Abstract
Lumnitzera littorea and Lumnitzera racemosa are mangrove species distributed widely along the Indonesian coasts. Besides their ecological importance, both are of interest owing to their wealth of natural products, some of which constitute potential sources for medicinal applications. We aimed to discover and characterize new anti-infective compounds, based on population-level sampling of both species from across the Indonesian Archipelago. Root metabolites were investigated by TLC, hyphenated LC-MS/MS and isolation, the internal transcribed spacer (ITS) region of rDNA was used for genetic characterization. Phytochemical characterization of both species revealed an unusual diversity in sulfated constituents with 3,3’,4’-tri-O-methyl-ellagic acid 4-sulfate representing the major compound in most samples. None of these compounds was previously reported for mangroves. Chemophenetic comparison of L. racemosa populations from different localities provided evolutionary information, as supported by molecular phylogenetic evidence. Samples of both species from particular locations exhibited anti-bacterial potential (Southern Nias Island and East Java against Gram-negative bacteria, Halmahera and Ternate Island against Gram-positive bacteria). In conclusion, Lumnitzera roots from natural mangrove stands represent a promising source for sulfated ellagic acid derivatives and further sulfur containing plant metabolites with potential human health benefits.
1. Introduction
Mangrove forests represent a unique habitat that comprises salt-tolerant plant species (mostly trees), predominantly bordering tropical and subtropical coastlines [1]. Besides their ecological significance, mangrove plant species have a wide variety of economic uses, such as construction material, fodder or textiles [2,3]. In addition, many mangrove plant species possess medicinal value and have been used traditionally in several regions of the world [4,5,6,7]. Due to the tidal influence, mangrove soils contain high levels of sulfate which is connected to the occurrence of sulfate-reducing microbial communities [8]. Nevertheless, the investigation of bioactive natural sulfur compounds in mangrove species was so far completely neglected.
The most diverse mangrove systems on earth can be found in Southeast Asian seas [9]. Comprising five main and more than 17,500 smaller islands, Indonesian mangroves cover around 30% of the total mangrove area of the world [10]. Lumnitzera littorea (Jack) Voigt and L. racemosa Willd., two true mangrove species belonging to the plant family Combretaceae (Myrtales), are distributed widely across the Indonesian coastline. In Africa, where L. racemosa also naturally occurs at the eastern coast, other members of Combretaceae (Combretum, Terminalia) are widely used for medicinal purposes due to their anti-microbial [11,12,13,14,15,16], anti-fungal [17,18], antioxidant and anti-inflammatory activities [16]. In Asia, L. racemosa from Taiwan was reported to contain antihypertensive tannins [19]. Furthermore, hepatoprotective, antioxidant [20,21,22,23], antibacterial [24,25], anti-angiogenic, anti-inflammatory [26], anti-cancer [23,27], and anti-coagulant effects [23] were described in L. racemosa from different parts of Asia. In leaves and twigs of L. racemosa mainly flavonoids and triterpenes [22,28] as well as phenolic acids and their derivatives, such as gallic acid and related compounds—galloyl sugars, ellagic acid, 3,3’,4-tri-O-methylellagic acid, neolignans and tannic acid—were found [22,26]. The extract of L. littorea leaves from Malaysia was reported to possess anti-microbial potential [29]. The leaf n-hexane extract of this species yielded triterpenes and sterols [30] whereas the twigs of L. littorea were described to contain macrocyclic lactones (represented by corniculatolide derivatives) and 6,7-dimethoxycoumarin [31].
Medicinally active compounds from mangroves are not always produced by the plant itself, but often by associated microorganisms such as endophytic fungi [32,33,34]. For example, the extracts of endophytic fungi isolated from leaves of ten mangrove species from Thailand, including L. littorea, showed some cytotoxic activity against cancer cell lines [35]. In line with the agenda of discovering new anti-infective and neuroactive constituents while at the same time promoting the protection and sustainable development of mangrove ecosystems, our work was focused on two mangrove species, namely Lumnitzera littorea and L. racemosa. Our aim was to (1) investigate the diversity of natural products present in the roots of L. littorea and L. racemosa from Indonesia, (2) evaluate selected biological effects, and (3) investigate the phylogenetic relationships of the two species as well as the chemophenetic patterns of their natural products across Indonesia. Therefore, we combined a molecular phylogeny of Lumnitzera based on the internal transcribed spacer (ITS) region with phytochemical analyses by hyphenated chromatographic and tandem mass spectrometric techniques. Chromatographic separation using reversed phase HPLC connected to high resolution ESI-MS that allow the determination of accurate mass and elemental composition represents a suitable technique to identify sulfur containing metabolites [36]. For the calculation of the molecular composition of sulfur-containing compounds, the small negative mass defect of sulfur isotopes and the isotopic pattern of sulfur distinct from that of carbon, nitrogen and hydrogen can be applied [36]. Since sulfur in addition to the most abundant isotope 32S (95%) possesses a 34S isotope (4.2%), also the larger as usual M+2 peak contributes to the determination of sulfur in compounds or fragments. Nevertheless, for complete structure elucidation, compounds have to be isolated and characterized by NMR. Phylogenetic approaches can be useful for identifying plant lineages with potential medicinal properties [37,38], the interpretation of chemical profiles [39], and might be a powerful tool for discovering novel compounds or novel compound variants [40,41,42], including antibiotic sources [43,44].
2. Materials and Methods
2.1. Plant Material
Leaf and root material of Lumnitzera littorea (Jack) Voigt and Lumnitzera racemosa Willd. was collected from 27 locations across the Indonesian archipelago (Table 1). Voucher specimens of the plants are deposited at Herbarium Bogoriense (BO), Indonesian Institute of Sciences (LIPI). Root samples for phytochemical analyses were cleaned and air-shadow-dried in the field, then kept in resealable zipper storage bags until being used for further treatment. For phylogenetic analyses, fresh leaves from the same plants were collected and dried in silica gel in resealable zipper storage bags.

Table 1.
Samples from L. littorea (no. 1–12) and L. racemosa (no. 13-31) used for phytochemical and DNA analyses. Abbreviations: J.M. = Jeprianto Manurung, Fr = Fine root, Rb = Root bark, NS = North Sumatra, EK = East Kalimantan, SS = Southeast Sulawesi, NS = North Sulawesi, CS = Central Sulawesi, ENT = East Nusa Tenggara.
2.2. Root Sample Extraction
Air-dried samples (root bark and fine roots) were ground using a ball mill (Retsch, MM400) for two minutes. In extraction experiments with n-hexane, ethyl acetate and methanol, the highest extract amount could be obtained with methanol. Therefore, 100 mg of each sample were vortexed with 5 mL methanol in Eppendorf tubes before sonication for an hour. All samples were centrifuged for fifteen minutes using a Megafuge 1.0R (Unity Lab Services) to gain pure solutions. The extracts were aliquoted for analytical investigations and bioassay screening. The crude extracts were directly used for TLC and low-resolution ESI-MS analyses. For LCMS measurements, 250 µL of each extract were purified by an SPE cartridge (RP18, Chromabond, Macherey-Nagel, Düren, Germany), using methanol as eluent, and the concentration was afterwards adjusted to 1 mg/mL.
2.3. General Experimental Procedures
Thin layer chromatography (TLC) analyses were done with silica gel 60 F254 (Merck, Darmstadt, Germany) using the solvent system CHCl3:MeOH 6:4. Compound spots were visualized using long-wavelength UV light (366 nm), short-wavelength UV light (254 nm) and spraying with vanillin-H2SO4 reagent, followed by heating. As sample LR15 in TLC displayed stronger spots compared to the others, preparative TLC (thickness 0.5 mm) was performed using the same conditions. The major bands were scraped off and extracted to verify the compound identity by MS investigations.
Low-resolution ESI-MS spectra were obtained from a Sciex API-3200 instrument (Applied Biosystems, Concord, ON, Canada) combined with an HTC-XT autosampler (CTC Analytics, Zwingen, Switzerland).
1H and 13C NMR spectra were recorded on an Agilent DD2 400 NMR spectrometer at 399.917 and 100.570 MHz, respectively. Chemical shifts are reported relative to TMS (1H NMR) or peaks of solvent (13C, CD3OD 49.0 ppm and DMSO-d6 39.5 ppm). For samples with low concentration, 1H and 13C NMR spectra were recorded on a Bruker Avance Neo 500 NMR spectrometer at 500.234 and 125.797 MHz, respectively, using a 5 mm prodigy probe with the TopSpin 4.0.7 spectrometer software. 2D NMR spectra were recorded on an Agilent VNMRS 600 MHz NMR spectrometer using standard CHEMPACK 8.1 pulse sequences (1H, 13C gHSQCAD and 1H, 13C gHMBCAD) implemented in Varian VNMRJ 4.2 spectrometer software.
Preparative HPLC was performed using an Agilent 1260/1290 system equipped with a quaternary pump and a diode array (DAD) detector (Agilent, VL+). Chromatographic separation was performed using a Macherey-Nagel Chromcart C18ec column (ID 4.6 mm, length 150 mm, particle size 5 µm) using bidest. water (TKA ultrapure water system) and methanol (Roth, Rotisolv HPLC Gradient Grade) as eluents.
2.4. UHPLC-ESI-QqTOF-MS and MS/MS
Samples (2 µL) were loaded on an EC 150/2 Nucleoshell RP 18 column (C18-phase, ID 2 mm, length 150 mm, particle size 2.7 µm, Macherey Nagel, Düren, Germany) under isocratic conditions (5% eluent B, 2 min), and separated using a linear gradient from 5% to 95% eluent B in 17 min. Separation was performed on an ACQUITY UPLC I-Class UHPLC System (Waters GmbH, Eschborn, Germany) with a flow rate of 0.4 mL/min and 40 °C column temperature. Eluents A and B were 0.3 mmol/L aq. ammonium formate and acetonitrile, respectively. The column effluent was introduced on-line into a TripleTOF 6600 QqTOF mass spectrometer equipped with a DuoSpray ESI/APCI ion source, operating in negative ion SWATH (Sequential Windowed Acquisition of All Theoretical Fragment Ion Mass Spectra) mode and controlled by the Analyst TF 1.7.1 software (AB Sciex GmbH, Darmstadt, Germany). The TOF scans (MS experiments) were acquired in the m/z range of 65 to 1250 (accumulation time 100 ms) with an ion spray voltage of −4.5 kV and 450 °C source temperature. For precursor selection, totally 38 SWATH windows (total m/z range of 65–1250) of 26 m/z were used. Nebulizer and drying gases were set to 60 and 70 psi, respectively, whereas the curtain gas was set to 55 psi. Declustering (DP) and collision (CE) potentials were −35 and −10 V, respectively. The product ion spectra (tandem mass spectra, MS/MS) were acquired in the high sensitivity mode (accumulation time 20 ms) in the m/z range of 65–1250 using unit Q1 resolution with mass resolution above 30,000. Collision potential (CE) was set to −35 V, whereas collision energy spread (CES) was 15 V. The data were evaluated by Peak View 1.2.0.3 software (AB Sciex GmbH, Darmstadt, Germany).
2.5. RP-UHPLC-ESI-LIT-Orbitrap-MS
Samples (2 µL) were loaded on an ACQUITY UPLC reversed-phase BEH column (C18-phase, ID 1 mm, length 50 mm, particle size 1.7 µm, Waters GmbH, Eschborn, Germany) under isocratic conditions (95% A + 5% eluent B, 2 min), and separated using a linear gradient from 5% to 95% eluent B in 10 min using water and acetonitrile (both containing 0.1% v/v formic acid) as eluents A and B, respectively. The separations were performed with a Dionex Ultimate 3000 UHPLC System (Thermo Fisher Scientific, Bremen, Germany) at the flow rate of 400 µL/min and column temperature of 40 °C. The column effluents were transferred on-line into a hybrid LTQ-Orbitrap Elite mass spectrometer (Thermo Fisher Scientific, Bremen, Germany), equipped with a heated electrospray ionization (HESI) source at 300 °C and operated in the negative ion mode. The analysis was performed under ion spray (IS) voltage of 3.8 kV, with nebulizer and auxiliary gases set to 10 and 5 psig, respectively. The capillary temperature was set to 325 °C. The spectra were acquired at the mass to charge ratio (m/z) range of 120–2000 and resolution of 30,000. Tandem mass spectra were acquired with isolation width of 0.5–2 m/z and collision induced dissociation mode (30–35% normalized intensity), activation time 10 ms and activation frequency 250. Spectra evaluation was performed in Xcalibur 2.2 software (Thermo Fisher Scientific).
2.6. Extraction and Isolation
62.58 g roots of LR7 were exhaustively extracted with methanol to give 3.86 g of crude extract after evaporation of the solvent.
1.5 g crude extract was separated on a silica gel column with an increasing polar gradient, started with pure chloroform, followed by 2.5%, 5%, 10% and 50% methanolic chloroform and a final elution with pure methanol (volume of each step: 250 mL). Based on the TLC profiles, the fractions were combined into 21 main fractions. Fraction 5, eluted with 2.5% MeOH, could be identified as 3,3’4’-tri-O-methylellagic acid (16) (12 mg, Rf = 0.92 in CHCl3/MeOH (3:1, v/v) on SG60). Fraction 18 (252.6 mg) eluted with 50% MeOH was further separated on a Sephadex LH20 column eluted with MeOH followed by repeated CC on a reverse phase column (C18) using H2O:MeOH (60:40, v/v) as eluents to give 3,3’4’-tri-O-methylellagic acid 4-sulfate (15) (3.9 mg, Rf = 0.33 in MeOH/H2O (2:3, v/v) on RP18) and mixtures of compounds 2, 3 and 4. Final purification was performed by preparative HPLC. Compound 2 (0.6 mg, Rf = 0.70 in MeOH/H2O (2:3, v/v) on RP18) was purified using a water (A)/methanol (B) gradient system (0–1.5 min, 20% B; 1.5–14 min, 20–50% B, 14–16 min 50–100% B (isocratic for 8 min)) and a flow rate of 0.8 mL/min at 25 °C, absorbance detection at 210 to 254 nm (Rt = 6.605 min). Compound 3 (3.2 mg, Rf = 0.15 in MeOH/H2O (2:3, v/v) on RP18) was obtained using the following gradient: 0–18 min, 40% B; 18–20 min, 40–100% B (isocratic for 10 min), and a flow rate of 0.6 mL/min at 15 °C, absorbance detection at 210 to 254 nm (Rt = 15.41 min).
1.35 g crude extract were partitioned by liquid-liquid-extraction between water and ethyl acetate. The ethyl acetate phase (419.8 mg) was further separated using a Sephadex LH20 column eluting with MeOH (h: 37.5 cm, d: 2.5 cm). Based on TLC profiles seven main fractions were combined. Fraction 6 could be identified as ellagic acid (6) (7.5 mg, Rf = 0.08 in H2O/MeOH (3:2, v/v + 1% formic acid) on RP18). Rechromatography of fraction 5 (16.2 mg) on Sephadex LH20 with MeOH (h: 76 cm, d: 2.5 cm) resulted in the isolation of 3,4-di-O-methylellagic acid (13) (4.7 mg, Rf = 0.02 in H2O/MeOH (3:2, v/v + 1% formic acid) on RP18).
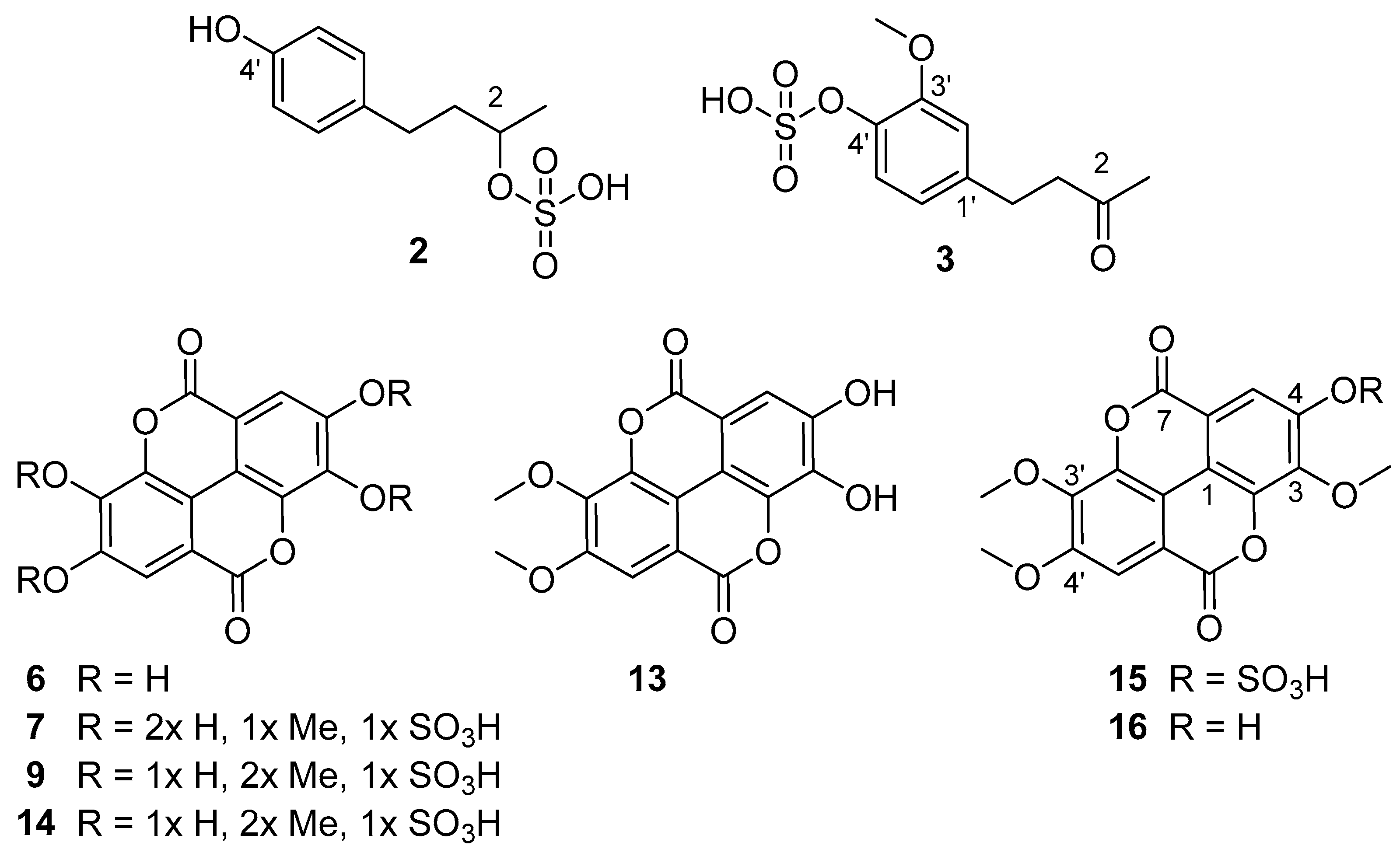
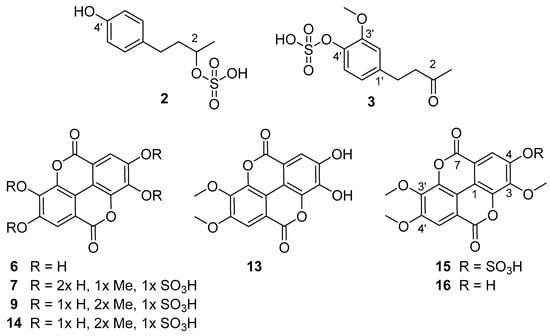
4-(4-Hydroxyphenyl)-2-butanol 2-sulfate (2): white amorphous compound; 1H NMR (400 MHz, methanol-d4) δ 7.03 (d-like, J = 8.4 Hz, 2H, H-2’/6’), 6.67 (d-like, J = 8.4 Hz, 2H, H-3’/5’), 4.46 (m, 1H, H-2), 2.55-2.70 (m, 2H, H-4), 1.89 (m, 1H, H-3a), 1.75 (m, 1H, H-3b), 1.33 (d, J = 6.3 Hz, 3H, H-1). 13C NMR (δ determined from cross peaks in HSQC and *HMBC experiments, methanol-d4) δ 156.4* (C-4’), 134.6* (C-1’), 130.3 (C-2’/6’), 116.1 (C-3’/5’), 76.8 (C-2), 40.5 (C-3), 31.8 (C-4), 21.2 (C-1). 2D-NMR see Table S2.; HRESIMS m/z 245.0484 [M-H]- (calcd for C10H13SO5, 245.0489).
4-(4-Sulfoxy-3-methoxyphenyl)-butan-2-one (3): yellow oily compound; 1H NMR (400 MHz, methanol-d4) δ 6.77 (d, J = 2.0 Hz, 1H, H-2’), 6.68 (d, J = 8.0 Hz, 1H, H-5’), 6.61 (dd, J = 8.0, 2.0 Hz, 1H, H-6’), 3.82 (s, 3H, 3’-OMe), 2.76 (s, 4H, H-3’/4’), 2.11 (s, 3H, H-1). 13C NMR (126 MHz, methanol-d4) δ 211.5 (C-2), 149.0 (3’), 145.8 (C-4’), 134.0 (C-1’), 121.7 (C-6’), 116.2 (C-5’), 113.1 (C-2’), 56.4 (3’-OMe), 46.3 (C-3), 30.5 (C-4), 30.1 (C-1). 2D-NMR see Table S3.; HRESIMS m/z 273.0435 [M-H]- (calcd for C11H13SO6 273.0438).
Ellagic acid (6): 1H NMR (400 MHz, DMSO-d6) δ 7.46 (s, 2H, H5/H5′), HRESIMS m/z 300.9996 [M-H]− (calcd for C14H6O8 300.9990).
3,4-O-Dimethylellagic acid (13): yellowish amorphous compound, 1H NMR (600 MHz, DMSO-d6) δ 7.53 (s, 1H, H5), 7.09 (s, 1H, H5′), 3.99 (s, 4H, 3-OMe), 3.96 (s, 3H, 4-OMe). 13C NMR from HMBC (600/150 MHz, DMSO-d6) δ 160.22 (C-7), 159.15 (C-7’), 156.46 (C-4‘), 152.73 (C-4), 151.92 (C-3‘), 141.79 (C-2), 140.04 (C-3), 134.51 (C-2‘), 115.00 (C-1), 114.97 (C-1‘), 113.09 (C-6‘), 112.95 (C6). 105.83 (C-5), 104.63 (C-5‘), 60.83 (3-OMe), 56.32 (4-OMe). 2D-NMR see Table S4. HRESIMS m/z 329.0370 [M-H]− (calcd for C16H10O8 329.0303).
3,3’4’-Tri-O-methylellagic acid 4-sulfate (15): white yellowish amorphous compound; 1H NMR (400 MHz, DMSO-d6) δ 8.24 (s, 1H, H-5), 7.66 (s, 1H, H-5′), 4.12 (s, 3H, 3-OMe), 4.06 (s, 3H, 3′-OMe), 4.02 (s, 3H, 4′-OMe). 13C NMR (126 MHz, DMSO-d6) δ 158.46 (C-7′), 158.31 (C-7), 154.37 (C-4′), 147.64 (C-4), 143.32 (C-3), 141.34 (C-3’), 140.89 (C-2’), 140.86 (C-2), 117.61 (C-5), 114.13 (C-1′), 112.96 (C-1), 112.83 (C-6′), 111.52 (C-6), 107.50 (C-5′), 61.47 (3-OMe), 61.32 (3′-OMe), 56.75 (4′-OMe). 2D-NMR see Table S5; HRESIMS m/z 423.0024 [M-H]− (calcd for C17H11SO11 423.0028)
3,3′,4-Tri-O-methylellagic acid (16): white yellowish amorphous compound. 1H NMR (400 MHz, DMSO-d6) δ 7.61 (s, 1H, H-5′), 7.52 (s, 1H, H-5), 4.06 (s, 3H, 3-OMe), 4.04 (s, 3H, 3′-OMe), 4.00 (s, 3H, 4′-OMe); 13C NMR (126 MHz, DMSO-d6) δ 158.44 (C-7′), 158.24 (C-7), 153.62 (C-4′), 152.96 (C-4), 141.37 (C-2′), 140.85 (C-3′), 140.67 (C-2), 140.18 (C-3), 113.33 (C-6′), 112.39 (C-6), 111.74 (C-1′), 111.67 (C-5), 110.80 (C-1), 107.33 (C-5′), 61.19 (3′-OMe), 60.83 (3-OMe), 56.60 (C-4′). 2D NMR see Table S6; ESI-HRMS m/z 343.0423 [M-H]- (calcd for C17H11O8 343.0459).
2.7. DNA Extraction, Polymerase Chain Reaction, and Sequencing
Genomic DNA was isolated from leaf samples using the Nucleo Spin Plant II Kit (Macherey-Nagel GmbH & Co. KG, Dueren, Germany), with minor modifications, using buffer PL2 and adding 20 µL RNAse, 30 µL mercaptoethanol and PVP (2%). The yield of DNA extraction was measured using a Qubit® 3.0 Fluorometer (Thermo Fischer Scientific, Waltham, MA, USA) and DNA bands were visualized with SYBR® Safe DNA gel stain (Thermo Fisher Scientific) on 1% agarose gels (1× TAE buffer solution) using a GenoPlex VWRⓇ gel documentation system with GenoCapture version 7.12.07.0 (Synoptics Ltd., Cambridge, UK). We performed polymerase chain reaction (PCR) of the ITS region, which comprises the ITS1 spacer, 5.8S rRNA gene, and ITS2 spacer using primers 17SE_m (5’-CGGTGAAGTGTTCGGATCG- 3’) and 26SE_m (5’-CGCTCGCCGTTACTAGGG-3’) [45], with reaction volumes of 25 µL, including 2 µL of genomic DNA, 0.3 µL of each primer, 0.5 µL BSA, 1µL DMSO and 20.9 µL, and 1 × Dream TaqGreen Master Mix, on a Labcycler Gradient PCR machine (SensoQuest GmbH, Germany). The initial denaturation step was set at 95 °C for 2 min, followed by 35 cycles of denaturation at 95 °C for 1 min, primer annealing at 53 °C for 1 min, extension at 72 °C for 2 min, followed by final extension at 72 °C for 10 min. PCR products were purified using a Nucleo Spin Gel and PCR clean-up kit (Macherey-Nagel GmbH & Co. KG, Dueren, Germany). The purified DNA samples were then measured using an Eppendorf Biophotometer to adjust the DNA concentration before Sanger sequencing at LGC Genomics GmbH, Berlin, Germany, using the same primers as mentioned above.
2.8. Phylogenetic Analyses
All sequences were aligned using the MUSCLE algorithm as implemented in Geneious 6.1.8 [46] and corrected by hand. Phylogenetic analyses were done using Maximum Likelihood (ML) with RAxML [47,48], and Bayesian inference with MrBayes v3.2.7 [49]. We used the GTR + Γ substitution model in all analyses to ensure comparability between the Maximum Likelihood and Bayesian analyses. In RaxML, we inferred the maximum likelihood tree with 100 non-parametric bootstrap replicates. In MrBayes, we ran the inference for 5 million generations (sampling every 5000 generations) with four runs and four chains each. The appropriateness of sampling (all Effective Sample Sizes (ESS) >200) was checked in Tracer v1.7.1 [50], before building a majority-rule consensus tree (with 2 million generations excluded as burn-in). In both analyses, Laguncularia racemose (L.) C.F.Gaertn. was set as outgroup.
2.9. Anti-infective Bioassays
Crude extracts (50 and 500 µg/mL) were tested in triplicate for antibacterial activity against the Gram-negative Aliivibrio fischeri and the Gram-positive Bacillus subtilis following the procedure described by dos Santos et al. [51]. Chloramphenicol (100 µM) was used as positive control and induced the complete inhibition of bacterial growth. The results are presented as relative values (% inhibition) in comparison to the negative control (bacterial growth in medium containing 1% DMSO without test compound = 0% inhibition). Negative values indicate an increase of bacterial growth, which is common with testing extracts containing further nutrients by nature.
The anthelmintic bioassay for all extracts (500 µg/mL) was performed in triplicate according to the method developed by Thomson and coworkers [52] using the model organism Caenorhabditis elegans (Bristol N2 wild-type strain) that previously was shown to correlate with anthelmintic activity against parasitic trematodes. The solvent DMSO (2%) and the standard anthelmintic drug ivermectin (10 μg/mL, 100% dead worms after 30 min incubation) were used as negative and positive controls, respectively. The number of dead and living nematodes in each sample was counted using the microscope Olympus CKX41. Results are given as percentage of dead worms.
The cytotoxic activity of selected samples (LL3, LL11, LR5, LR15) was evaluated at the concentrations of 0.05 and 50 mg/mL against the human prostate cancer cell line PC3 and the colon adenocarcinoma cell line HT-29 by determining cell viability in MTT and CV assays as described previously [51]. Digitonin (125 g/mL) was used as positive control. The results are given as percentage of control values without treatment (=100%).
3. Results and Discussion
3.1. Phytochemcial Analyses
The metabolite profiles of roots from 12 accessions of Lumnitzera littorea and 19 accessions of L. racemosa collected across Indonesia were investigated by TLC (Figure 1) and liquid chromatography, coupled on-line to mass spectrometry (LC-MS) or tandem mass spectrometry (LC-MS/MS). For structure verification, selected compounds were isolated and investigated by nuclear magnetic resonance spectroscopy (NMR).
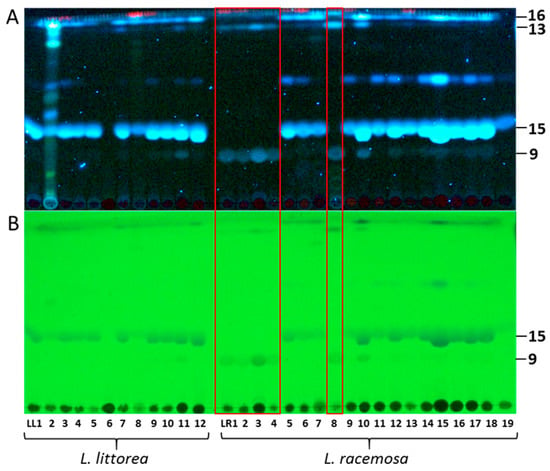
Figure 1.
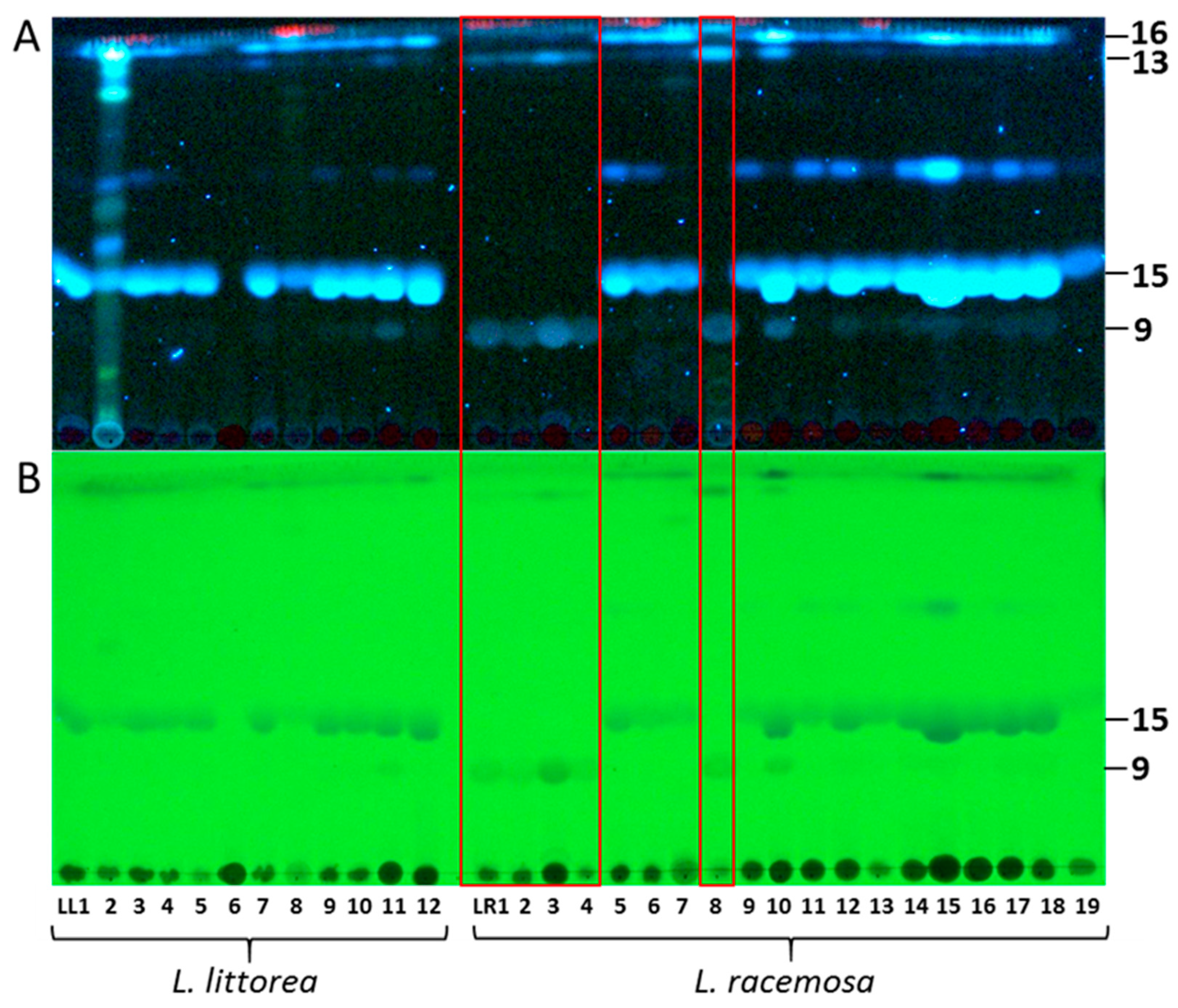
TLC profiles of root extracts from Lumnitzera littorea and L. racemosa showing the occurrence of dimethylellagic acid sulfate (9), 3,4-dimethylellagic acid (13), 3,3’,4’-trimethylellagic acid 4-sulfate (15) and 3,3’,4’-trimethylellagic acid (16): (A) Detection of bands at 366 nm (fluorescence), (B) Detection of bands at 254 nm (fluorescence quench). The red box highlights the samples LR1, LR2, LR3, LR4 and LR8 forming “Clade 2” in the phylogenetic analysis (Figure 4).
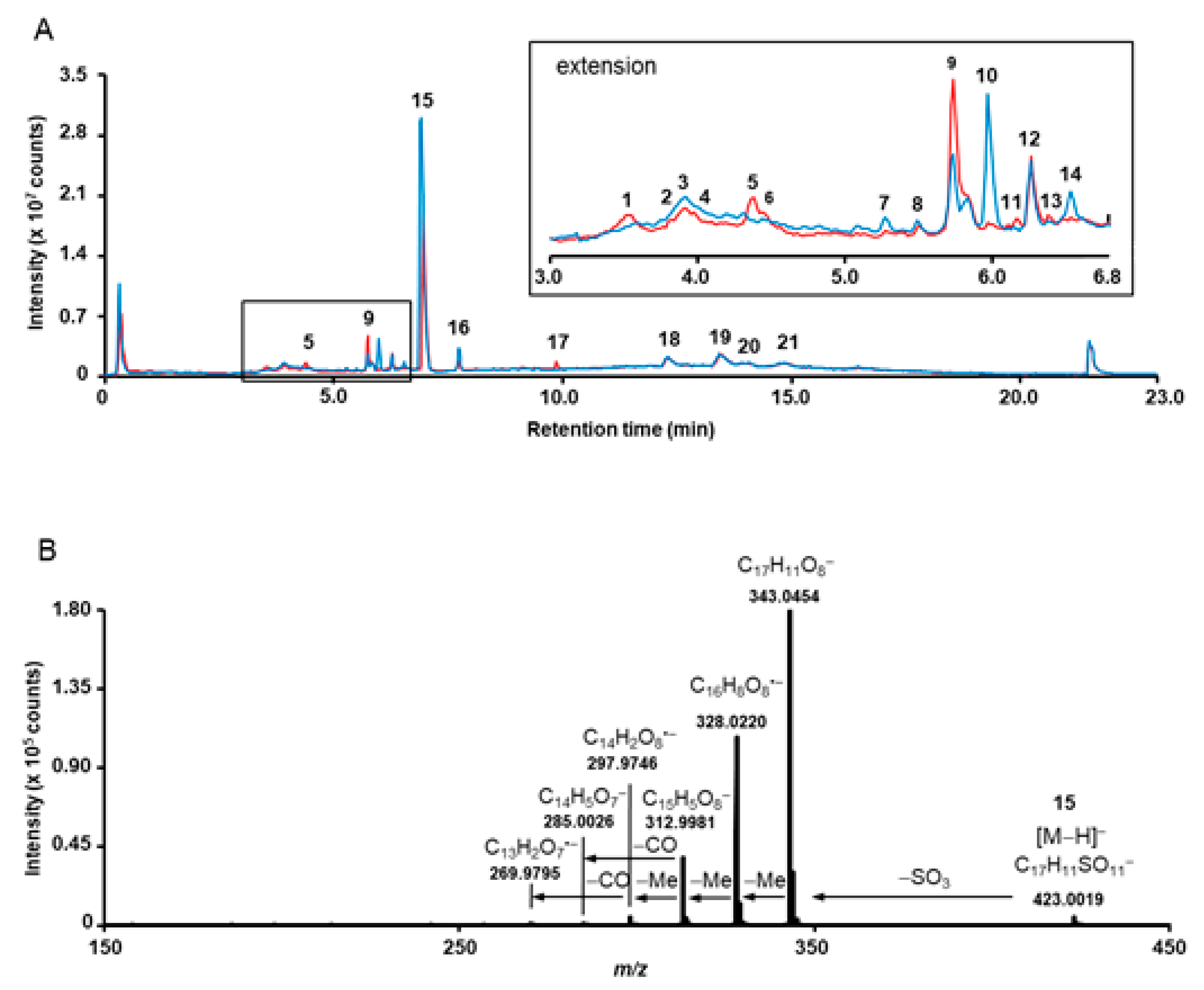
Quadrupole-time of flight mass spectrometry (QqTOF-MS) allowed annotation of 21 individual metabolites in the total ion current chromatograms (TICs) of L. littorea (LL11) and L. racemosa (LR15) root extracts (Table 2, Figure 2). The majority of the analytes could be detected in both species, while compound 17 could only be found in L. littorea, and constituents 7 and 10 are predominantly occurring in L. racemosa (Figure 2). These compounds were successfully cross-annotated in the root extracts obtained from other accessions by ultra-high-performance liquid chromatography – quadrupole mass spectrometry (RP-UHPLC-Q-MS) with detection in UV and visible (UV-VIS) spectra (Table S1, Figure S2).

Table 2.
Metabolites annotated in roots of Lumnitzera littorea and L. racemosa by reversed phase ultra-high-performance chromatography-tandem mass spectrometry (RP-UHPLC-MS/MS). The annotated metabolites are numbered according to peak numbers in Figure 2. Individual tandem mass spectra are shown in Figure S1.
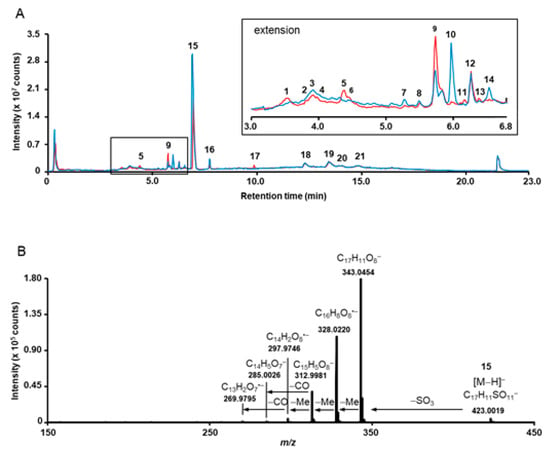
Figure 2.
Overlay of the total ion chromatograms (TICs), acquired for the methanolic extracts obtained from Lumnitzera littorea (red, LL11) and L. racemosa (blue, LR15) roots (A) and tandem mass spectrum, acquired for the m/z 423.0 corresponding to 3,3’,4’-tri-O-methylellagic acid 4-sulfate (15) (B). The insert in A represent the chromatogram section in the tR range 3.0–6.8 min. The individual annotated metabolites are numbered with rising retention time according to Table 2.
The major metabolites detected in samples from both species were ellagic acid derivatives (Figure 1, Figure 2 and Figure 3, Table 2), mostly O-methylated at different positions. Commonly, ellagic acid derivatives, including ellagitannins, are widespread in Combretaceae: Ellagic acid and several methylated derivatives were previously detected in leaves and twigs of L. racemosa [26,30] and in related species such as, e.g., in Terminalia species which are widely used in traditional medicine [53], in Pteleopsis hylodendron Mildbr. [54] and in Combretum alfredii Hance [55]. In our samples, the broad bands in TLCs along with multiple signals in specific extracted ion chromatograms (XICs) acquired in RP-UHPLC-QqTOF-MS experiments, indicate the presence of several positional isomers, i.e., compounds characterized by the same molecular weight, but featured with different substitution patterns. Such O-methylated species can be assigned by characteristic losses of 15 u, accompanying formation of an odd [M-H-15]− ion [53]. The corresponding signals are characteristic for methyl substitution both in cyclic [56] and aromatic (often in combination with a carbonyl loss) systems [57]. High intensities of such signals in tandem mass spectra of phenolic compounds typically indicate methylation as part of a methoxy group (OCH3) [58].
Surprisingly, most of the O-methylated ellagic acid derivatives contain a sulfate moiety (Figure 3). This moiety can be easily identified by a characteristic loss of 80 u from the deprotonated ions, corresponding to the cleavage of sulfur trioxide (SO3) under CID conditions resulting in the formation of characteristic [M−H−SO3]− fragments (for example m/z 315.0140 and 329.0296 for 7 and 9, respectively, Table 2, Supplementary Figure S1). This neutral loss is well-known for sulfated phenolic compounds [59] which not only occur naturally in plants (reviewed by Correira de Silva and co-workers [60]), and marine organism [61,62] but are also quite typical as an important group of polyphenol metabolites dedicated for kidney-clearance from human plasma [63]. In addition to the characteristic loss of 80 u, the tandem mass spectra of sulfated compounds feature diagnostic signals in the low m/z range, which can be attributed to SO3− (m/z 79.9567) and HSO4− (m/z 96.9594) (Table 2 and Supplementary Figure S1). However, to the best of our knowledge, sulfated ellagic acid derivatives were not reported in mangrove species before.
Remarkably, further sulfur containing compounds could be detected (compounds 1–5, 8, 11, 18–21, Table 1, Figure S1). In compounds 20 and 21 the sulfur appears to be integrated in another form than as sulfate, based on the molecular formula and the MS/MS spectra. However, the exact structures of these compounds could not be assigned based on the acquired SWATH tandem mass spectra. Compounds 8 (m/z 487.0179) and 10 (m/z 551.1027) share common fragments with ellagic acid (6) and trimethylellagic acid (16), respectively, suggesting a structural relationship. Indeed, the mass difference of 208 u between 10 and 16 is, most likely, due to moieties localized outside the aromatic core. Its elemental composition (C7H12O7) might indicate a probable presence of a sugar acid (hexahydroxyheptanoic acid) or a substituted sugar in the structure (Supplementary Figure S1–11).
The most prominent compound 15 present in both species was isolated and identified as 3,3’,4’-tri-O-methylellagic acid 4-sulfate by 1D and 2D NMR measurements (Table S5) and comparison to published data [64]. Compound 16 (Table S6) was verified as the corresponding 3,3’,4-tri-O-methylellagic acid without sulfation [65]. During separation, the sulfation of this compound is reflected in a clear enhancement of polarity visible by lower Rf value on normal phase TLC (Figure 1) and reduced retention time on reversed phase column (Figure 2). 1D and 2D NMR allowed the elucidation of compound 13 as 3,4-di-O-methylellagic acid (Table S4). The substitution pattern was established by HMBC correlations and a ROESY correlation between 4-OMe and H-5. According to its chromatographic behavior compound 9 can be assigned as the derived sulfate. Compound 6 was verified as ellagic acid by 1H NMR and comparison of MS data to a reference standard. Furthermore, we could obtain several sulfated phenylbutane derivatives including 4-(4-hydroxyphenyl)-2-butanol 2-sulfate (2, Table S2) previously isolated from roots of Rheum maximowiczii [66]. The downfield shift of H-2 and C-2 compared to the aglycon indicates sulfation at this position. However, due to the low amount of isolated compound we could not determine the configuration at C-2. Compound 3 was identified as the previously undescribed 4-(4-sulfoxy-3-methoxy phenyl)-butan-2-one (zingerone sulfate). NMR data (Table S3) are in good agreement with data published for the basic skeleton 4-(4-hydroxy- 3-methoxyphenyl)-butan-2-one [67]. Compound 4 represents according to the MS fragmentation pattern most likely the corresponding zingerol sulfate.
Sulfated phenolics are rare natural products in plants and were not described before for Combretaceae. There are only a few reports about the occurrence of sulfated ellagic acid derivatives in plants, e.g., 3-O-methylellagic acid 4-sulfate and 3,3′-di-O-methylellagic acid 4-sulfate were detected previously in Jaboticaba wood from Myrciaria cauliflora (Mart.) O. Berg (Myrtaceae) [68]. The first compound was found to exhibit antioxidant and anti-inflammatory activities in cigarette smoke extract-exposed small airway epithelial cells and may be useful for the treatment of COPD [68]. The major metabolite 3,3’,4’-tri-O-methylellagic acid 4-sulfate (15) found in this study in most of the Lumnitzera samples was reported to occur in rhizomes of Geum rivale L, [64] and roots of Potentilla candidans Humb. & Bonpl. Ex Nestl. [69], both members of the Rosaceae family.
So far, the biological function of sulfated secondary phenolics in plants is unknown. The presence of a sulfate moiety enhances the water solubility of compounds and increases ion strength in the containing tissue. The accumulation of sulfates may be organ-specific. In case of the investigated Lumnitzera species, they are found in the roots, which normally are in contact with sea water, while they were not reported from studies on leaves and twigs [26,29,30,31]. In this study, both species showed similar patterns of sulfated metabolites despite L. littorea is predominantly occurring at well-drained sites with less salinity, and typically growing as a tree, while L. racemosa is more resistant to saline conditions and occurs at the margin of bare salt pans [70] often growing as a shrub. It was postulated before that the occurrence of sulfated flavonoids in plants is an ecological trait rather than a systematic feature [64,71] and that these compounds might play a role in adaptation to water-stress in salty soils. Furthermore, natural products with a sulfated scaffold have emerged as antifouling agents with low or nontoxic effects to the environment [72] Thus, a similar function can be assumed for the detected sulfated compounds in Lumnitzera roots.
In general, marine organisms contain a significant number of phenolic metabolites which occur in sulfated form [61,62,73]. Given the high concentration of sulfate in sea water, mangrove soils contain high levels of sulfate and thus, sulfate is easily available for plants. Sulfate-reducing bacteria influence iron, phosphorus and sulfur availability in anoxic mangrove sediments and mangrove species zonation across the intertidal zone [74]. Anaerobic sulfate reducing microbial communities are involved in sulfur cycling in these soils and in the decomposition of mangrove-derived soil organic matter [8,75].
3.2. Phylogenetic Analyses
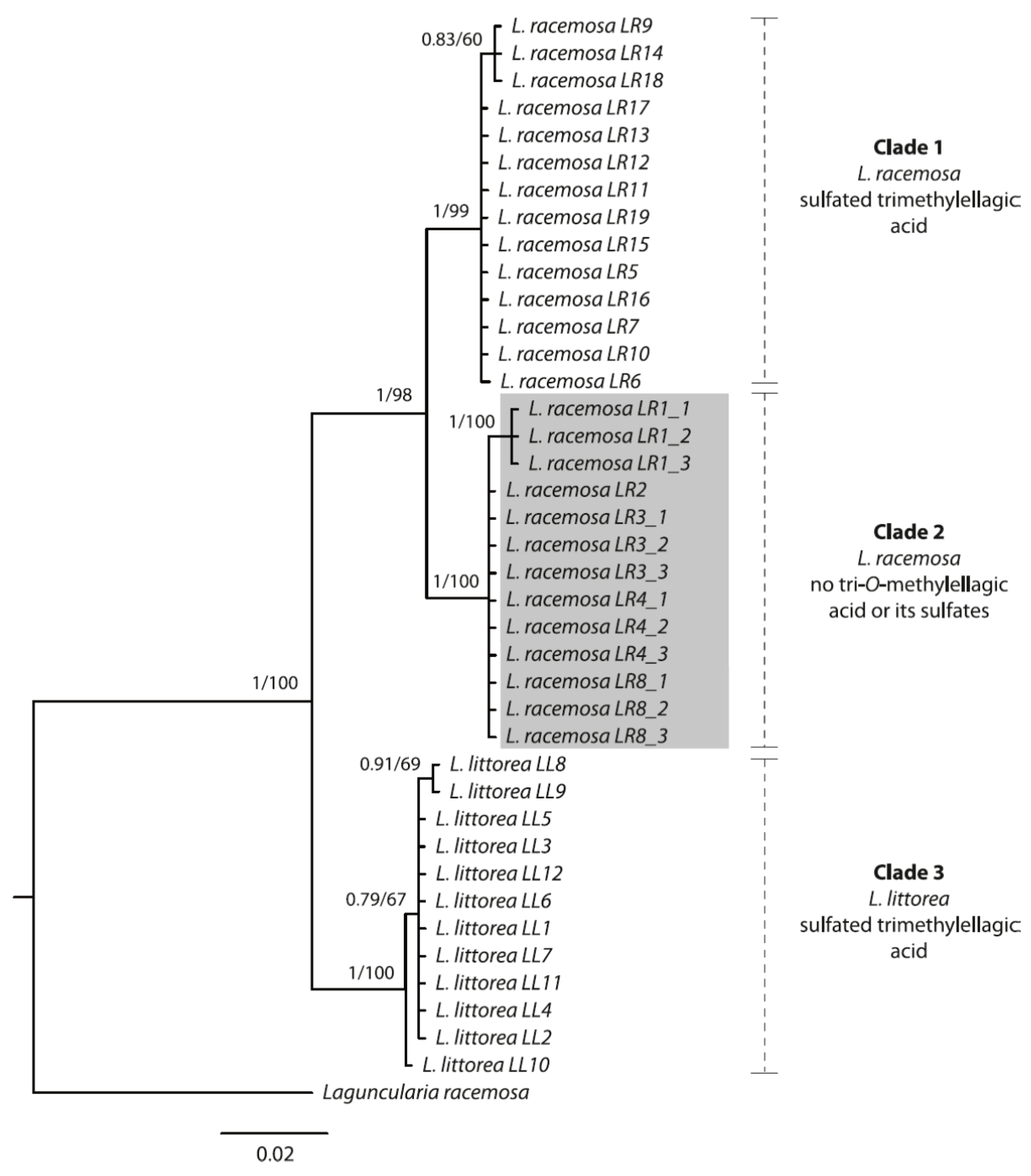
The variation of the metabolite pattern across different populations could be supported by molecular phylogenetic evidence. As Bayesian and ML analyses yielded topologically identical trees, we here only show the results from the Bayesian analysis (with support values from both; Figure 4).
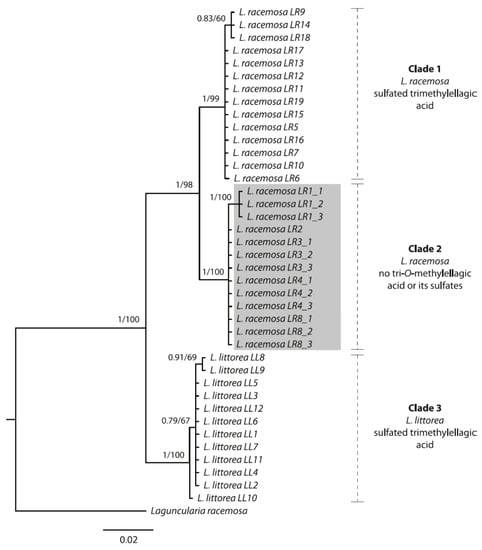
Figure 4.
Bayesian majority-rule consensus tree based on the ITS rDNA region of Lumnitzera littorea and L. racemosa samples collected from various locations in Indonesia. Numbers above branches show Bayesian posterior probability values and corresponding bootstrap support values from the maximum likelihood analysis. Grey: highly supported clade, forming a different chemotype.
Both species form well-supported monophyletic groups. Notably, within L. racemosa, samples from populations LR1, LR2, LR3, LR4 and LR8 form a well-supported clade (Clade 2, Figure 4), which is characterized by sharing the same TLC and mass spectrometry metabolic profile, thus suggesting a different chemotype. These samples completely lacked sulfated and nonsulfated trimethylellagic acids, but were dominated by dimethylellagic acid and its sulfate. In contrast, the phylogenetic tree does not show a geographical pattern, at least not on the level where we have appropriate resolution. In order to confirm these results, we sequenced a second individual from each of these specific populations, except for LR2 (for which only one plant had been found at the location). On the one hand, the infraspecific, interpopulational variation in metabolic profiles calls for caution when selecting cultivated mangrove plants for the purpose of metabolic profiling for medicinal purposes (compare to [76]). On the other hand, the nuclear ITS region seems to constitute a useful guide for selecting individuals for cultivation, at least for Lumnitzera and some other mangrove plant species (this study, and [76]).
3.3. Evaluation of Anti-Infective Properties
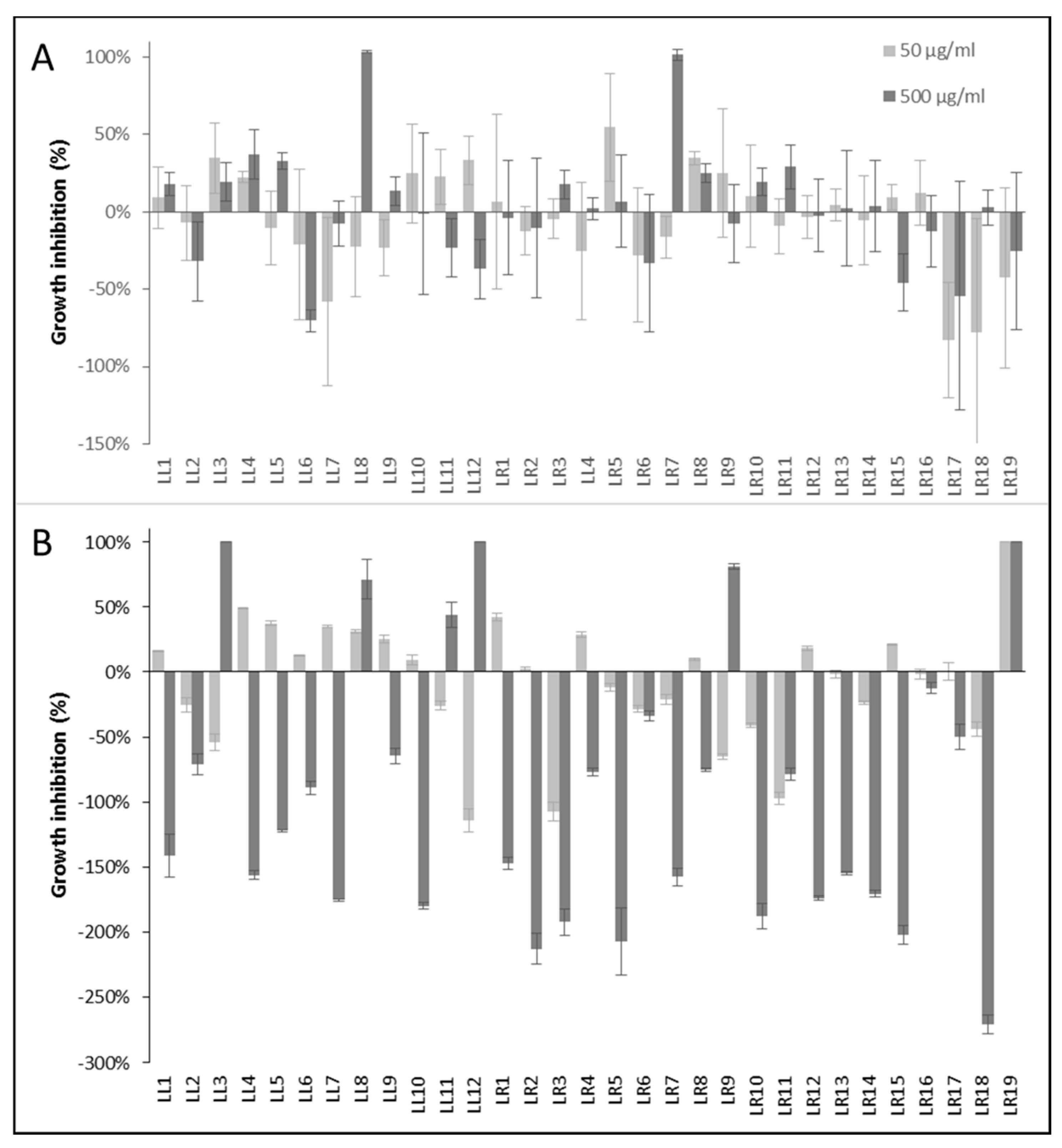
To evaluate the anti-infective potential of the Lumnitzera root extracts, the antibacterial and anthelmintic activities were determined using nonpathogenic model organism as test systems. The samples from different locations varied significantly in their antibacterial activities (Figure 5).
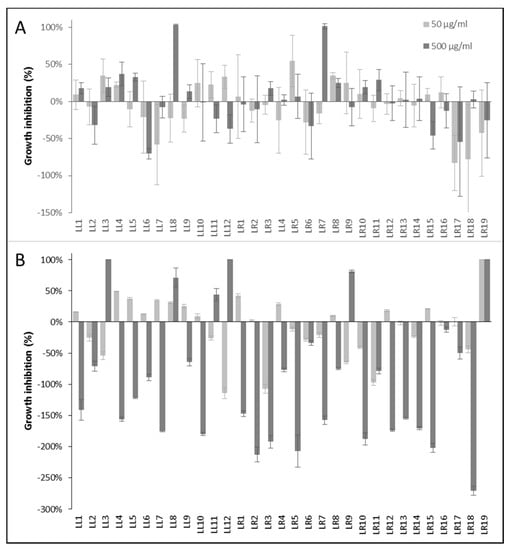
Figure 5.
Antibacterial activity of Lumnitzera littorea and L. racemosa extracts against (A) Gram-positive Bacillus subtilis and (B) Gram-negative Aliivibrio fischeri. Chloramphenicol (100 µM) was used as positive control and induced the complete inhibition of bacterial growth. Negative values indicate growth enhancement.
Only two extracts (LL8 and LR7) completely inhibited the growth of the Gram-positive Bacillus subtilis at 500 µg/mL after 16 h (Figure 5A). The activity against Gram-negative bacteria showed a different pattern (Figure 5B). Here, samples LL3, LL12 and LR19 completely inhibited the growth of the test organism Allivibrio fischeri at a concentration of 500 µg/mL after 24 h, LL8 and LR9 induced 71% and 81% inhibition, respectively. Especially the extract LR19 exhibited a noteworthy antibacterial potential against the Gram-negative test organism since it induced 100% growth inhibition already at the lower test concentration of 50 µg/mL. At this point, the assignment of the antibacterial properties to a certain (group of) constituents is not possible. The selective antibacterial activity of different accessions of only a few samples of the same plant species suggests, however, that the effects might be connected to associated microorganisms rather than intrinsic plant-produced metabolites.
None of the Lumnitzera crude extracts showed anthelmintic activity against the nematode Caenorhabditis elegans (Figure S3). This is in contrast to studies on mangrove plant species from other families. For example, anthelmintic activity was found for leaf and stem extracts of Acanthus ilicifolius L. (Acanthaceae) [77,78].
Two samples from each species (LL3, LL11, LR5, LR15) were exemplarily tested for their cytotoxic activity against the human prostate cancer cell line PC3 and the colon adenocarcinoma cell line HT-29. The investigated samples did not influence the viability of the cancer cell lines at a concentration of 0.05 µg/mL, however, completely inhibited the cell growth at a concentration of 50 µg/mL indicating a moderate cytotoxic potential (Figure S4).
Ellagic acid is known to possess a wide range of biological activities based on its antioxidant and chemopreventive potential including antimicrobial, anti-inflammatory, neuroprotective, antihepatotoxic, anticholestatic, antifibrogenic, anticarcinogenic, cytotoxic, and antiviral effects [79,80]. Several of these multiple activities may, however, be related to general tanning properties of polyphenolics, rather than being specific effects [81,82]. Furthermore, ellagic acid and related compounds are potent Aldose reductase inhibitors that could play an important role in the management of diabetic complications [69]. Methylation often reduces these effects whereas the introduction of a sulfate group increases the inhibitory activity [69], but reduces membran permeability. In contrast, the only moderate cytotoxicity of ellagic acid against cancer cell lines is further decreased by the presence of a sulfate moiety [64]. The potassium salts of 3,3’-dimethylellagic acid 4-sulfate and 3,3’,4’-trimethylellagic acid 4-sulfate were also found to exhibit moderate antimicrobial potential against Gram-positive Bacillus subtilis and Staphylococcus aureus with MIC values in the range of 22.5–50.8 µg/mL [83]. These compounds were, however, not effective against Gram-negative Escherichia coli. [83].
In our investigations the antibacterial activity is likely not directly connected to the sulfated ellagic acid derivatives or the other sulfated metabolites. These metabolites occur across all samples, but the antibacterial effects are limited to samples from particular locations (Figure 5). Nevertheless, Lumnitzera roots are a promising source for pharmacologically interesting sulfated ellagic acid derivatives and further sulfated plant metabolites.
4. Conclusions
In our study, a series of unusual sulfated constituents was characterized in root samples from the mangrove species Lumnitzera littorea and L. racemosa (Combretaceae). Thus, most of the methylated ellagic acid derivatives isolated from both species possess a sulfate moiety. However, L. racemosa samples from North Sumatra (LR1), Aceh (LR2, LR3), East Kalimantan (LR4), and Maluku (LR8), completely lack sulfated and non-sulfated trimethylellagic acid, but instead are dominated by dimethylellagic acid and its sulfate. This phytochemical pattern is corroborated by phylogenetic data, where these specific samples form a well-supported clade in the ITS tree. Interestingly, the occurrence of antimicrobial activity and sulfated ellagic acid derivatives are not connected. Although the ellagic acid derivatives are present within all samples, the antibacterial effects are limited to samples from particular locations in Indonesia, suggesting that other compounds from the plant or root-associated microorganisms might be responsible. The moderate cytotoxic effect, in contrast, can be attributed to the occurrence of the ellagic acid derivatives. In summary, Lumnitzera roots represent a potentially promising source for sulfated ellagic acid derivatives and further sulfur containing plant metabolites.
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/separations8060082/s1, Figure S1: MS/MS spectra of compounds 1–21, Figure S2: PDA chromatograms of root extracts from L. littorea (LL1-LL12) and L. racemosa (LR1-LR19), Figure S3: Anthelmintic activity of root extracts from Lumnitzera littorea (LL1-LL12) and L. racemosa (LR1-LR19), Figure S4: Cytotoxic activity of root extracts from L. littorea (LL3, LL11) and L. racemosa (LR5, LR15) against PC3 and HT29 cells, Figure S5: NMR and MS spectra of compound 2, Figure S6: NMR and MS spectra of compound 3, Figure S7: NMR and MS spectra of compound 6, Figure S8: NMR and MS spectra of compound 13, Figure S9: NMR and MS spectra of compound 15, Figure S10: NMR and MS spectra of compound 16, Table S1: Peak areas (TIC) of main compounds detected in root extracts of L. littorea (LL1-LL12) and L. racemosa (LR1-LR19), Table S2: 2D NMR data of compound 2, Table S3: 2D NMR data of compound 3, Table S4: 2D NMR data of compound 13, Table S5: 2D NMR data of compound 15, Table S6: 2D NMR data of compound 16.
Author Contributions
Conceptualization, J.M., A.N.M.-R. and L.A.W.; methodology, K.F., A.F., A.N.M.-R. and J.S.; validation, K.F., A.F., A.N.M.-R. and J.S.; formal analysis, J.M., K.F., J.K. and J.S.; investigation, J.M. and J.K.; resources, A.N.M.-R.; data curation, K.F., A.N.M.-R. and J.S.; writing—original draft preparation, J.M. and K.F.; writing—review and editing, A.N.M.-R., J.M., J.K., J.S., K.F., A.F., L.A.W. and A.A.; visualization, J.M., K.F., J.K., A.F. and J.S.; supervision, A.N.M.-R., L.A.W, J.S., K.F. and A.A.; project administration, A.N.M.-R. and L.A.W.; funding acquisition, J.M., A.N.M.-R. and L.A.W. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by BMBF (grants no. 16GW0120K and 16GW0123 to Alexandra Muellner-Riehl and Ludger Wessjohann) within the frame of the project “Indonesian Plant Biodiversity and Human Health – BIOHEALTH”. Jeprianto Manurung is funded by a PhD grant from the German Academic Exchange Service (DAAD project no. 91653688), hosted by Alexandra Muellner-Riehl. Jan Schnitzler was supported by the German Centre for Integrative Biodiversity Research (iDiv) Halle-Jena-Leipzig funded by the German Research Foundation (DFG—FZT 118). Jonas Kappen and Katrin Franke also received funds within the frame of the project ProCognito by EFRE and the state Saxony-Anhalt (ZS/2018/11/95581).
Institutional Review Board Statement
This work contains no human or animal studies.
Informed Consent Statement
This work contains no human studies. The plant studies were done according to Prior Informed Consent with LIPI as Indonesian representative.
Data Availability Statement
Plant material: Plant material is deposited at LIPI, Bogor, Indonesia. Chemical data: All primary data and reference compounds are stored at the IPB primary data storage for 10+ years, and in the compound depository to the extent available or stable. Pending availability, detailed data can be shared upon request. DNA data: All newly generated ITS sequences for this study have been deposited in GenBank (https://www.ncbi.nlm.nih.gov/genbank/) and are available from there.
Acknowledgments
The authors would like to thank Lina Juswara, Hetty I.P. Normakristagaluh, Ruliyana Susanti, and Witjaksono (all Research Center for Biology, Indonesian Institute of Sciences, LIPI) for administrative support regarding material collection; Fajria Novari (Ministry of Environment and Forestry Republic of Indonesia) for help with plant material export permits; and all local forestry officers at the different sampling locations for logistic support. We are grateful to Anke Dettmer, Mthandazo Dube, and Martina Lerbs (all IPB Halle) for the performance of antibacterial, anthelmintic and cytotoxic bioassays, respectively. Thanks to Andrea Porzel and Gudrun Hahn (IPB Halle) for NMR measurements. We acknowledge support from Leipzig University for Open Access Publishing.
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
References
- Kathiresan, K.; Bingham, B.L. Biology of mangroves and mangrove ecosystems. Adv. Mar. Biol. 2001, 40, 81–251. [Google Scholar]
- Noor, Y.R.; Khazali, M.; Suryadiputra, I.N.N. Panduan Pengenalan Mangrove di Indonesia; Ditjen PHKA; Wetlands International Indonesia Programme: Bogor, India, 2006; ISBN 9799589908. [Google Scholar]
- Giesen, W.; Wulffraat, S.; Zieren, M.; Scholten, L. Mangrove Guidebook for Southeast Asia; FAO Regional Office for Asia and the Pacific, Wetlands International: Bangkok, Thailand, 2007; ISBN 974-7946-85-8. [Google Scholar]
- Bandaranayake, W.M. Traditional and medicinal uses of mangroves. Mangroves Salt Marshes 1998, 2, 133–148. [Google Scholar] [CrossRef]
- Kovacs, J.M. Assessing mangrove use at the local scale. Landsc. Urban Plan. 1999, 43, 201–208. [Google Scholar] [CrossRef]
- Pattanaik, C.; Reddy, C.S.; Dhal, N.K.; Das, R. Utilisation of mangrove forests in Bhitarkanika wildlife sanctuary, Orissa. Indian J. Tradit. Knowl. 2008, 7, 598–603. [Google Scholar]
- Govindasamy, C.; Kannan, R. Pharmacognosy of mangrove plants in the system of unani medicine. Asian Pac. J. Trop. Dis. 2012, 2, S38–S41. [Google Scholar] [CrossRef]
- Balk, M.; Keuskamp, J.A.; Laanbroek, H.J. Potential for sulfate reduction in mangrove forest soils: Comparison between two dominant species of the Americas. Front. Microbiol. 2016, 7, 1855. [Google Scholar] [CrossRef]
- Spalding, M.; Blasco, F.; Field, C.D. World Mangrove Atlas; International Society for Mangrove Ecosystems: Okinawa, Japan; World Conservation Monitoring Centre: Cambridge, UK; International Tropical Timber Organization: Yokohama, Japan, 1997; ISBN 4906584039. [Google Scholar]
- Giri, C.; Ochieng, E.; Tieszen, L.L.; Zhu, Z.; Singh, A.; Loveland, T.; Masek, J.; Duke, N. Status and distribution of mangrove forests of the world using earth observation satellite data. Glob. Ecol. Biogeogr. 2011, 20, 154–159. [Google Scholar] [CrossRef]
- Fyhrquist, P.; Mwasumbi, L.; Hæggström, C.-A.; Vuorela, H.; Hiltunen, R.; Vuorela, P. Ethnobotanical and antimicrobial investigation on some species of Terminalia and Combretum (Combretaceae) growing in Tanzania. J. Ethnopharmacol. 2002, 79, 169–177. [Google Scholar] [CrossRef]
- Martini, N.D.; Katerere, D.R.P.; Eloff, J.N. Biological activity of five antibacterial flavonoids from Combretum erythrophyllum (Combretaceae). J. Ethnopharmacol. 2004, 93, 207–212. [Google Scholar] [CrossRef] [PubMed]
- Eloff, J.N.; Famakin, J.O.; Katerere, D.R.P. Isolation of an antibacterial stilbene from Combretum woodii (Combretaceae) leaves. Afr. J. Biotechnol. 2005, 4, 1167–1171. [Google Scholar]
- Eloff, J.N.; Famakin, J.O.; Katerere, D.R.P. Combretum woodii (Combretaceae) leaf extracts have high activity against Gram-negative and Gram-positive bacteria. Afr. J. Biotechnol. 2005, 4, 1161–1166. [Google Scholar]
- Eloff, J.N.; Katerere, D.R.; McGaw, L.J. The biological activity and chemistry of the southern African Combretaceae. J. Ethnopharmacol. 2008, 119, 686–699. [Google Scholar] [CrossRef]
- Aderogba, M.A.; Kgatle, D.T.; McGaw, L.J.; Eloff, J.N. Isolation of antioxidant constituents from Combretum apiculatum subsp. apiculatum. S. Afr. J. Bot. 2012, 79, 125–131. [Google Scholar] [CrossRef]
- Masoko, P.; Eloff, J.N. The diversity of antifungal compounds of six South African Terminalia species (Combretaceae) determined by bioautography. Afr. J. Biotechnol. 2005, 4, 1425–1431. [Google Scholar]
- Masoko, P.; Picard, J.; Eloff, J.N. The antifungal activity of twenty-four southern African Combretum species (Combretaceae). S. Afr. J. Bot. 2007, 73, 173–183. [Google Scholar] [CrossRef]
- Lin, T.-C.; Hsu, F.-L.; Cheng, J.-T. Antihypertensive Activity of Corilagin and Chebulinic Acid, Tannins from Lumnitzera racemosa. J. Nat. Prod. 1993, 56, 629–632. [Google Scholar] [CrossRef]
- Ravikumar, S.; Gnanadesigan, M. Hepatoprotective and antioxidant activity of a mangrove plant Lumnitzera racemosa. Asian Pac. J. Trop. Biomed. 2011, 1, 348–352. [Google Scholar] [CrossRef]
- Thao, N.P.; Bui, T.T.L.; Chau, N.D.; Bui, H.T.; Kim, E.J.; Kang, H.K.; Lee, S.H.; Jang, H.D.; Nguyen, T.C.; van Nguyen, T.; et al. In vitro evaluation of the antioxidant and cytotoxic activities of constituents of the mangrove Lumnitzera racemosa Willd. Arch. Pharm. Res. 2015, 38, 446–455. [Google Scholar] [CrossRef]
- Darwish, A.G.G.; Samy, M.N.; Sugimoto, S.; Abdel-Salam, H.; Matsunami, K. Effects of Hepatoprotective compounds from the leaves of Lumnitzera racemosa on acetaminophen-induced liver damage in vitro. Chem. Pharm. Bull. 2016, 64, 360–365. [Google Scholar] [CrossRef] [PubMed]
- Paul, T.; Ramasubbu, S. The antioxidant, anticancer and anticoagulant activities of Acanthus ilicifolius L. roots and Lumnitzera racemosa Willd. leaves, from southeast coast of India. J. Appl. Pharm. Sci. 2017, 7, 081–087. [Google Scholar] [CrossRef]
- D’Souza, L.; Wahidulla, S.; Devi, P. Antibacterial phenolics from the mangrove Lumnitzera racemosa. Indian J. Mar. Sci. 2010, 39, 294–298. [Google Scholar]
- Abeysinghe, P.D. Antibacterial activity of aqueous and ethanol extracts of mangrove species collected from Southern Sri Lanka. Asian J. Pharm. Biol. Res. 2012, 2, 79–83. [Google Scholar]
- Yu, S.-Y.; Wang, S.-W.; Hwang, T.-L.; Wei, B.-L.; Su, C.-J.; Chang, F.-R.; Cheng, Y.-B. Components from the leaves and twigs of mangrove Lumnitzera racemosa with anti-angiogenic and anti-inflammatory effects. Mar. Drugs 2018, 16, 404. [Google Scholar] [CrossRef]
- Eswaraiah, G.; Peele, K.A.; Krupanidhi, S.; Indira, M.; Kumar, R.B.; Venkateswarulu, T.C. GC–MS analysis for compound identification in leaf extract of Lumnitzera racemosa and evaluation of its in vitro anticancer effect against MCF7 and HeLa cell lines. J. King Saud Univ. Sci. 2020, 32, 780–783. [Google Scholar] [CrossRef]
- Gosh, M.S.; Gouri, P. Chemical investigation of some mangrove species: Part VIII. Lumnitzera racemosa. J. Indian Chem. Soc. 1980, 57, 568–570. [Google Scholar]
- Saad, S.; Taher, M.; Susanti, D.; Qaralleh, H.; Rahim, N.A.B.A. Antimicrobial activity of mangrove plant (Lumnitzera littorea). Asian Pac. J. Trop. Biomed. 2011, 4, 523–525. [Google Scholar] [CrossRef]
- Le Nguyen, T.T.; Pham, T.T.; Hansen, P.E.; Nguyen, P.K.P. In vitro α-glucosidase inhibitory activity of compounds isolated from mangrove Lumnitzera littorea leaves. Sci. Technol. Dev. J. 2019, 22. [Google Scholar] [CrossRef]
- Wongsomboon, P.; Maneerat, W.; Pyne, S.G.; Vittaya, L.; Limtharakul, T.R. 12-Hydroxycorniculatolide a from the mangrove tree, Lumnitzera littorea. Nat. Prod. Commun. 2018, 13, 1327–1328. [Google Scholar] [CrossRef]
- Debbab, A.; Aly, A.H.; Proksch, P. Mangrove derived fungal endophytes—A chemical and biological perception. Fungal Divers. 2013, 61, 1–27. [Google Scholar] [CrossRef]
- Ancheeva, E.; Daletos, G.; Proksch, P. Lead compounds from mangrove-associated microorganisms. Mar. Drugs 2018, 16, 319. [Google Scholar] [CrossRef]
- Deng, Q.; Li, G.; Sun, M.; Yang, X.; Xu, J. A new antimicrobial sesquiterpene isolated from endophytic fungus Cytospora sp. from the Chinese mangrove plant Ceriops tagal. Nat. Prod. Res. 2018, 555–564. [Google Scholar] [CrossRef] [PubMed]
- Chaeprasert, S.; Piapukiew, J.; Whalley, A.J.; Sihanonth, P. Endophytic fungi from mangrove plant species of Thailand: Their antimicrobial and anticancer potentials. Bot. Mar. 2010, 53, 9. [Google Scholar] [CrossRef]
- Raab, A.; Feldmann, J. Biological sulphur-containing compounds—Analytical challenges. Anal. Chim. Acta 2019, 1079, 20–29. [Google Scholar] [CrossRef]
- Saslis-Lagoudakis, C.H.; Savolainen, V.; Williamson, E.M.; Forest, F.; Wagstaff, S.J.; Baral, S.R.; Watson, M.F.; Pendry, C.A.; Hawkins, J.A. Phylogenies reveal predictive power of traditional medicine in bioprospecting. Proc. Natl. Acad. Sci. USA 2012, 109, 15835–15840. [Google Scholar] [CrossRef]
- Holzmeyer, L.; Hartig, A.-K.; Franke, K.; Brandt, W.; Muellner-Riehl, A.N.; Wessjohann, L.A.; Schnitzler, J. Evaluation of plant sources for antiinfective lead compound discovery by correlating phylogenetic, spatial, and bioactivity data. Proc. Natl. Acad. Sci. USA 2020, 117, 12444–12451. [Google Scholar] [CrossRef]
- Leonhardt, S.D.; Rasmussen, C.; Schmitt, T. Genes versus environment: Geography and phylogenetic relationships shape the chemical profiles of stingless bees on a global scale. Proc. Biol. Sci. 2013, 280, 20130680. [Google Scholar] [CrossRef]
- Schmitt, I.; Barker, F.K. Phylogenetic methods in natural product research. Nat. Prod. Rep. 2009, 26, 1585–1602. [Google Scholar] [CrossRef]
- Adamek, M.; Alanjary, M.; Ziemert, N. Applied evolution: Phylogeny-based approaches in natural products research. Nat. Prod. Rep. 2019, 36, 1295–1312. [Google Scholar] [CrossRef]
- Mawalagedera, S.M.U.P.; Callahan, D.L.; Gaskett, A.C.; Rønsted, N.; Symonds, M.R.E. Combining evolutionary enference and metabolomics to identify plants with medicinal potential. Front. Ecol. Evol. 2019, 7, 471. [Google Scholar] [CrossRef]
- Abdelmohsen, U.R.; Pimentel-Elardo, S.M.; Hanora, A.; Radwan, M.; Abou-El-Ela, S.H.; Ahmed, S.; Hentschel, U. Isolation, phylogenetic analysis and anti-infective activity screening of marine sponge-associated actinomycetes. Mar. Drugs 2010, 8, 399–412. [Google Scholar] [CrossRef]
- Prasad, M.A.; Zolnik, C.P.; Molina, J. Leveraging phytochemicals: The plant phylogeny predicts sources of novel antibacterial compounds. Future Sci. OA 2019, 5, FSO407. [Google Scholar] [CrossRef]
- Grudinski, M.; Pannell, C.M.; Chase, M.W.; Ahmad, J.A.; Muellner-Riehl, A.N. An evaluation of taxonomic concepts of the widespread plant genus Aglaia and its allies across Wallace’s Line (tribe Aglaieae, Meliaceae). Mol. Phylogenet. Evol. 2014, 73, 65–76. [Google Scholar] [CrossRef] [PubMed]
- Kearse, M.; Moir, R.; Wilson, A.; Stones-Havas, S.; Cheung, M.; Sturrock, S.; Buxton, S.; Cooper, A.; Markowitz, S.; Duran, C.; et al. Geneious Basic: An integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 2012, 28, 1647–1649. [Google Scholar] [CrossRef]
- Silvestro, D.; Michalak, I. raxmlGUI: A graphical front-end for RAxML. Org. Divers. Evol. 2012, 12, 335–337. [Google Scholar] [CrossRef]
- Stamatakis, A. RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 2014, 30, 1312–1313. [Google Scholar] [CrossRef] [PubMed]
- Ronquist, F.; Teslenko, M.; van der Mark, P.; Ayres, D.L.; Darling, A.; Höhna, S.; Larget, B.; Liu, L.; Suchard, M.A.; Huelsenbeck, J.P. MrBayes 3.2: Efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 2012, 61, 539–542. [Google Scholar] [CrossRef]
- Rambaut, A.; Drummond, A.J.; Xie, D.; Baele, G.; Suchard, M.A. Posterior summarization in Bayesian phylogenetics using tracer 1.7. Syst. Biol. 2018, 67, 901–904. [Google Scholar] [CrossRef]
- Dos Santos, C.H.C.; de Carvalho, M.G.; Franke, K.; Wessjohann, L. Dammarane-type triterpenoids from the stem of Ziziphus glaziovii Warm. (Rhamnaceae). Phytochemistry 2019, 162, 250–259. [Google Scholar] [CrossRef] [PubMed]
- Thomsen, H.; Reider, K.; Franke, K.; Wessjohann, L.A.; Keiser, J.; Dagne, E.; Arnold, N. Characterization of constituents and anthelmintic properties of Hagenia abyssinica. Sci. Pharm. 2012, 80, 433–446. [Google Scholar] [CrossRef]
- Singh, A.; Bajpai, V.; Kumar, K.; Sharma, K.; Kumar, B. Profiling of gallic and ellagic acid derivatives in different plant parts of Terminalia Arjuna by HPLC-ESI-QTOF-MS/MS. Nat. Prod. Comm. 2016, 11, 239–244. [Google Scholar] [CrossRef]
- Atta-Ur-Rahman; Ngounou, F.N.; Choudhary, M.I.; Malik, S.; Makhmoor, T.; Nur-E-Alam, M.; Zareen, S.; Lontsi, D.; Ayafor, J.F.; Sondengam, B.L. New antioxidant and antimicrobial ellagic acid derivatives from Pteleopsis hylodendron. Planta Med. 2001, 67, 335–339. [Google Scholar] [CrossRef]
- Bai, M.; Wu, L.-J.; Cai, Y.; Wu, S.-Y.; Song, X.-P.; Chen, G.-Y.; Zheng, C.-J.; Han, C.-R. One new lignan derivative from the Combretum alfredii Hance. Nat. Prod. Res. 2017, 31, 1022–1027. [Google Scholar] [CrossRef] [PubMed]
- Bathe, U.; Frolov, A.; Porzel, A.; Tissier, A. CYP76 oxidation network of abietane diterpenes in Lamiaceae reconstituted in yeast. J. Agric. Food Chem. 2019, 67, 13437–13450. [Google Scholar] [CrossRef]
- Song, R.; Lin, H.; Zhang, Z.; Li, Z.; Xu, L.; Dong, H.; Tian, Y. Profiling the metabolic differences of anthraquinone derivatives using liquid chromatography/tandem mass spectrometry with data-dependent acquisition. Rapid Commun. Mass Spectrom. 2009, 23, 537–547. [Google Scholar] [CrossRef] [PubMed]
- Frolov, A.; Henning, A.; Böttcher, C.; Tissier, A.; Strack, D. An UPLC-MS/MS method for the simultaneous identification and quantitation of cell wall phenolics in Brassica napus seeds. J. Agric. Food Chem. 2013, 61, 1219–1227. [Google Scholar] [CrossRef]
- Geng, C.-A.; Chen, H.; Chen, X.-L.; Zhang, X.-M.; Lei, L.-G.; Chen, J.-J. Rapid characterization of chemical constituents in Saniculiphyllum guangxiense by ultra fast liquid chromatography with diode array detection and electrospray ionization tandem mass spectrometry. Int. J. Mass Spectrom. 2014, 361, 9–22. [Google Scholar] [CrossRef]
- Correia-da-Silva, M.; Sousa, E.; Pinto, M.M.M. Emerging sulfated flavonoids and other polyphenols as drugs: Nature as an inspiration. Med. Res. Rev. 2014, 34, 223–279. [Google Scholar] [CrossRef]
- Subhashini, P.; Dilipan, E.; Thangaradjou, T.; Papenbrock, J. Bioactive natural products from marine angiosperms: Abundance and functions. Nat. Prod. Bioprospect. 2013, 3, 129–136. [Google Scholar] [CrossRef]
- Kurth, C.; Welling, M.; Pohnert, G. Sulfated phenolic acids from Dasycladales siphonous green algae. Phytochemistry 2015, 117, 417–423. [Google Scholar] [CrossRef] [PubMed]
- Mullen, W.; Borges, G.; Lean, M.E.J.; Roberts, S.A.; Crozier, A. Identification of metabolites in human plasma and urine after consumption of a polyphenol-rich juice drink. J. Agric. Food Chem. 2010, 58, 2586–2595. [Google Scholar] [CrossRef]
- Owczarek, A.; Różalski, M.; Krajewska, U.; Olszewska, M. Rare ellagic acid sulphate derivatives from the rhizome of Geum rivale L.—Structure, cytotoxicity, and validated HPLC-PDA Assay. Appl. Sci. 2017, 7, 400. [Google Scholar] [CrossRef]
- Khac, D.D.; Tran-Van, S.; Campos, A.M.; Jean-Yves Lallemand, J.Y.; Fetizon, M. Ellagic compounds from Diplopanax stachyathus. Phytochemistry 1990, 29, 251–256. [Google Scholar] [CrossRef]
- Shikishima, Y.; Takaishi, Y.; Honda, G.; Ito, M. Phenylbutanoids and stilbene derivatives of Rheum maximowiczii. Phytochemistry 2001, 56, 377–381. [Google Scholar] [CrossRef]
- Pabst, A.; Barron, D.; Adda, J.; Schreier, P. Phenylbutan-2-one β-d-glucosides from raspberry fruit. Phytochemistry 1990, 29, 3853–3858. [Google Scholar] [CrossRef]
- Zhao, D.-K.; Shi, Y.-N.; Petrova, V.; Yue, G.G.L.; Negrin, A.; Wu, S.-B.; D’Armiento, J.M.; Lau, C.B.S.; Kennelly, E.J. Jaboticabin and related polyphenols from Jaboticaba (Myrciaria cauliflora) with anti-inflammatory activity for Chronic Obstructive Pulmonary Disease. J. Agric. Food Chem. 2019, 67, 1513–1520. [Google Scholar] [CrossRef] [PubMed]
- Terashima, S.; Shimizu, M.; Nakayama, H.; Ishikura, M.; Ueda, Y.; Imai, K.; Suzui, A.; Morita, N. Terashima et al 1990: Studies on aldose reductase Inhibitors from Medicinal plant of "Sinfito" Potentilla candicans, and further synthesis of their related compounds. Chem. Pharm. Bull. 1990, 38, 2733–2736. [Google Scholar] [CrossRef]
- Tomlinson, P.B.; Bunt, J.S.; Primack, R.B.; Duke, N.C. Lumnitzera rosea (Combretaceae)—Its status and floral morphology. J. Arnold Arbor. 1978, 59, 342–351. [Google Scholar]
- Harborne, J.B. Flavonoid sulphates: A new class of sulphur compounds in higher plants. Phytochemistry 1975, 14, 1147–1155. [Google Scholar] [CrossRef]
- Almeida, J.R.; Correia-da-Silva, M.; Sousa, E.; Antunes, J.; Pinto, M.; Vasconcelos, V.; Cunha, I. Antifouling potential of nature-inspired sulfated compounds. Sci. Rep. 2017, 7, 42424. [Google Scholar] [CrossRef]
- Filho, L.C.K.; Picão, B.W.; Silva, M.L.A.; Cunha, W.R.; Pauletti, P.M.; Dias, G.M.; Copp, B.R.; Bertanha, C.S.; Januario, A.H. Bioactive aliphatic sulfates from marine invertebrates. Mar. Drugs 2019, 17, 527. [Google Scholar] [CrossRef]
- Sherman, R.E.; Fahey, T.J.; Howarth, R.W. Soil-plant interactions in a neotropical mangrove forest: Iron, phosphorus and sulfur dynamics. Oecologia 1998, 115, 553–563. [Google Scholar] [CrossRef] [PubMed]
- Kristensen, E.; Holmer, M.; Bussarawit, N. Benthic metabolism and sulfate reduction in a Southeast Asian mangrove swamp. Mar. Ecol. Prog. Ser. 1991, 73, 93–103. [Google Scholar] [CrossRef]
- Glasenapp, Y.; Korth, I.; Nguyen, X.-V.; Papenbrock, J. Sustainable use of mangroves as sources of valuable medicinal compounds: Species identification, propagation and secondary metabolite composition. S. Afr. J. Bot. 2019, 121, 317–328. [Google Scholar] [CrossRef]
- Husori, D.I.; Sumardi, H.T.; Gemasih, S.; Ningsih, S.R. In vitro anthelmintic activity of Acanthus ilicifolius leaves extracts on Ascardia galli and Pheretima posthuma. J. Appl. Pharm. Sci. 2018, 8, 164–167. [Google Scholar] [CrossRef]
- Sardar, P.K.; Dev, S.; Al Bari, M.A.; Paul, S.; Yeasmin, M.S.; Das, A.K.; Biswas, N.N. Antiallergic, anthelmintic and cytotoxic potentials of dried aerial parts of Acanthus ilicifolius L. Clin. Phytosci. 2018, 4, 894. [Google Scholar] [CrossRef]
- Vattem, D.A.; Shetty, K. Biological functionality of ellagic acid: A review. J. Food Biochem. 2005, 29, 234–266. [Google Scholar] [CrossRef]
- García-Niño, W.R.; Zazueta, C. Ellagic acid: Pharmacological activities and molecular mechanisms involved in liver protection. Pharmacol. Res. 2015, 97, 84–103. [Google Scholar] [CrossRef]
- Zhu, M.; Phillipson, J.D.; Greengrass, P.M.; Bowery, N.E.; Cai, Y. Plant Polyphenols: Biologically active compounds or non-selective binder protein? Phytochemistry 1997, 44, 441–447. [Google Scholar] [CrossRef]
- Michels, K.; Heinke, R.; Schöne, P.; Kuipers, O.P.; Arnold, N.; Wessjohann, L.A. A fluorescence-based bioassay for antibacterials and its application in screening natural product extracts. J. Antibiot. 2015, 68, 734–740. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.-K.; Xu, J.-K.; Zhang, X.-Q.; Yao, X.-S.; Ye, W.-C. Chemical constituents with antibacterial activity from Euphorbia sororia. Nat. Prod. Res. 2008, 22, 353–359. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).