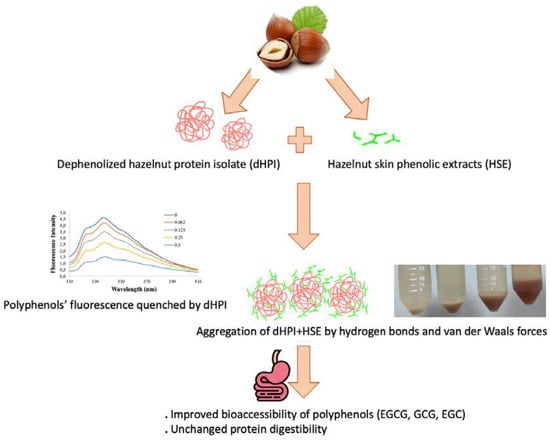
Interactions between Hazelnut (Corylus avellana L.) Protein and Phenolics and In Vitro Gastrointestinal Digestibility
Abstract
1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Preparation of Hazelnut Skin Extracts (HSE)
2.3. Preparation of Protein Isolate from Hazelnut Meal
2.4. Amino Acid Profile of the Hazelnut Protein Isolate
2.5. Preparation of Protein–Phenolic Complex Solutions
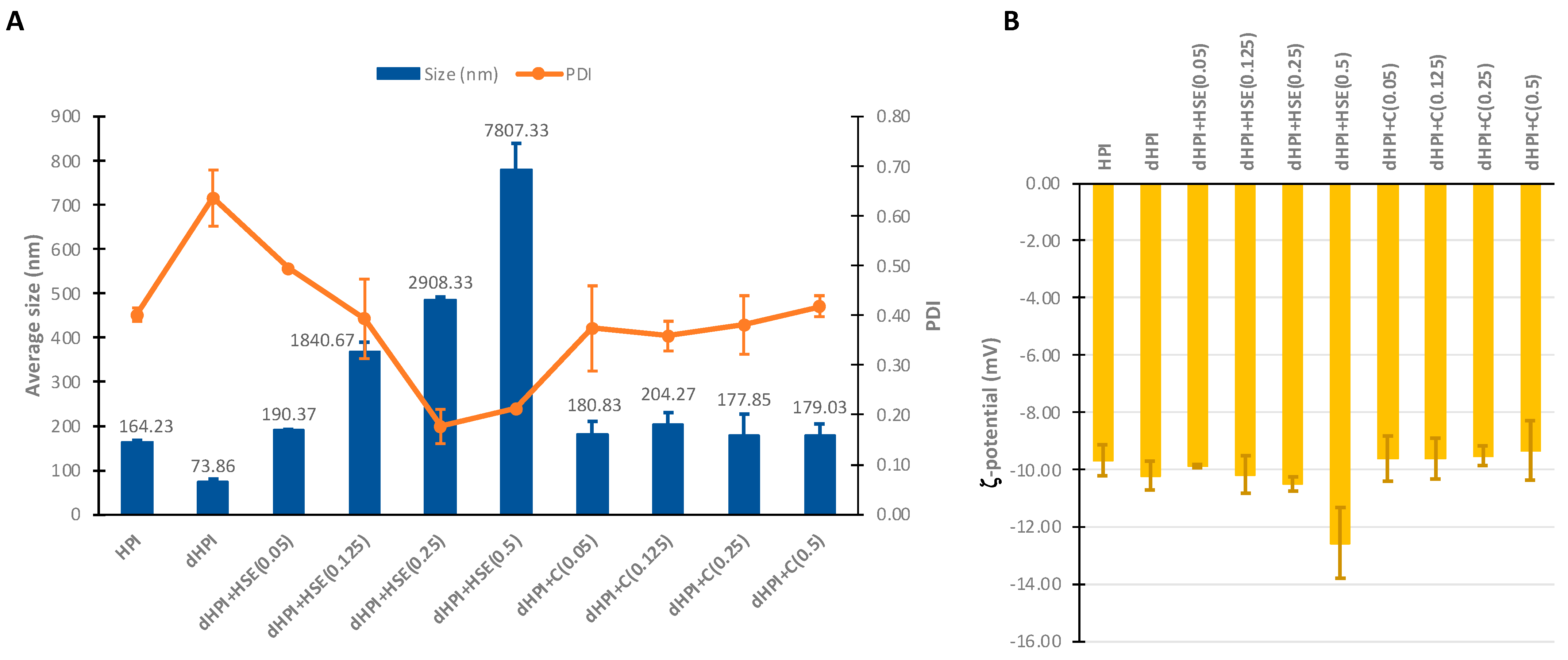
2.5.1. Particle Size, Size Distribution, and ζ-Potential Determination
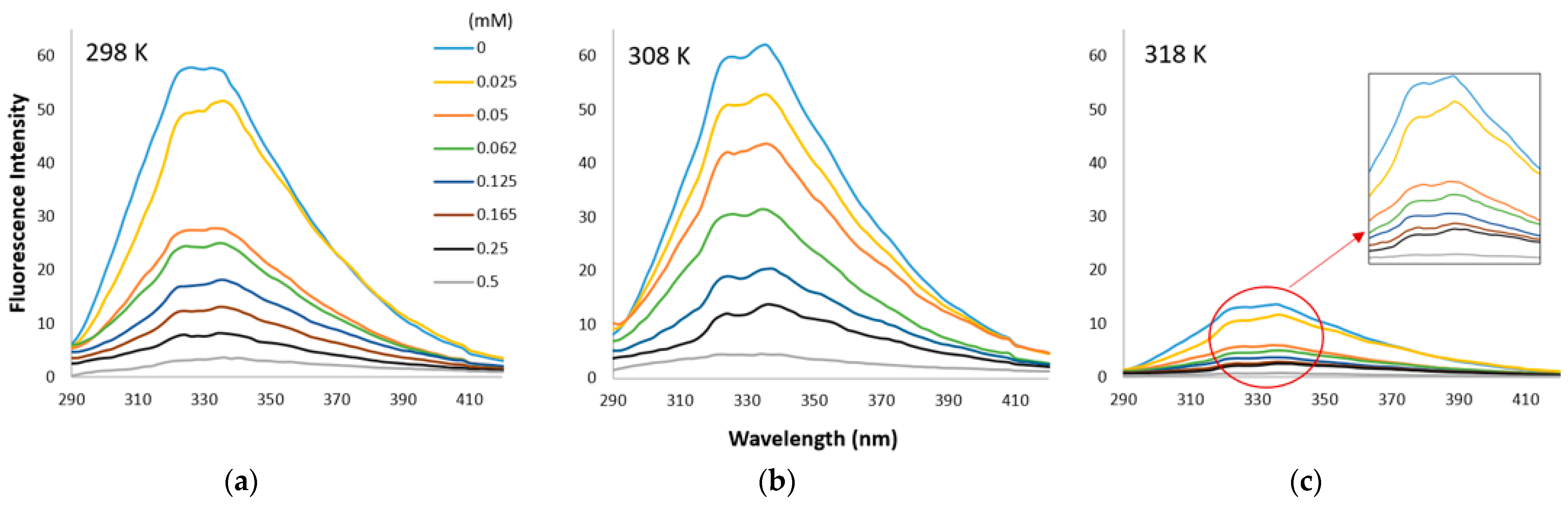
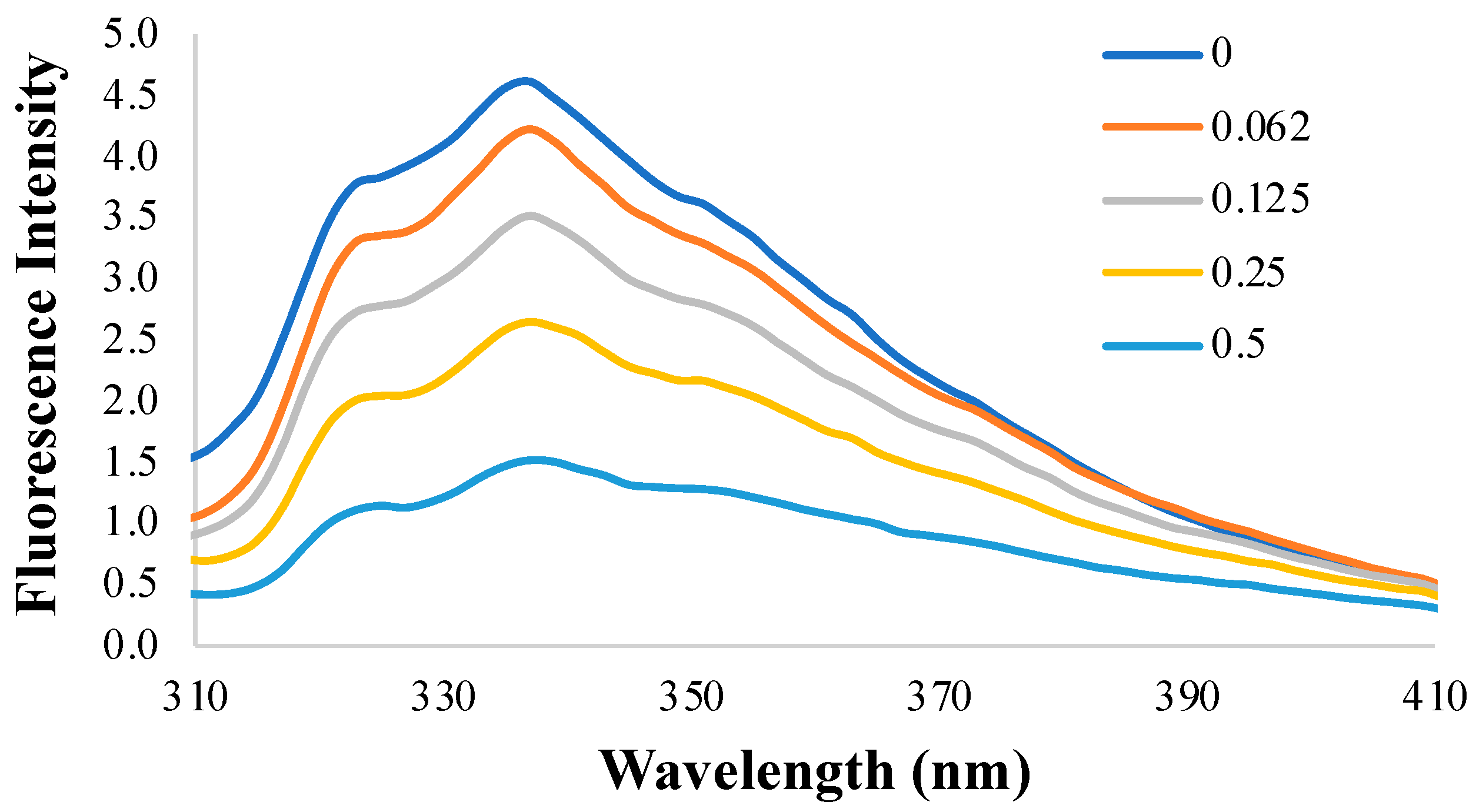
2.5.2. Fluorescence Quenching
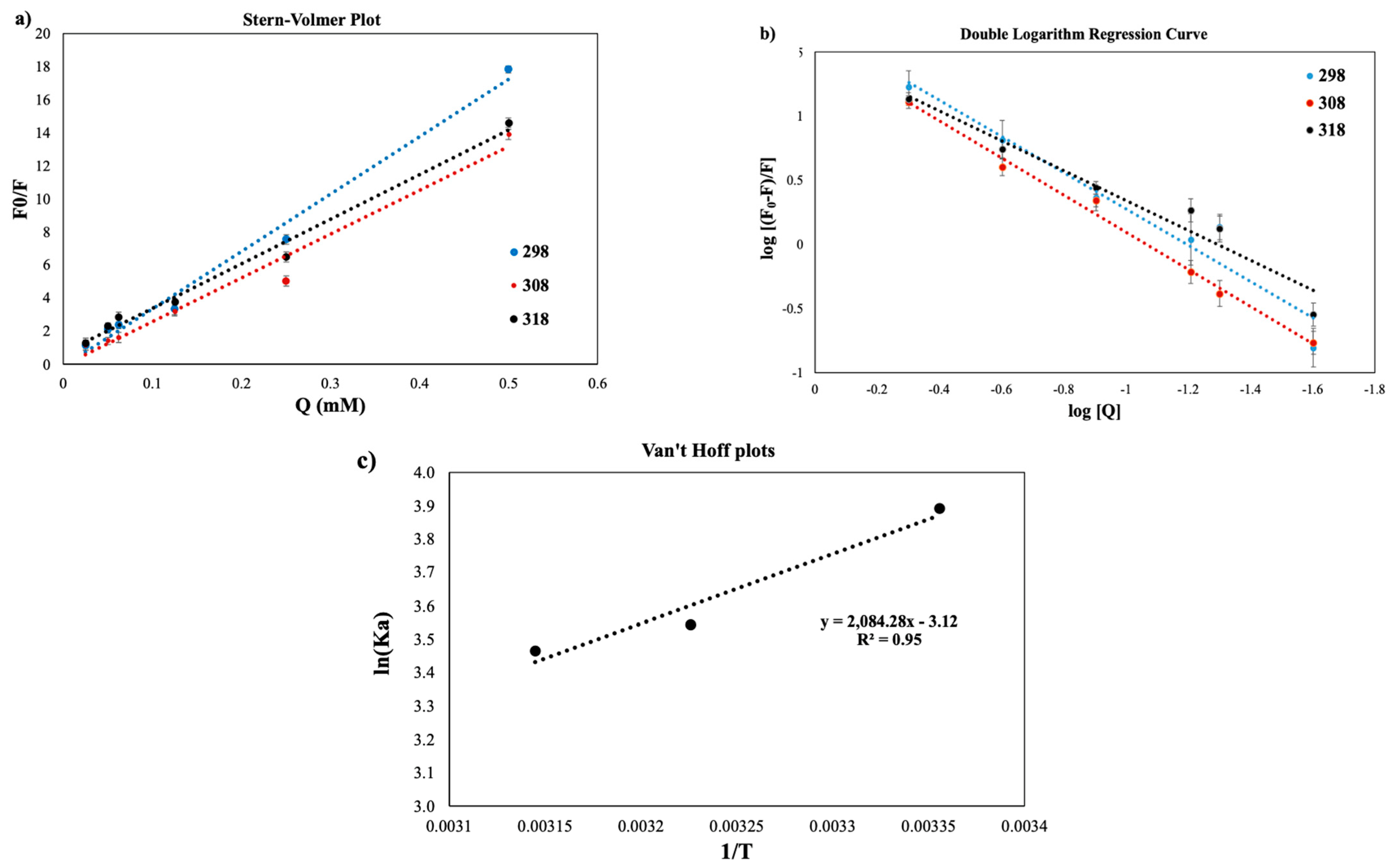
Stern–Volmer Equation
Thermodynamic Parameters
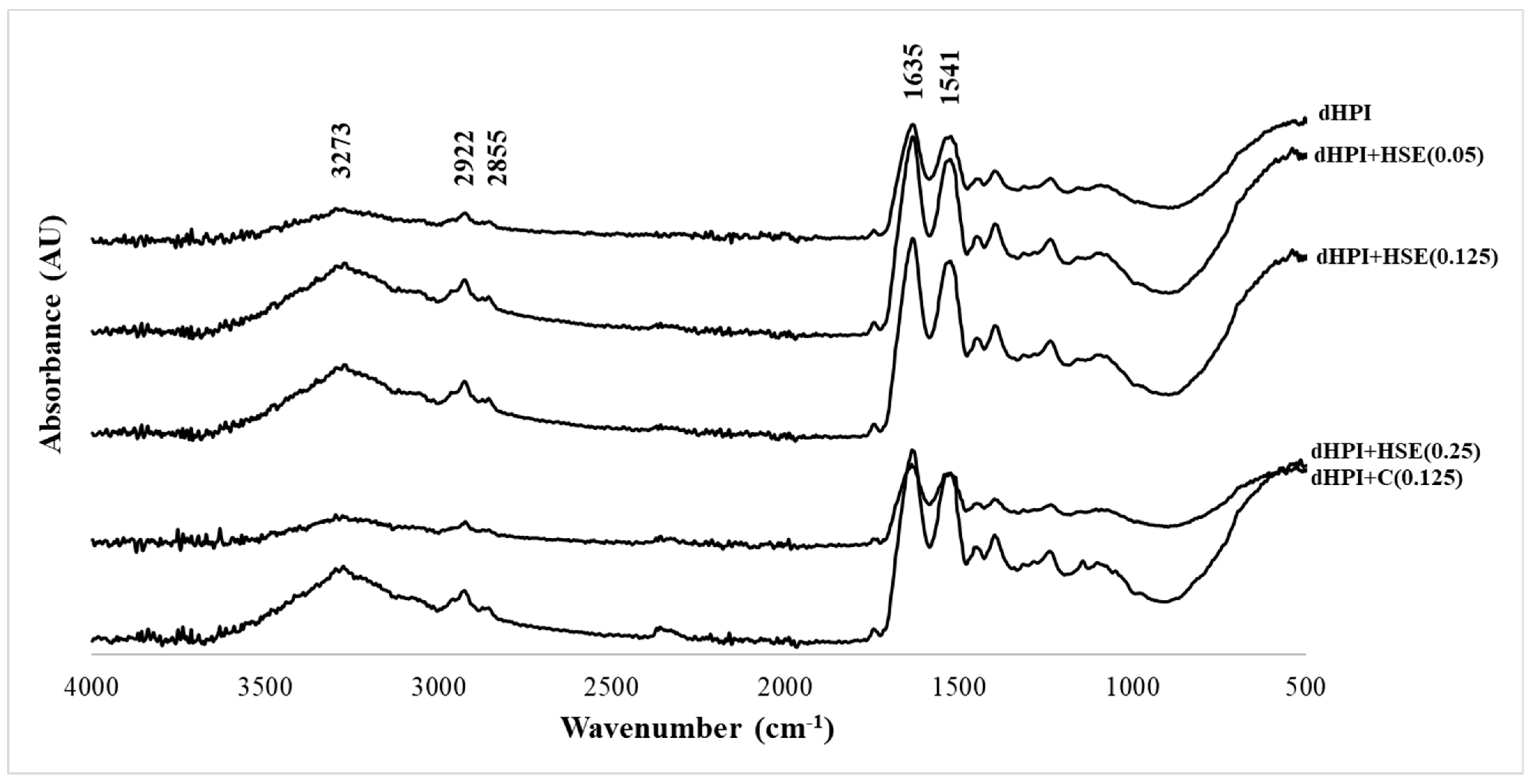
2.5.3. Fourier Transform Infrared (FTIR) Spectroscopy
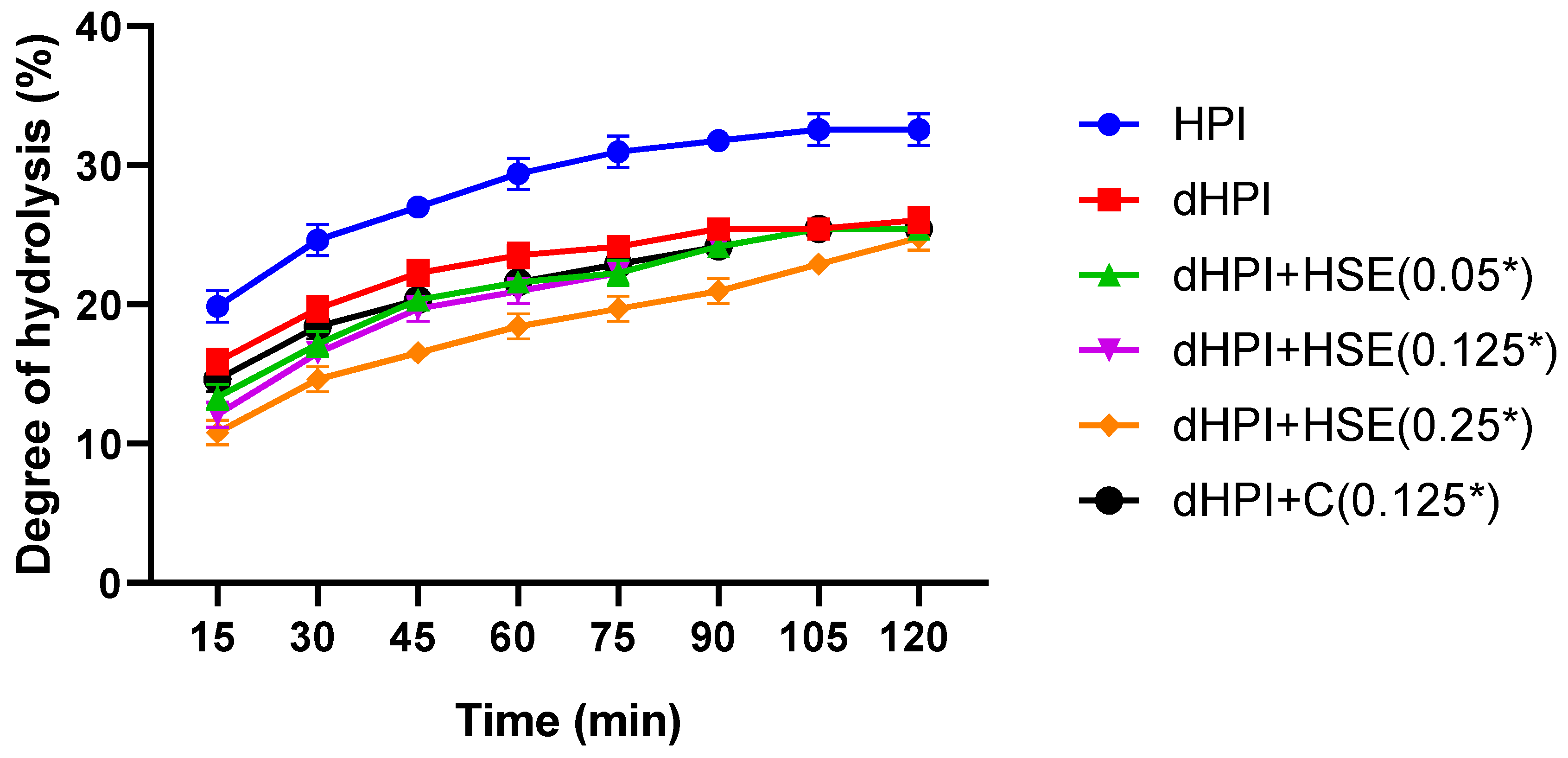
2.6. Assessment of Hazelnut Protein Digestibility by Pancreatin
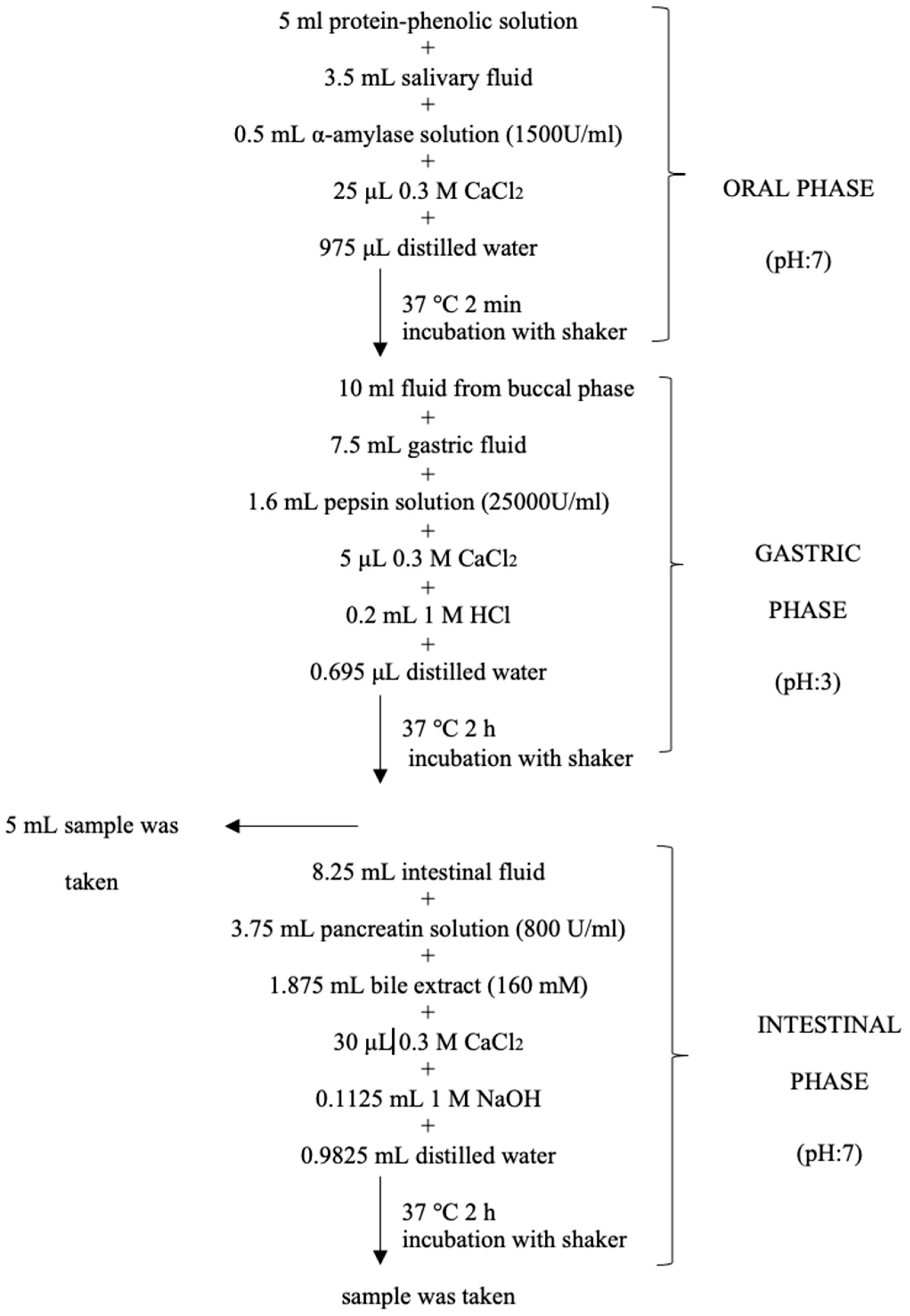
2.7. Simulated In Vitro Gastrointestinal Digestion for HSE Bioaccessibility
2.7.1. Spectrophotometric Analyses
2.7.2. HPLC-DAD Analysis of Phenolic Compounds
2.8. Statistical Analysis
3. Results and Discussion
3.1. Effect of Dephenolization on Protein Purity, Protein Recovery, and Amino Acid Profile
3.2. Average Particle Size, Size Distribution, and ζ-Charge
3.3. Fluorescence Quenching
3.3.1. Stern–Volmer Plots
3.3.2. Thermodynamic Parameters
3.4. Fourier Transform Infrared (FTIR) Spectroscopy
3.5. Effect of Phenolics on the Digestibility of Hazelnut Proteins
3.6. Simulated In Vitro Gastrointestinal Digestion
3.6.1. Spectrophotometric Analyses
3.6.2. HPLC-DAD Analysis of Phenolic Compounds
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- FAOSTAT. Crops and Livestock Products. Available online: https://www.fao.org/faostat/en/#data/QCL (accessed on 21 October 2022).
- USDA. Food Composition Database. Available online: https://fdc.nal.usda.gov/fdc-app.html#/ (accessed on 21 October 2022).
- Saricaoglu, F.T.; Gul, O.; Besir, A.; Atalar, I. Effect of High Pressure Homogenization (HPH) on Functional and Rheological Properties of Hazelnut Meal Proteins Obtained from Hazelnut Oil Industry by-Products. J. Food Eng. 2018, 233, 98–108. [Google Scholar] [CrossRef]
- Rohn, S. Possibilities and Limitations in the Analysis of Covalent Interactions between Phenolic Compounds and Proteins. Food Res. Int. 2014, 65, 13–19. [Google Scholar] [CrossRef]
- Ozdal, T.; Capanoglu, E.; Altay, F. A Review on Protein–Phenolic Interactions and Associated Changes. Food Res. Int. 2013, 51, 954–970. [Google Scholar] [CrossRef]
- Su, S.; Wan, Y.; Guo, S.; Zhang, C.; Zhang, T.; Liang, M. Effect of Peptide–Phenolic Interaction on the Antioxidant Capacity of Walnut Protein Hydrolysates. Int. J. Food Sci. Technol. 2018, 53, 508–515. [Google Scholar] [CrossRef]
- Labuckas, D.O.; Maestri, D.M.; Perelló, M.; Martínez, M.L.; Lamarque, A.L. Phenolics from Walnut (Juglans regia L.) Kernels: Antioxidant Activity and Interactions with Proteins. Food Chem. 2008, 107, 607–612. [Google Scholar] [CrossRef]
- Grosso, A.L.; Riveros, C.; Asensio, C.M.; Grosso, N.R.; Nepote, V. Improving Walnuts’ Preservation by Using Walnut Phenolic Extracts as Natural Antioxidants through a Walnut Protein-Based Edible Coating. J. Food Sci. 2020, 85, 3043–3051. [Google Scholar] [CrossRef]
- Rawel, H.M.; Rohn, S. Nature of Hydroxycinnamate-Protein Interactions. Phytochem. Rev. 2010, 9, 93–109. [Google Scholar] [CrossRef]
- Jakobek, L. Interactions of Polyphenols with Carbohydrates, Lipids and Proteins. Food Chem. 2015, 175, 556–567. [Google Scholar] [CrossRef]
- Zhang, H.; Yu, D.; Sun, J.; Liu, X.; Jiang, L.; Guo, H.; Ren, F. Interaction of Plant Phenols with Food Macronutrients: Characterisation and Nutritional–Physiological Consequences. Nutr. Res. Rev. 2014, 27, 1–15. [Google Scholar] [CrossRef]
- AOAC. Official Methods of Analysis of the Association of Official Analytical Chemists; The Association: Arlington, VA, USA, 1990; Volume 2. [Google Scholar]
- Saad, N.; Mohd Esa, N.; Ithnin, H.; Husna Shafie, N. Optimization of Optimum Condition for Phytic Acid Extraction from Rice Bran. Afr. J. Plant Sci. 2011, 5, 168–176. [Google Scholar]
- Tatar, F.; Tunç, M.T.; Kahyaoglu, T. Turkish Tombul Hazelnut (Corylus avellana L.) Protein Concentrates: Functional and Rheological Properties. J. Food Sci. Technol. 2015, 52, 1024–1031. [Google Scholar] [CrossRef] [PubMed]
- AOCS. Official Methods and Recommended Practices of the American Oil Chemists’ Society, 15th ed.; Helrich, K., Ed.; AOCS Press: Arlington, VA, USA, 1990; ISBN 0-935584-42-0. [Google Scholar]
- Harrysson, H.; Hayes, M.; Eimer, F.; Carlsson, N.G.; Toth, G.B.; Undeland, I. Production of Protein Extracts from Swedish Red, Green, and Brown Seaweeds, Porphyra Umbilicalis Kützing, Ulva Lactuca Linnaeus, and Saccharina Latissima (Linnaeus) J. V. Lamouroux Using Three Different Methods. J. Appl. Phycol. 2018, 30, 3565–3580. [Google Scholar] [CrossRef]
- Günal-Köroğlu, D.; Turan, S.; Capanoglu, E. Interaction of Lentil Protein and Onion Skin Phenolics: Effects on Functional Properties of Proteins and in Vitro Gastrointestinal Digestibility. Food Chem. 2022, 372, 130892. [Google Scholar] [CrossRef] [PubMed]
- Adrar, N.; Bahadori, F.; Ceylan, F.D.; Topçu, G.; Bedjou, F.; Capanoglu, E. Stability Evaluation of Interdigitated Liposomes Prepared with a Combination of 1,2-Distearoyl-Sn-Glycero-3-Phosphocholine and 1,2-Dilauroyl-Sn-Glycero-3-Phosphocholine. J. Chem. Technol. Biotechnol. 2021, 96, 2537–2546. [Google Scholar] [CrossRef]
- Bernat, N.; Cháfer, M.; Rodríguez-García, J.; Chiralt, A.; González-Martínez, C. Effect of High Pressure Homogenisation and Heat Treatment on Physical Properties and Stability of Almond and Hazelnut Milks. LWT-Food Sci. Technol. 2015, 62, 488–496. [Google Scholar] [CrossRef]
- Papadopoulou, A.; Green, R.J.; Frazier, R.A. Interaction of Flavonoids with Bovine Serum Albumin: A Fluorescence Quenching Study. J. Agric. Food Chem. 2004, 53, 158–163. [Google Scholar] [CrossRef] [PubMed]
- Lakowicz, J.R. Principles of Fluorescence Spectroscopy, 3rd ed.; Springer: Baltimore, MD, USA, 2006; ISBN 978-0-387-46312-4. [Google Scholar]
- Zhang, Y.; Chen, S.; Qi, B.; Sui, X.; Jiang, L. Complexation of Thermally-Denatured Soybean Protein Isolate with Anthocyanins and Its Effect on the Protein Structure and in Vitro Digestibility. Food Res. Int. 2018, 106, 619–625. [Google Scholar] [CrossRef]
- Jia, J.; Gao, X.; Hao, M.; Tang, L. Comparison of Binding Interaction between β-Lactoglobulin and Three Common Polyphenols Using Multi-Spectroscopy and Modeling Methods. Food Chem. 2017, 228, 143–151. [Google Scholar] [CrossRef]
- Seedher, N.; Agarwal, P. Complexation of Fluoroquinolone Antibiotics with Human Serum Albumin: A Fluorescence Quenching Study. J. Lumin 2010, 130, 1841–1848. [Google Scholar] [CrossRef]
- Ross, P.D.; Subramanian, S. Thermodynamics of Protein Association Reactions: Forces Contributing to Stability. Biochemistry 1981, 20, 3096–3102. [Google Scholar] [CrossRef]
- Shi, M.; Huang, L.Y.; Nie, N.; Ye, J.H.; Zheng, X.Q.; Lu, J.L.; Liang, Y.R. Binding of Tea Catechins to Rice Bran Protein Isolate: Interaction and Protective Effect during in Vitro Digestion. Food Res. Int. 2017, 93, 1–7. [Google Scholar] [CrossRef]
- Berdutina, A.V.; Neklyudov, A.D.; Ivankin, A.I.; Karpo, B.S.; Mitaleva, S.I. Comparison of Proteolytic Activities of the Enzyme Complex from Mammalian Pancreas and Pancreatin. Appl. Biochem. Microbiol. 2000, 36, 363–367. [Google Scholar] [CrossRef]
- Karamać, M.; Kosińska-Cagnazzo, A.; Kulczyk, A. Use of Different Proteases to Obtain Flaxseed Protein Hydrolysates with Antioxidant Activity. Int. J. Mol. Sci. 2016, 17, 1027. [Google Scholar] [CrossRef] [PubMed]
- Moreno, C.; Mojica, L.; de Mejía, E.G.; Camacho Ruiz, R.M.; Luna-Vital, D.A. Combinations of Legume Protein Hydrolysates Synergistically Inhibit Biological Markers Associated with Adipogenesis. Foods 2020, 9, 1678. [Google Scholar] [CrossRef]
- Daliri, H.; Ahmadi, R.; Pezeshki, A.; Hamishehkar, H.; Mohammadi, M.; Beyrami, H.; Heshmati, M.K.; Ghorbani, M. Quinoa Bioactive Protein Hydrolysate Produced by Pancreatin Enzyme- Functional and Antioxidant Properties. LWT 2021, 150, 111853. [Google Scholar] [CrossRef]
- Çağlar, A.F.; Göksu, A.G.; Çakır, B.; Gülseren, İ. Tombul Hazelnut (Corylus avellana L.) Peptides with DPP-IV Inhibitory Activity: In Vitro and in Silico Studies. Food Chem. X 2021, 12, 100151. [Google Scholar] [CrossRef] [PubMed]
- Adler-Nissen, J. Determination of the Degree of Hydrolysis of Food Protein Hydrolysates by Trinitrobenzenesulfonic Acid. J. Agric. Food Chem. 1979, 27, 1256–1262. [Google Scholar] [CrossRef]
- Nielsen, P.M.; Petersen, D.; Dambmann, C. Improved Method for Determining Food Protein Degree of Hydrolysis. J. Food Sci. 2001, 66, 642–646. [Google Scholar] [CrossRef]
- Thurlkill, R.L.; Grimsley, G.R.; Scholtz, J.M.; Pace, C.N. PK Values of the Ionizable Groups of Proteins. Protein Sci. 2006, 15, 1214–1218. [Google Scholar] [CrossRef]
- Minekus, M.; Alminger, M.; Alvito, P.; Ballance, S.; Bohn, T.; Bourlieu, C.; Carriere, F.; Boutrou, R.; Corredig, M.; Dupont, D.; et al. A Standardised Static in Vitro Digestion Method Suitable for Food-an International Consensus. Food Funct. 2014, 5, 1113–1124. [Google Scholar] [CrossRef]
- Turkmen, N.; Sari, F.; Velioglu, Y.S. Effects of Extraction Solvents on Concentration and Antioxidant Activity of Black and Black Mate Tea Polyphenols Determined by Ferrous Tartrate and Folin–Ciocalteu Methods. Food Chem. 2006, 99, 835–841. [Google Scholar] [CrossRef]
- Apak, R.; Güçlü, K.; Özyürek, M.; Karademir, S.E. Novel Total Antioxidant Capacity Index for Dietary Polyphenols and Vitamins C and E, Using Their Cupric Ion Reducing Capability in the Presence of Neocuproine: CUPRAC Method. J. Agric. Food Chem. 2004, 52, 7970–7981. [Google Scholar] [CrossRef] [PubMed]
- Kumaran, A.; Karunakaran, J.R. Antioxidant and Free Radical Scavenging Activity of an Aqueous Extract of Coleus Aromaticus. Food Chem. 2006, 97, 109–114. [Google Scholar] [CrossRef]
- Capanoglu, E.; Beekwilder, J.; Boyacioglu, D.; Hall, R.; de Vos, R. Changes in Antioxidant and Metabolite Profiles during Production of Tomato Paste. J. Agric. Food Chem. 2008, 56, 964–973. [Google Scholar] [CrossRef]
- Jing, H.E.; Xing, Y.F.; Huang, B.O.; Yi-Zheng Zhang, A.; Zeng, C.M. Tea Catechins Induce the Conversion of Preformed Lysozyme Amyloid Fibrils to Amorphous Aggregates. J. Agric. Food Chem. 2009, 57, 11391–11396. [Google Scholar] [CrossRef]
- Chu, Q.; Bao, B.; Wu, W. Mechanism of Interaction between Phenolic Compounds and Proteins Based on Non-Covalent and Covalent Interactions. Med. Res. 2018, 2, 180014. Available online: http://www.medicineresearch.org/EN/Y2018/V2/I3/180014 (accessed on 21 October 2022).
- Dai, S.; Lian, Z.; Qi, W.; Chen, Y.; Tong, X.; Tian, T.; Lyu, B.; Wang, M.; Wang, H.; Jiang, L. Non-Covalent Interaction of Soy Protein Isolate and Catechin: Mechanism and Effects on Protein Conformation. Food Chem. 2022, 384, 132507. [Google Scholar] [CrossRef]
- Ghribi, A.M.; Gafsi, I.M.; Sila, A.; Blecker, C.; Danthine, S.; Attia, H.; Bougatef, A.; Besbes, S. Effects of Enzymatic Hydrolysis on Conformational and Functional Properties of Chickpea Protein Isolate. Food Chem. 2015, 187, 322–330. [Google Scholar] [CrossRef]
- Mohan, A.; Rajendran, S.R.C.K.; He, Q.S.; Bazinet, L.; Udenigwe, C.C. Encapsulation of Food Protein Hydrolysates and Peptides: A Review. RSC Adv. 2015, 5, 79270–79278. [Google Scholar] [CrossRef]
- Cao, Y.; Xiong, Y.L. Interaction of Whey Proteins with Phenolic Derivatives Under Neutral and Acidic PH Conditions. J. Food Sci. 2017, 82, 409–419. [Google Scholar] [CrossRef]
- Chen, Z.; Wang, C.; Gao, X.; Chen, Y.; Santhanam, R.K.; Wang, C.; Xu, L.; Chen, H. Interaction Characterization of Preheated Soy Protein Isolate with Cyanidin-3-O-Glucoside and Their Effects on the Stability of Black Soybean Seed Coat Anthocyanins Extracts. Food Chem. 2019, 271, 266–273. [Google Scholar] [CrossRef] [PubMed]
- Al-Shabib, N.A.; Khan, J.M.; Malik, A.; Rehman, M.T.; AlAjmi, M.F.; Husain, F.M.; Hisamuddin, M.; Altwaijry, N. Molecular Interaction of Tea Catechin with Bovine β-Lactoglobulin: A Spectroscopic and in Silico Studies. Saudi Pharm. J. 2020, 28, 238–245. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; He, W.; Chen, S.; Chen, J.; Zeng, M.; Qin, F.; He, Z. Interactions of Digestive Enzymes and Milk Proteins with Tea Catechins at Gastric and Intestinal PH. Int. J. Food Sci. Technol. 2017, 52, 247–257. [Google Scholar] [CrossRef]
- Wu, X.; Wu, H.; Liu, M.; Liu, Z.; Xu, H.; Lai, F. Analysis of Binding Interaction between (−)-Epigallocatechin (EGC) and β-Lactoglobulin by Multi-Spectroscopic Method. Spectrochim. Acta A Mol. Biomol. Spectrosc. 2011, 82, 164–168. [Google Scholar] [CrossRef] [PubMed]
- Hasni, I.; Bourassa, P.; Hamdani, S.; Samson, G.; Carpentier, R.; Tajmir-Riahi, H.A. Interaction of Milk α- and β-Caseins with Tea Polyphenols. Food Chem. 2011, 126, 630–639. [Google Scholar] [CrossRef]
- Kanakis, C.D.; Hasni, I.; Bourassa, P.; Tarantilis, P.A.; Polissiou, M.G.; Tajmir-Riahi, H.A. Milk β-Lactoglobulin Complexes with Tea Polyphenols. Food Chem. 2011, 127, 1046–1055. [Google Scholar] [CrossRef]
- Yılmaz, H.; Lee, S.; Chronakis, I.S. Interactions of β-Lactoglobulin with Bovine Submaxillary Mucin vs. Porcine Gastric Mucin: The Role of Hydrophobic and Hydrophilic Residues as Studied by Fluorescence Spectroscopy. Molecules 2021, 26, 6799. [Google Scholar] [CrossRef]
- Acharya, D.P.; Sanguansri, L.; Augustin, M.A. Binding of Resveratrol with Sodium Caseinate in Aqueous Solutions. Food Chem. 2013, 141, 1050–1054. [Google Scholar] [CrossRef]
- Tang, B.; Huang, Y.; Ma, X.; Liao, X.; Wang, Q.; Xiong, X.; Li, H. Multispectroscopic and Docking Studies on the Binding of Chlorogenic Acid Isomers to Human Serum Albumin: Effects of Esteryl Position on Affinity. Food Chem. 2016, 212, 434–442. [Google Scholar] [CrossRef]
- Petrucci, R.H.; Herring, F.G.; Madura, J.D.; Bissonnette, C. (Eds.) Spontaneous Change: Entropy and Gibbs Energy. In General Chemistry: Principles and Modern Applications; Pearson: Toronto, ON, Canada, 2011; pp. 819–862. ISBN 978-0-13-206452-1. [Google Scholar]
- Li, X.; Wang, S. Study on the Interaction of (+)-Catechin with Human Serum Albumin Using Isothermal Titration Calorimetry and Spectroscopic Techniques. New J. Chem. 2014, 39, 386–395. [Google Scholar] [CrossRef]
- Li, X.; Hao, Y. Probing the Binding of (+)-Catechin to Bovine Serum Albumin by Isothermal Titration Calorimetry and Spectroscopic Techniques. J. Mol. Struct. 2015, 1091, 109–117. [Google Scholar] [CrossRef]
- Dogan, A.; Siyakus, G.; Severcan, F. FTIR Spectroscopic Characterization of Irradiated Hazelnut (Corylus avellana L.). Food Chem. 2007, 100, 1106–1114. [Google Scholar] [CrossRef]
- Ferraro, V.; Madureira, A.R.; Sarmento, B.; Gomes, A.; Pintado, M.E. Study of the Interactions between Rosmarinic Acid and Bovine Milk Whey Protein α-Lactalbumin, β-Lactoglobulin and Lactoferrin. Food Res. Int. 2015, 7, 450–459. [Google Scholar] [CrossRef]
- Han, L.; Lu, K.; Zhou, S.; Zhang, S.; Xie, F.; Qi, B.; Li, Y. Development of an Oil-in-Water Emulsion Stabilized by a Black Bean Protein-Based Nanocomplex for Co-Delivery of Quercetin and Perilla Oil. LWT 2021, 138, 110644. [Google Scholar] [CrossRef]
- Li, D.; Zhao, Y.; Wang, X.; Tang, H.; Wu, N.; Wu, F.; Yu, D.; Elfalleh, W. Effects of (+)-Catechin on a Rice Bran Protein Oil-in-Water Emulsion: Droplet Size, Zeta-Potential, Emulsifying Properties, and Rheological Behavior. Food Hydrocoll. 2020, 98, 105306. [Google Scholar] [CrossRef]
- Cirkovic Velickovic, T.D.; Stanic-Vucinic, D.J. The Role of Dietary Phenolic Compounds in Protein Digestion and Processing Technologies to Improve Their Antinutritive Properties. Compr. Rev. Food Sci. Food Saf. 2018, 17, 82–103. [Google Scholar] [CrossRef]
- Ni, H.; Hayes, H.; Stead, D.; Liu, G.; Yang, H.; Li, H.; Raikos, V. Interaction of Whey Protein with Polyphenols from Salal Fruits (Gaultheria Shallon) and the Effects on Protein Structure and Hydrolysis Pattern by Flavourzyme®. Int. J. Food Sci. Technol. 2020, 55, 1281–1288. [Google Scholar] [CrossRef]
- Wang, X.Y.; Tang, C.H. Physicochemical and Antioxidant Properties of Buckwheat Protein Isolates with Different Polyphenolic Content Modified by Limited Hydrolysis with Trypsin–ProQuest. Food Technol. Biotechnol. 2012, 50, 17–24. [Google Scholar]
- Xie, Y.; Kosińska, A.; Xu, H.; Andlauer, W. Milk Enhances Intestinal Absorption of Green Tea Catechins in in Vitro Digestion/Caco-2 Cells Model. Food Res. Int. 2013, 53, 793–800. [Google Scholar] [CrossRef]
- Green, R.J.; Murphy, A.S.; Schulz, B.; Watkins, B.A.; Ferruzzi, M.G. Common Tea Formulations Modulate in Vitro Digestive Recovery of Green Tea Catechins. Mol. Nutr. Food Res. 2007, 51, 1152–1162. [Google Scholar] [CrossRef]
- Chen, Z.-Y.; Yan Zhu, Q.; Fan Wong, Y.; Zhang, Z.; Yin Chung, H. Stabilizing Effect of Ascorbic Acid on Green Tea Catechins. J. Agric. Food Chem. 1998, 46, 2512–2516. [Google Scholar] [CrossRef]
- Zhu, Q.Y.; Huang, Y.; Tsang, D.; Chen, Z.Y. Regeneration of α-Tocopherol in Human Low-Density Lipoprotein by Green Tea Catechin. J. Agric. Food Chem. 1999, 47, 2020–2025. [Google Scholar] [CrossRef] [PubMed]
- Shim, S.M.; Yoo, S.H.; Ra, C.S.; Kim, Y.K.; Chung, J.O.; Lee, S.J. Digestive Stability and Absorption of Green Tea Polyphenols: Influence of Acid and Xylitol Addition. Food Res. Int. 2012, 45, 204–210. [Google Scholar] [CrossRef]
| Defatted Meal | Dephenolized Meal | |
|---|---|---|
| Protein content (%) | 46.9 ± 1.0 | 28.1 ± 0.6 |
| Crude proteins isolated (%) | 39.5 ± 1.1 | 13.0 ± 1.0 |
| Purity of isolates (%) | 78.7 ± 1.3 | 98.4 ± 1.0 |
| Recovery (%, pure basis) | 76.7 ± 6.1 | 45.6 ± 4.5 |
| GLYCINE | 38.9 ± 0.8 | ASPARGINE | 88.2 ± 0.7 |
| ALANINE | 35.8 ± 1.1 | LYSINE | 18.9 ± 0.5 |
| SERINE | 41.8 ± 0.4 | GLUTAMINE | 184.8 ± 3.4 |
| PROLINE | 28.4 ± 1.0 | METHIONINE | 4.7 ± 5.3 |
| VALIN | 27.5 ± 1.4 | HISTIDINE | 17.3 ± 3.2 |
| THREONINE | 25.3 ± 0.3 | PHENYLALANINE | 38.8 ± 1.7 |
| CYSTEINE | nd | ARGININE | 94.3 ± 11.5 |
| ISOLEUCINE | 53.9 ± 2.2 | TYROSINE | 19.9 ± 0.1 |
| LEUCINE | 54.6 ± 3.0 |
| Temperature (K) | Stern-Volmer Constants | Double-Log Plot | ΔG (kJ mol−1) | ΔH (J mol−1) | ΔS (J K−1 mol−1) | ||||
|---|---|---|---|---|---|---|---|---|---|
| Ksv (1014 L mol−1) | Kq (106 L mol−1) | R2 | KA (105 L mol−1) | n | R2 | ||||
| 298 | 34.696 | 34.696 | 0.9855 | 49.9114 | 1.4216 | 0.9523 | −9671.72 | −18,944.28 | −31.12 |
| 308 | 26.548 | 26.548 | 0.9732 | 34.5939 | 1.4431 | 0.9927 | −9298.33 | ||
| 318 | 27.001 | 27.001 | 0.9881 | 31.9890 | 1.1618 | 0.9485 | −9049.4 | ||
| dHPI | dHPI+HSE (0.05) | dHPI+HSE (0.125) | dHPI+HSE (0.25) | dHPI+C (0.125) | |
|---|---|---|---|---|---|
| α-helix | 17.92 | 32.88 | 28.78 | 26.76 | 36.88 |
| β-sheet | 60.78 | 54.10 | 49.29 | 28.23 | 54.62 |
| β-turn | 17.38 | 1.78 | 24.10 | 11.79 | 1.63 |
| Random Coil | 3.92 | 11.23 | 11.44 | 33.22 | 6.87 |
| Initial Phase | Gastric Phase | Intestinal Phase | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| TPC | RI | DPPH | CUPRAC | TPC | RI | DPPH | CUPRAC | TPC | RI | DPPH | CUPRAC | |
| C | 1040.4 ± 61.5 aB | 100 | 143.0 ± 0.5 aA | 1222.0 ± 66.5 bB | 760.7 ± 3.3 bC | 73.1 | 131.4 ± 0.9 aB | 1067.6 ± 23.2 bC | 1416.1 ± 6.8 aA | 136.1 | 94.0 ± 5.9 bC | 2285.6 ± 90.5 aA |
| HSE | 714.4 ± 54.3 cA | 100 | 142.6 ± 0.1 aA | 705.0 ± 8.9 cA | 357.8 ± 22.4 cC | 50.1 | 128.5 ± 1.3 aB | 321.2 ± 22.7 cC | 559.0 ± 33.5 cB | 78.2 | 105.8 ± 7.7 bC | 638.8 ± 26.1 cB |
| HPI+C | 982.1 ± 66.6 bA | 94.4 | 130.6 ± 2.1 bB | 1945.6 ± 54.5 aAB | 891.8 ± 32.1 aB | 85.7 | 128.7 ± 0.4 aB | 2175.0 ± 195.2 aA | 1024.0 ± 63.9 bA | 98.4 | 181.0 ± 11.9 aA | 1849.3 ± 35.7 bB |
| dHPI+HSE | 317.9 ± 26.4 dB | 44.5 | 104.6 ± 12.4 cA | 522.5 ± 4.4 dB | 256.5 ± 16.7 dC | 35.9 | 62.9 ± 7.0 bB | 364.5 ± 19.4 cC | 488.6 ± 29.4 dA | 68.4 | 106.8 ± 10.6 bA | 763.3 ± 8.1 cA |
| Compounds (mg/g) | C | dHPI+C | HSE | dHPI+HSE | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Initial | Gastric | Intestinal | Initial | Gastric | Intestinal | Initial | Gastric | Intestinal | Initial | Gastric | Intestinal | |
| Gallic acid | nd | nd | nd | nd | nd | nd | 1.0 ± 0.3 cA | 1.1 ± 0.2 bA | 1.1 ± 0.1 aB | nd | 0.9 ± 0.0 bB | 1.5 ± 0.1 aA |
| Protocatechuic acid | nd | nd | nd | nd | nd | nd | 1.0 ± 0.7 aA | 0.8 ± 0.0 aB | 1.0 ± 0.1 aA | nd | 0.8 ± 0.0 aA | 0.6 ± 0.0 bB |
| Gallocatechin gallate | nd | nd | 10.4 ± 0.6 aC | nd | nd | 32.1 ± 2.6 aA | 0.2 ± 0.0 cA | 0.9 ± 0.0 bA | 9.9 ± 0.6 aD | nd | 0.3 ± 0.0 bB | 29.7 ± 2.8 aB |
| (-)-Epigallocatechin | nd | nd | 18.5 ± 1.6 aA | nd | nd | 14.2 ± 1.2 aB | 2.4 ± 0.2 aA | 2.2 ± 0.1 bA | 0.6 ± 0.0 cD | nd | 1.5 ± 0.1 bB | 4.5 ± 0.3 aC |
| Catechin | 2417.3 ± 102.4 aA | 2144.4 ± 87.3 bA | 917.8 ± 23.9 cA | nd | 1837.5 ± 64.9 aB | 885.8 ± 17.4 bB | 4.0 ± 0.8 bB | 2.6 ± 0.2 cC | 4.9 ± 0.3 aC | nd | 2.8 ± 0.3 aC | 1.9 ± 0.1 bD |
| Epicatechin | nd | nd | 22.9 ± 2.5 aB | nd | nd | 151.5 ± 8.4 aA | 2.0 ± 0.1 bA | 1.3 ± 0.1 cA | 2.4 ± 0.2 aD | nd | 1.0 ± 0.0 bB | 3.2 ± 0.2 aC |
| (-)-Epigallocatechin gallate | nd | nd | 11.1 ± 0.2 aB | nd | nd | nd | 2.1 ± 0.3 bA | 0.6 ± 0.0 cA | 8.3 ± 0.6 aC | nd | 0.5 ± 0.0 bB | 41.8 ± 2.6 aA |
| Phlorizin | nd | nd | nd | nd | nd | nd | 0.4 ± 0.0 aA | 0.3 ± 0.0 bA | nd | nd | 0.3 ± 0.0 bB | 0.6 ± 0.0 aA |
| Quercetin 3-O-rhamnoside | nd | nd | nd | nd | nd | nd | 1.0 ± 0.0 aA | 0.8 ± 0.0 bA | 0.7 ± 0.0 cB | nd | 0.5 ± 0.0 bB | 0.9 ± 0.0 aA |
| Quercetin | nd | nd | nd | nd | nd | nd | 0.1 ± 0.0 aA | nd | nd | nd | nd | nd |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ceylan, F.D.; Yılmaz, H.; Adrar, N.; Günal Köroğlu, D.; Gultekin Subasi, B.; Capanoglu, E. Interactions between Hazelnut (Corylus avellana L.) Protein and Phenolics and In Vitro Gastrointestinal Digestibility. Separations 2022, 9, 406. https://doi.org/10.3390/separations9120406
Ceylan FD, Yılmaz H, Adrar N, Günal Köroğlu D, Gultekin Subasi B, Capanoglu E. Interactions between Hazelnut (Corylus avellana L.) Protein and Phenolics and In Vitro Gastrointestinal Digestibility. Separations. 2022; 9(12):406. https://doi.org/10.3390/separations9120406
Chicago/Turabian StyleCeylan, Fatma Duygu, Hilal Yılmaz, Nabil Adrar, Deniz Günal Köroğlu, Busra Gultekin Subasi, and Esra Capanoglu. 2022. "Interactions between Hazelnut (Corylus avellana L.) Protein and Phenolics and In Vitro Gastrointestinal Digestibility" Separations 9, no. 12: 406. https://doi.org/10.3390/separations9120406
APA StyleCeylan, F. D., Yılmaz, H., Adrar, N., Günal Köroğlu, D., Gultekin Subasi, B., & Capanoglu, E. (2022). Interactions between Hazelnut (Corylus avellana L.) Protein and Phenolics and In Vitro Gastrointestinal Digestibility. Separations, 9(12), 406. https://doi.org/10.3390/separations9120406