to observation [
31]. WFDI is a non-contact, label-free optical microscopy that can obtain quantitative profiles of red blood cell thickness and measurements of nanometer-scale membrane fluctuations, which correlate with their stiffness. Red blood cells from individuals with SCA were found to be significantly stiffer than healthy ones. Furthermore, the technique has been shown to be sensitive enough to distinguish classes of RBCs in SCA, including sickle-shaped RBCs with apparently normal morphology, compared with more rigid, crescent-shaped RBCs. It is expected that this approach will be useful for the diagnosis of SCA and for determining the efficacy of therapeutic agents.
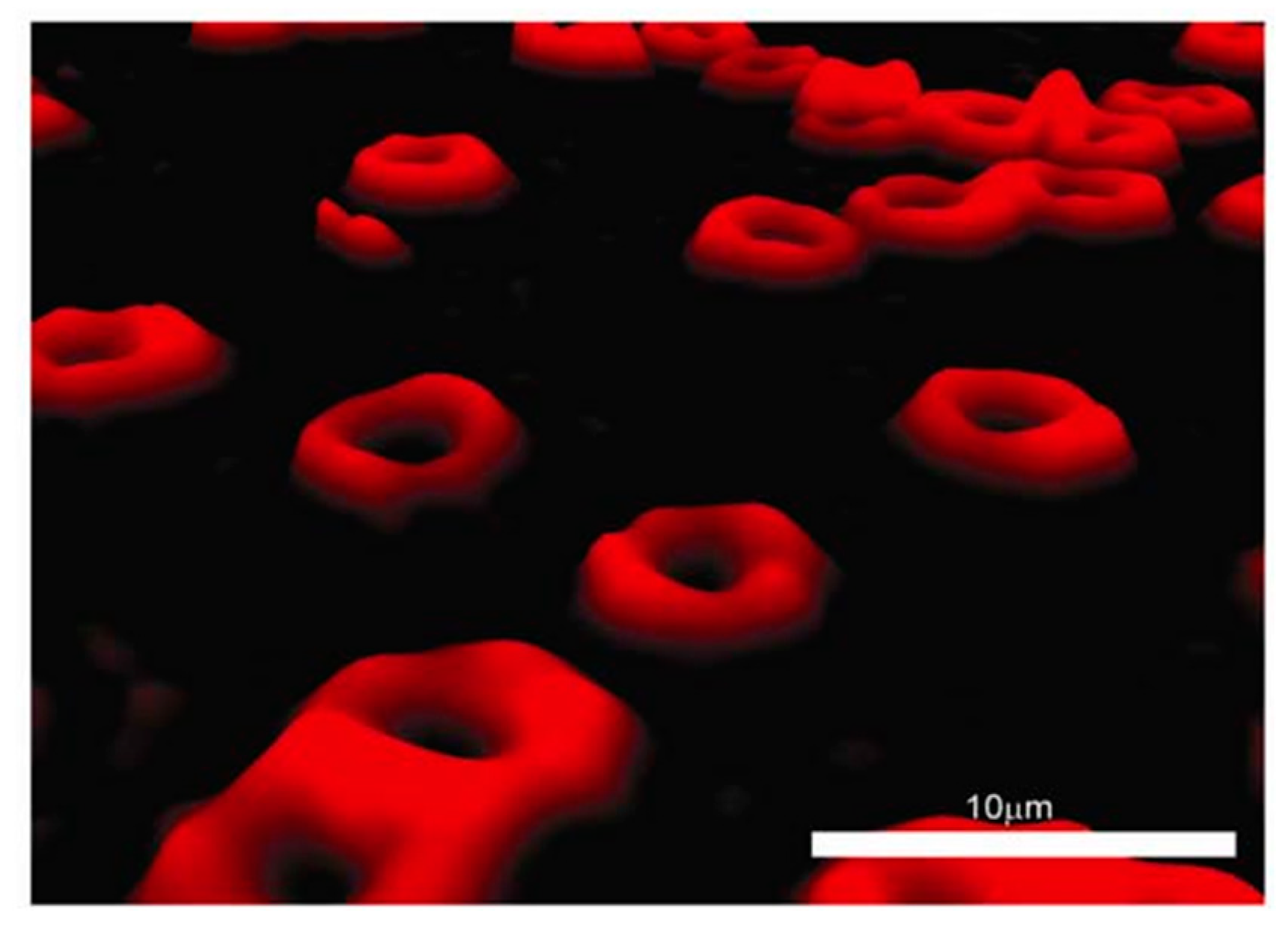
Erythrocytes are an attractive cell type in the human body for different studies. They move throughout the blood system to deliver oxygen to all organs in the body. The shape and the volume of erythrocytes can be used for clinical diagnosis purposes. For these reasons, a number of studies report the advantages of digital holographic microscopy for three-dimensional visualization of erythrocytes [
32,
33].
Figure 6 shows visualization of human erythrocytes using a digital holographic microscope.
Diatoms are among the dominant phytoplankton in the oceans, and their external silicon derivatives, resembling artificial photonic crystals, are expected to play an active role in light processing. Digital holography enables the study of the interaction with light of the
Coscinodiscus wailesii cell wall, reconstructing the trapping of light inside the cytoplasm of cells that is difficult to access by standard microscopy [
34]. The complete characterization of the propagating beam, in terms of phase and intensity, removes the long-standing ambiguity about the origin of light. The data are discussed in light of the behavior of living cells in response to their environment.
Choi Y. S. and Lee S. J. [
35] applied DHM for 3D volumetric measurement of moving red blood cells. The 3D motion of human red blood cells in flow was visualized. It was demonstrated that only one pair of particle hologram images is necessary to obtain full 3D flow information, and this is a major advantage in analyzing the movement of individual red blood cells. Five focus functions were evaluated. Sample trajectories as well as 3D velocity profiles of RBCs in micro-tube flow are presented and measurement uncertainties are discussed.
Khmaladze A. et al. [
10] developed a dual-wavelength imaging technique for the quantitative investigation of the three-dimensional cell structure. The technique has nanometer axial resolution. 3D images of cells with subsequent measurement of cell thickness are presented. As a result, the accuracy and level of detail of the cell imaging presented here is superior to what had been developed previously.
Langehanenberg P. et al. [
36] proposed autofocusing in digital holographic phase-contrast microscopy with application to live-cell imaging. The best autofocus method for application in digital holographic microscopy was found by comparing four numerical methods.
Remmersmann C. et al. [
37] reported the optimization of a setup for temporal phase shift (TPS)-based digital holographic microscopy. Theoretical and experimental studies on possible phase errors in the digital reconstruction of holograms have been carried out. Coherent and partially coherent light sources are compared. Using LED and laser-based digital holographic microscopy, it was demonstrated that perturbations in the reconstructed amplitude and phase distributions can be reduced by using partially coherent light sources.
3.3. Imaging of Biological Tissues
Bokemeyer A. et al. [
38] reported a method for visualization of fibrotic changes of the submucosal layer of stenotic (C,D) compared to non-stenotic bowel tissue (A,B) of Crohn’s disease patients, using digital holographic microscopy (DHM). The authors show strong correlation of the amount of fibrosis as determined by immunofluorescence (IF), serving as a gold standard, with the refractive index (RI) values assessed by DHM. Histological evaluation of HematoxylinEosin-(HE)-staining image and the corresponding quantitative DHM phase contrast image visualize significant fibrotic changes of the submucosal layer of stenotic compared to non-stenotic bowel tissues (
Figure 7).
Inflammatory bowel diseases, including Crohn’s disease and ulcerative colitis, represent a great burden for patients due to the chronic course of the disease. Therefore, there is an urgent need to explore new potential drugs. Holographic microscopy (DHM) allows for quantitative phase contrast imaging without staining and provides an estimate of tissue density by measuring the optical path length delay and, accordingly, the refractive index. Colitis induced by dextran sodium sulphate was induced in wild-type mice and sections were examined by histological analyses and DHM [
39]. The authors proved that the average refractive index can be a precise marker to distinguish the different layers of the intestinal wall, in a way that the stroma is characterized by the highest value and the submucosa by the lowest. It is also shown that DHM permits detection of inflamed segments with a very good correlation between the severity of inflammation and the refractive index. In is shown that DHM offers a new diagnostic method for quantification of inflammation in models of colitis.
A digital holographic microscope for blood cell counting is presented by Riesenberg R. et al. [
40]. The microscope consists of an illumination chip, a micro-fluidic chip, and a CMOS detection chip. The design has been tested for blood cell diagnostics. A single hologram delivers a number of images for all Z-positions in the micro-fluidic channel, which is the basis of blood cell counting. Images of moving red blood cells were recorded, and subsequently used for clinical diagnosis.
Clinical laboratories are usually equipped with modern flow cytometers for rapid and accurate cell sorting. These cytometers use different biomarkers, which very often induce changes in cell morphology, and occasionally lead to cell death. Holographic microscopy is label-free and thus suitable for imaging of live moving cells. In [
41], an integrated, optical neural network is reported that has potential for ultrafast cell counting and sorting. The authors report up to 89% accuracy for classification of monocyte, granulocyte, and lymphocyte yields.
Digital linear holographic microscopy was used to record holograms of mammalian cells (HEC 293, B16, and E0771) in culture [
42]. The holograms were reconstructed using “4-Deep Octopus software”, using an FFT-based algorithm to obtain the phases. These were then used to determine the maximum phase shifts in individual cells. Addition of 0.5 mM H
2O
2 to the cell medium resulted in rapid rounding of cultured cells followed by cell membrane rupture.
Changes in cell morphology and cell membrane ruptures were observed in real time and were evident in the phase images. The results show that quantitative phase contrast images obtained by the digital holographic microscope can be used for the automated determination of cell viability and confluence in mammalian cell cultures in real time and without markers.
Volumetric observation is essential for understanding the details of complex biological phenomena. In [
43], a bright-field microscope that yields information about a specific 2D plane and a holographic microscope that yields information distributed in a 3D volume are integrated to obtain two complementary images simultaneously. The developed system has been successfully applied to capture the dynamics of T cell adhesion on inflamed endothelial layers, including their capture, rolling, crawling, and migration.
Kim M. K. [
44] reported a new digital holographic method that achieves axial imaging resolution by superimposing several numerically reconstructed hologram diffraction fields. The principle of digital holography is applied to imaging for objects with diffuse surfaces, one such example being a biological specimen. The head of an insect measuring several millimeters is imaged with an axial resolution of 120 µm and a lateral resolution of ~20 µm. A 3D digital model of the surface structure of the object is obtained with good accuracy.
Chalut K. et al. [
45] have developed a quantitative phase microscopy method with asynchronous digital holography. The system can perform quantitative phase measurements over periods of milliseconds. The use of acousto-optic modulators in each arm of the interferometer allows phase-shift interferometry to be obtained. This new system has been shown to be able to obtain quantitative phase images of live cells, in particular with a sample of red blood cells and a sample of smooth muscle cells.
Alm K. et al. [
33] applied DHM to study the differentiation of adherent 3T3L1 fibroblasts. DHM is a very appropriate technique to visualize the differentiation of cells because the cells under investigation remain undisturbed. The authors reported that the differentiation process changed the cell shapes (
Figure 8).
3.4. Imaging of Live Cells in Culture
A digital holographic microscope (DHM) for the quantitative study of cell dynamics was developed by Rappaz B. et al. [
46]. Live cell cultures are studied by recording the phase shift they cause on the transmitted wave front. The developed DHM makes it possible to observe the processes of cell dynamics. The application of the new method to study the dynamics of neurons in culture during hypotonic stress is reported. Such stress leads to a smaller phase, because it depends on the thickness and the refractive index of the specimen. To separate these two contributions, a procedure called the “separation procedure” is applied. This procedure allows information on both cellular morphometry and integral refractive index to be extracted from dynamic quantitative phase images. Quantitative, local measurement of cell thickness can be performed with an accuracy of 1 µm. Spatial averaging allows measurement of the average thickness of cell regions corresponding to the area of typical neuron bodies, i.e., 170 µm
2, with an accuracy of several tens of nanometers. On the other hand, the spatial variation of the integral refractive index is estimated to be 0.005 and the average integral refractive index can be measured with an accuracy of 0.0003. The cellular refractive index is a parameter, which is not well documented due to the intracellular content and multiple scattering processes in biological tissues.
Marquet P. et al. [
47] presented DHM studies of live cells in culture. They measured the length of the optical path in the cell to the nearest wavelength. The authors compared the obtained DHM images with analogues obtained using the widely used techniques of phase contrast and Nomarski differential interference. The authors report for the first time DHM imaging of the absolute phase distribution of live neurons in culture. Their study compares DHM with the well-known techniques of phase contrast (PhC) and differential interference contrast (DIC) microscopy, widely used in biology.
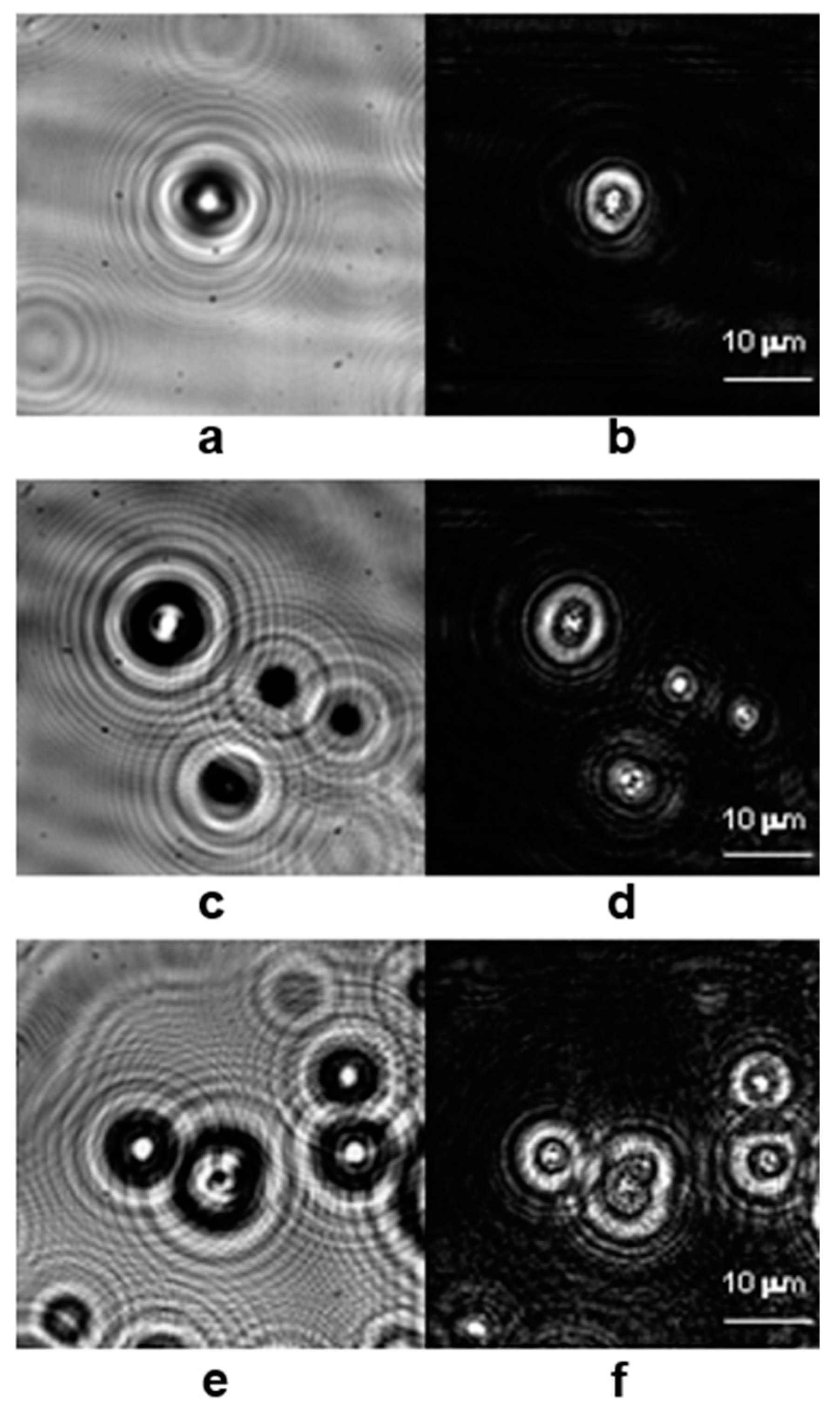
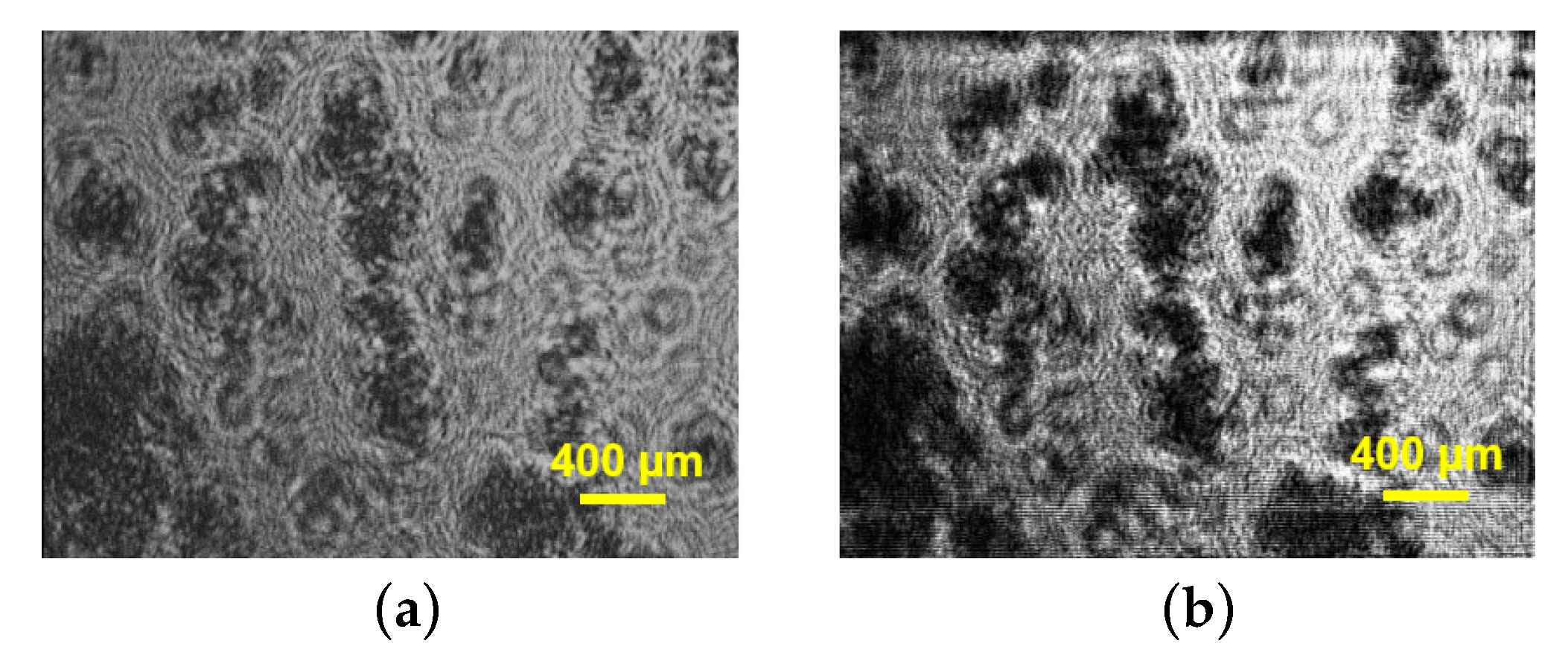
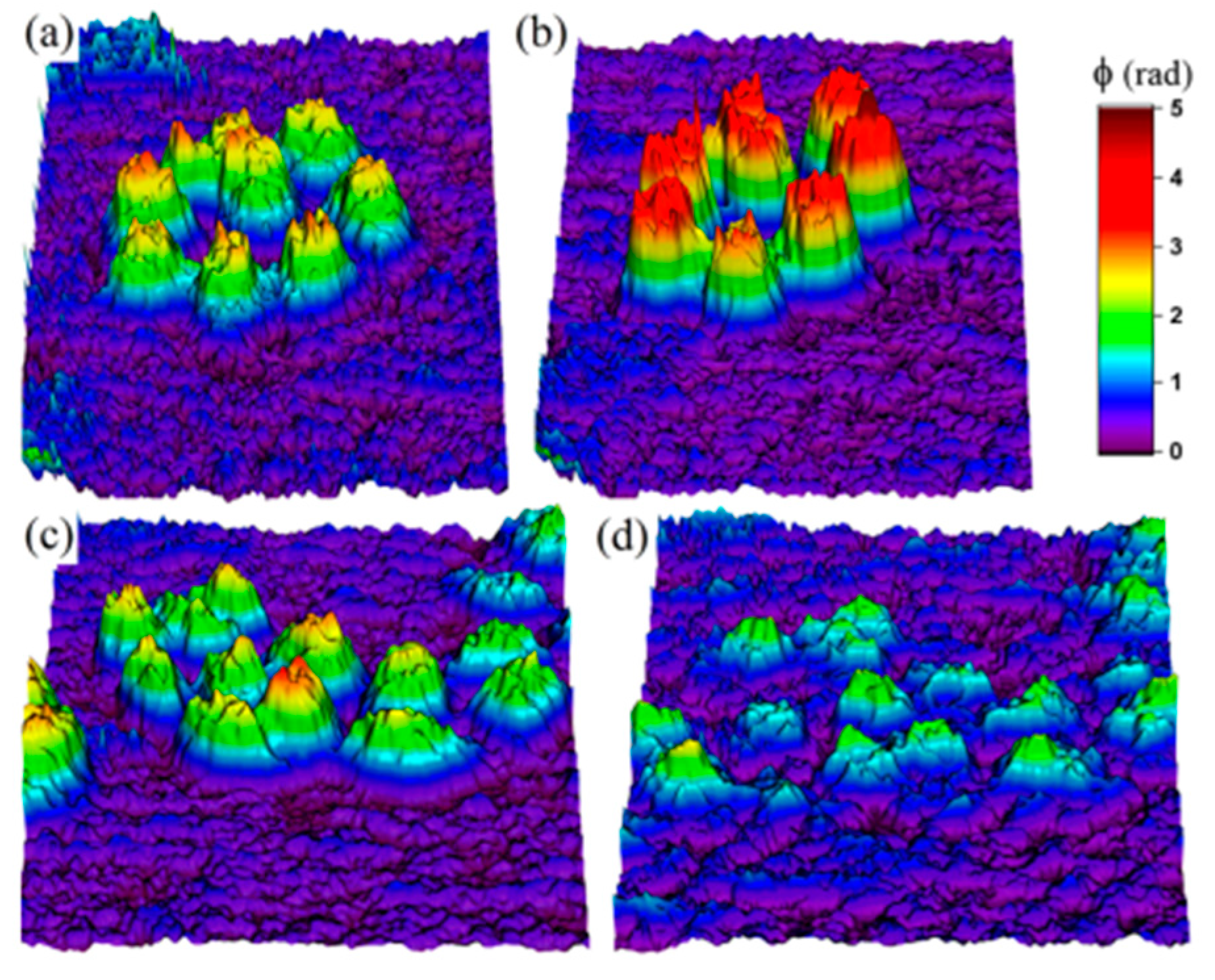
Halkoglu P. et al. applied DHM for the first time for metrological measurements of cell clusters in cell suspension cultures in vitro [
48]. Three different cell suspensions of
Fabiana imbricata Ruiz & Pav., designated A, D and MSD, were investigated [
48]. The reconstructed intensities presented in
Figure 4,
Figure 5 and
Figure 6 visualize the cell clusters in the investigated suspensions. The cell suspension cultures of
Fabiana imbricata Ruiz & Pav. consist of cell aggregates that grow in a continuously shaken liquid medium. Small cell clusters with sizes between 120 and 180 µm were observed in the three suspensions examined. DHM is a very versatile technique for various biological applications. Digital holographic microscopy has been shown to be easy to apply to study the structure of cell aggregates in cell suspension cultures in vitro.
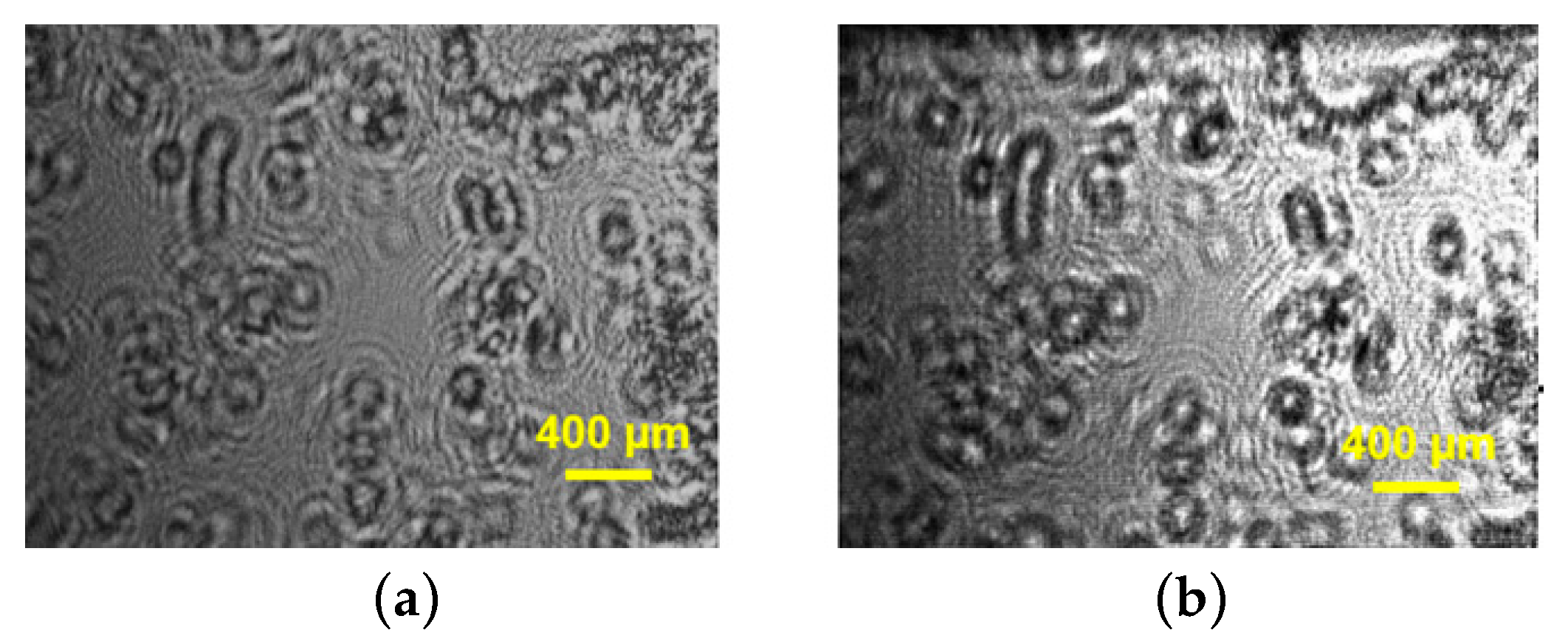
The reconstructed intensities in
Figure 9,
Figure 10 and
Figure 11 illustrate the possibility of direct observation of live cell suspension cultures. These experiments illustrate the capability of DHM for non-invasively visualizing and quantifying biological cells and cell clusters. That is why DHM can be successfully used for cell counting, measuring the size of cells and cell clusters, label-free viability analysis of adherent cell cultures, etc.
It is shown that DIHM can be used for biological studies of cell suspension cultures. Small cell aggregates having dimensions between 120 and 180 μm have been observed in all three suspensions. The cell aggregates in suspension D have the smallest dimensions (120–150 μm) while the biggest cell aggregates are characteristic for suspension A (140–180 μm). The results are promising for future use of DIHM for accurate, fast, and reliable determination of cell growth, which is of critical importance in plant cell and tissue culture. DIHM is a very attractive novel technique for application in biological research and in the agricultural sciences. Other life science and medical applications are also envisaged.
Scholz G. et al. [
49] also applied digital in-line holographic microscopy (DIHM) for basic cell culture investigation. The authors demonstrated counting of cells and movement tracking of living cells.
Recently, Hellesvik M. et al. [
50] exploited the potential of commercial digital holographic microscopy for studying 3D matrix cell culture assays. The authors benefited from the autofocus functionality of digital phase holographic imaging to obtain 3D information for cells migrating in a 3D environment. It was demonstrated that it is possible to quantitatively measure some properties of a cell culture assay, in particular growth of cell clumps over time, and single cell invasion out of cell clumps.
3.5. Visualization of the Effect of Drugs, Different Types of Stress and Nanoparticles on Cells and Tissues
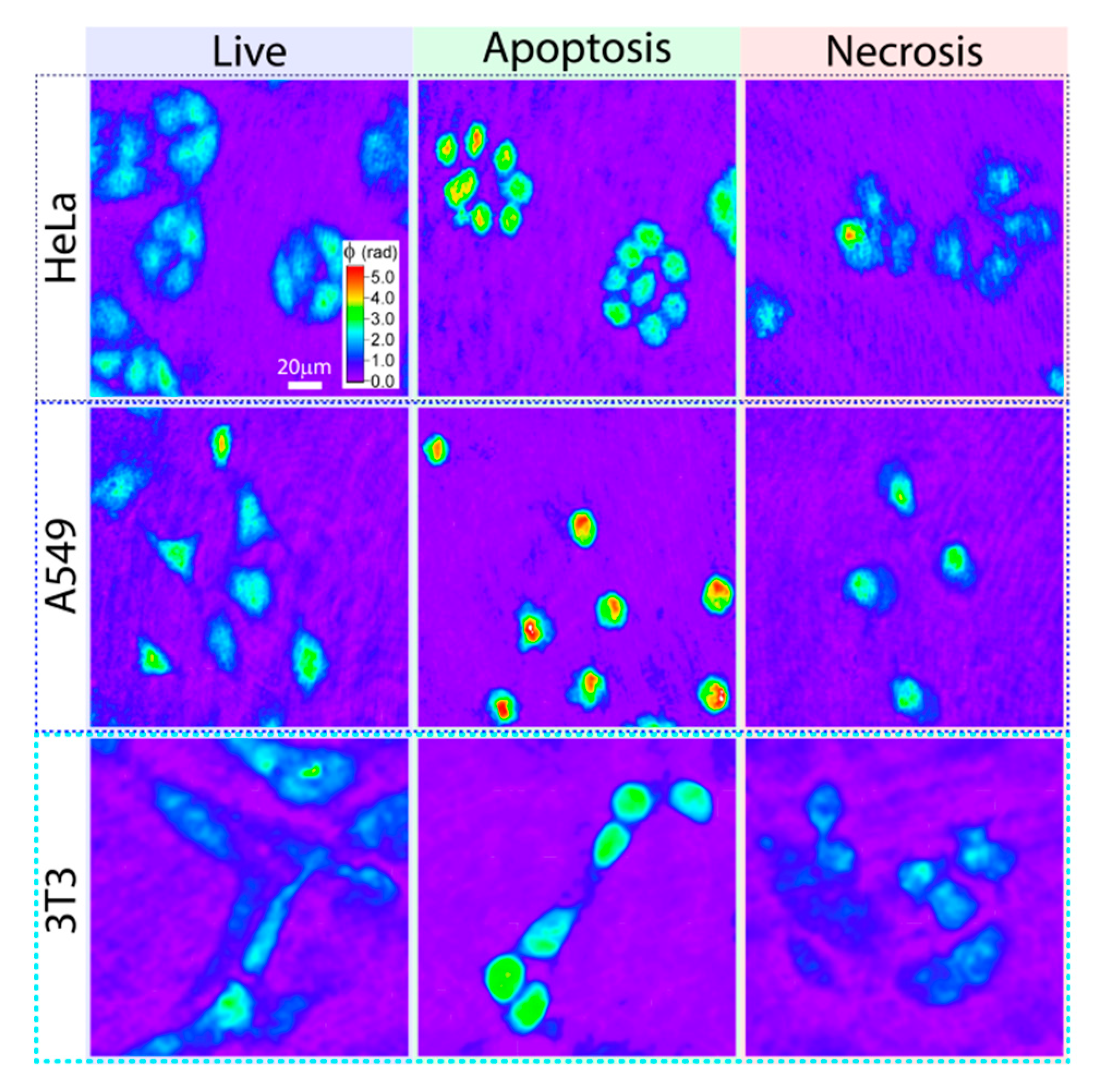
Belashov A. et al. [
51] demonstrated the ability of off-axis digital holography to distinguish between different states of cells: live, necrosis and apoptosis (
Figure 12). The accuracy of the method is about 78% among cells of different lines and in different states (live, apoptotic, necrotic). The described classifier operates with a database containing information on 10 optical parameters of individual cells, extracted from their phase images obtained using digital holography.
The optical setup presented by Parshall D. et al. [
52] offers high-resolution holographic imaging of microscopic objects. The amplitude image presents the changes in the intensity of the reference wave, while the phase noise comes predominantly from the optical surfaces in the imaging system. The authors report [
52] a great number of experiments that demonstrate the potential of digital holography for high-resolution imaging of microscopic biological samples. The main advantage of phase imaging digital holography is the measurement and observation of optical path changes in biological objects. The authors report biological applications of dual-wavelength phase imaging DHM. In addition, the authors also propose high-resolution three-wavelength phase imaging.
Jeong K. et al. [
53] showed that coherent digital holography records changes in biological samples at a depth of 10 optical thickness lengths inside the tissue. This opens perspectives for imaging changes at depths inaccessible to conventional survey approaches. The authors demonstrate that direct visualization of the effect of anticancer drugs on tissue is possible. In this way, it is possible to image and quantify the tissue and cellular reaction to drugs.
Kemper B. et al. [
54] demonstrated the capabilities of digital holographic microscopy to visualize drug-induced morphological changes. The authors report experiments with tumor cells exposed to an anticancer drug. Digital holograms of several cells were recorded continuously every 120 s for 16 h in an environment with stable temperature. DHM clearly showed that the drug first induced morphological changes (cell rounding), then the cell thickness increased. Then, for all samples, the final collapse of the cell is precisely detected by a significant decrease in phase contrast. The results show that digital holographic phase-contrast microscopy can be applied to quantitative long-term monitoring of living cells. The studies show new methods of label-free dynamic monitoring of changes in cell morphology to access new parameters, for example, to quantitatively monitor the time-dependent responses of cells to drugs.
Dubey V. et al. [
55] exploited the capability of DHM, using a low spatial coherence light source, to examine the effect of hydrogen peroxide (H
2O
2)-induced oxidative stress on sperm cells. Four different concentrations of H
2O
2 were tested (10 µM, 40 µM, 70 µM, 100 µM) in this oxidative stress study on sperm cells. The authors report gradual progressive motility loss (
Figure 13) for samples exposed to increasing H
2O
2 concentration, in comparison with the control sample.
More recently, Vom Werth K. et al. [
56] investigated the potential of DHM to monitor changes in cell morphology in response to bacterial stress. The reported research was focused on adaptive immune cells. Primary T cells are subjected to derivatives of certain gram-negative and gram-positive bacteria that are often found in patients with sepsis. The authors also examined the influence of different strains of the same species to identify strain-dependent differences. DHM was used to record changes in cell morphology over time. The results of these studies unequivocally show that the T cell response to bacterial stress is pathogen-dependent, and specific morphological changes can be detected using DHM.
Belashov A. et al. [
57] used digital holographic microscopy to investigate the changes in the morphological characteristics of cells of two cultured lines of cancer origin, HeLa and A549, induced by photodynamic treatment (PDT) with Radachlorin photosensitizer. The results show that the phase shift increased at low doses, which can be associated with apoptosis, while at high doses it decreased, which can be associated with necrosis.
Figure 14 presents 3D images of the phase plots of HeLa cells obtained before and after PDT. The authors reported that the two cell types responded differently to similar irradiation doses. The results obtained suggest that average phase shift of the transmitted wave front can be used for quantitative non-invasive cell death analysis. This conclusion was confirmed by comparison with fluorescent analysis with standard test assays made on the basis of confocal microscopy.
Warnasooriya N. et al. [
58] applied DHM to visualize gold nanoparticles in the environment of live cells. The paper describes a heterodyne DHM, which was applied to investigate 40 nm gold particles. A near-infrared laser diode was used as a light source. The system is based on the Mach-Zehnder interferometer. The accuracy of the measurements performed is ±5 nm. It was shown that a single digital hologram is sufficient to determine the position of a nanoparticle in a sample with 90-micrometer thickness. Thus, it was demonstrated that DHM has a significant advance towards the development of 3D microscopy in a living cell environment.
Label-free DHM is a very attractive technique for in vitro studies. Eder, K. et al. [
59] employed digital holographic microscopy to study the effects caused by three different types of organic nanoparticles in development for medical use. The results show that quantitative phase imaging with DHM is highly suitable to identify harmful or low-toxic nanomaterials.
4. Conclusions
In this paper, we review the research carried out in the area of digital holographic microscopy (DHM) for imaging of live cells. The novel microscopic technique is non-destructive and label-free and offers unmatched imaging capabilities for biological and bio-medical applications. It is shown that DHM is suitable for imaging and modelling of key metabolic processes in living cells, microbial communities or multicellular plant tissues. Live-cell imaging by DHM allows investigation of the dynamic processes underlying the function and morphology of cells.
DHM imaging is an attractive and advanced imaging method because digital holography yields a 3D image from a single recorded interferogram. This makes it possible for a dynamic microscope capable of fast 3D imaging to be developed. The attractive characteristics of DHM are a very high acquisition rate, non-contact, non-destructive, marker-free in vivo and in vitro imaging.
DHM can be applied to study the morphology of living, dividing and dying cells. DHM is a non-invasive, non-destructive and non-phototoxic method which allows the user to perform both qualitative and quantitative measurements of living cells over time. DHM will extend the imaging ability to label-free living cells, and dynamically changing phenomena in cells under the influence of drugs, different types of stress and nanoparticles. It is a very attractive technique for application in biological research, bio-medical studies and in the agricultural sciences.
Finally, a few suggestions for future development of DHM will be mentioned. Machine learning methods have emerged as a promising approach to holographic reconstruction. These methods could be used to achieve better quality of reconstructed images, if large amounts of data are available for a certain application. DHM at present is revolutionizing fields such as pathology, ophthalmology, and dermatology, enabling doctors to make more accurate diagnoses. An expansion of digital holographic microscopy applications in healthcare is expected in the coming years.