Hemp Chemotype Definition by Cannabinoids Characterization Using LC-ESI(+)-LTQ-FTICR MS and Infrared Multiphoton Dissociation
Abstract
:1. Introduction
2. Materials and Methods
2.1. Chemicals and Reagents
2.2. Preparation of Standard Solutions
2.3. Phytocannabinoids Extraction and Sample Preparation
2.4. LC-ESI-LTQ-FTICR MS Analyses
3. Results
3.1. Cannabinoids Profile by LC-ESI(+)-LTQ-FTICR MS Analysis and IRMPD/CID Mass Fragmentation Characterization
3.2. Quantitative Analysis of CBD, THC and CBD and Chemotype Definition
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hanuš, L.; Meyer, S.; Muñoz, E.; Taglialatela-Scafati, O.; Appendino, G. Phytocannabinoids: A unified critical inventory. Nat. Prod. Rep. 2016, 33, 1357–1392. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Crocq, M. History of cannabis and the endocannabinoid system. Dialogues Clin. Neurosci. 2020, 22, 223–228. [Google Scholar] [CrossRef] [PubMed]
- Marco, E.; Laviola, G. The endocannabinoid system in the regulation of emotions throughout lifespan: A discussion on therapeutic perspectives. J. Psychopharmacol. 2012, 26, 150–163. [Google Scholar] [CrossRef]
- Citti, C.; Linciano, P.; Russo, F.; Luongo, L.; Iannotta, M.; Maione, S.; Laganà, A.; Capriotti, A.; Forni, F.; Vandelli, M.; et al. A novel phytocannabinoid isolated from Cannabis sativa L. with an in vivo cannabimimetic activity higher than Δ9-tetrahydrocannabinol: Δ9-Tetrahydrocannabiphorol. Sci. Rep. 2019, 9, 20335. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gülck, T.; Møller, B. Phytocannabinoids: Origins and Biosynthesis. Trends Plant Sci. 2020, 25, 985–1004. [Google Scholar] [CrossRef] [PubMed]
- Montone, C.M.; Cerrato, A.; Botta, B.; Cannazza, G.; Capriotti, A.L.; Cavaliere, C.; Citti, C.; Ghirga, F.; Piovesana, S.; Laganà, A. Improved identification of phytocannabinoids using a dedicated structure-based workflow. Talanta 2020, 219, 121310. [Google Scholar] [CrossRef]
- Grotenhermen, F. Pharmacology of Cannabinoids. Neuroendocrinol. Lett. Nos. 2004, 25, 14–23. [Google Scholar]
- Moreno-Sanz, G.; Vera, C.F.; Sánchez-Carnerero, C.; Roura, X.N.; Baena, V.S.D.M. Biological Activity of Cannabis sativa L. Extracts Critically Depends on Solvent Polarity and Decarboxylation. Separations 2020, 7, 56. [Google Scholar] [CrossRef]
- ElSohly, M.; Radwan, M.; Gul, W.; Chandra, S.; Galal, A. Phytochemistry of Cannabis sativa L. In Progress in the Chemistry of Organic Natural Products; Springer: Berlin/Heidelberg, Germany, 2017; pp. 1–36. [Google Scholar]
- Linciano, P.; Citti, C.; Russo, F.; Tolomeo, F.; Lagann, A.; Capriotti, A.L.; Luongo, L.; Iannotta, M.; Belardo, C.; Maione, S.; et al. Identification of a new cannabidiol n-hexyl homolog in a medicinal cannabis variety with an antinociceptive activity in mice: Cannabidihexol. Sci. Rep. 2020, 10, 22019. [Google Scholar] [CrossRef]
- Dujourdy, L.; Besacier, F. A study of cannabis potency in France over a 25 years period (1992–2016). Forensic Sci. Int. 2017, 272, 72–80. [Google Scholar] [CrossRef]
- Vergara, D.; Gaudino, R.; Blank, T.; Keegan, B. Modeling cannabinoids from a large-scale sample of Cannabis sativa chemotypes. PLoS ONE 2020, 15, e0236878. [Google Scholar] [CrossRef] [PubMed]
- Maccarrone, M. Phytocannabinoids and endocannabinoids: Different in nature. Rend. Lincei. Sci. Fis. Nat. 2020, 31, 931–938. [Google Scholar] [CrossRef]
- Giupponi, L.; Leoni, V.; Carrer, M.; Ceciliani, G.; Sala, S.; Panseri, S.; Pavlovic, R.; Giorgi, A. Overview on italian hemp production chain, related productive and commercial activities and legislative framework. Ital. J. Agron. 2020, 15, 194–205. [Google Scholar] [CrossRef]
- Thichak, S.; Natakankitkul, S.; Chansakaow, S.; Chutipongvivate, S. Identification of Drug-Type and Fiber-Type of Hemp (Cannabis sativa L.) by Multiplex PCR. Chiang Mai J. Sci. 2011, 38, 608–618. [Google Scholar]
- United Nations Office on Drugs and Crime. ST/NAR/40: Recommended Methods for the Identification and Analysis of Cannabis and Cannabis Products; UNODC: New York, NY, USA, 2009; ISBN 978-92-1-148242-3. [Google Scholar]
- United Nations Office on Drugs and Crime. ST/NAR/48: Recommended Methods for the Identification and Analysis of Synthetic Cannabinoid Receptor Agonists in Seized Materials; Rev. 1.; UNODC: New York, NY, USA, 2013. [Google Scholar]
- Bianco, G.; Pascale, R.; Lelario, F.; Bufo, S.A.; Cataldi, T.R.I. Investigation of Glucosinolates by Mass Spectrometry. In Glucosinolates. Reference Series in Phytochemistry; Mérillon, J.M., Ramawat, K., Eds.; Springer: Cham, Switzerland, 2017; pp. 431–461. [Google Scholar] [CrossRef]
- Bianco, G.; Pascale, R.; Carbone, C.; Acquavia, M.; Cataldi, T.; Schmitt-Kopplin, P.; Buchicchio, A.; Russo, D.; Milella, L. Determination of soyasaponins in Fagioli di Sarconi beans (Phaseolus vulgaris L.) by LC-ESI-FTICR-MS and evaluation of their hypoglycemic activity. Anal. Bioanal. Chem. 2018, 410, 1561–1569. [Google Scholar] [CrossRef]
- Onzo, A.; Pascale, R.; Acquavia, M.; Cosma, P.; Gubitosa, J.; Gaeta, C.; Iannece, P.; Tsybin, Y.; Rizzi, V.; Guerrieri, A.; et al. Untargeted analysis of pure snail slime and snail slime-induced Au nanoparticles metabolome with MALDI FT-ICR MS. J. Mass Spectrom. 2021, 56, e4722. [Google Scholar] [CrossRef] [PubMed]
- Lelario, F.; Maria, S.D.; Rivelli, A.R.; Russo, D.; Milella, L.; Bufo, S.A.; Scrano, L. A Complete Survey of Glycoalkaloids Using LC-FTICR-MS and IRMPD in a Commercial Variety and a Local Landrace of Eggplant (Solanum melongena L.) and their Anticholinesterase and Antioxidant Activities. Toxins 2019, 11, 230. [Google Scholar] [CrossRef] [Green Version]
- Bianco, G.; Lelario, F.; Battista, F.G.; Bufo, S.A.; Cataldi, T.R.I. Identification of glucosinolates in capers by LC-ESI-hybrid linear ion trap with Fourier transform ion cyclotron resonance mass spectrometry (LC-ESI-LTQ-FTICR MS) and infrared multiphoton dissociation. J. Mass Spectrom. 2012, 47, 1160–1169. [Google Scholar] [CrossRef]
- Lelario, F.; Bianco, G.; Bufo, S.A.; Cataldi, T.R.I. Establishing the occurrence of major and minor glucosinolates in Brassicaceae by LC–ESI-hybrid linear ion-trap and Fourier-transform ion cyclotron resonance mass spectrometry. Phytochemistry 2012, 73, 74–83. [Google Scholar] [CrossRef]
- Agneta, R.; Rivelli, A.R.; Ventrella, E.; Lelario, F.; Sarli, G.; Bufo, S.A. Investigation of Glucosinolate Profile and Qualitative Aspects in Sprouts and Roots of Horseradish (Armoracia rusticana) Using LC-ESI–Hybrid Linear Ion Trap with Fourier Transform Ion Cyclotron Resonance Mass Spectrometry and Infrared Multiphoton Dissocia. J. Agric. Food Chem. 2012, 60, 7474–7482. [Google Scholar] [CrossRef]
- Onzo, A.; Acquavia, M.A.; Pascale, R.; Iannece, P.; Gaeta, C.; Nagornov, K.O.; Tsybin, Y.O.; Bianco, G. Metabolic profiling of Peperoni di Senise PGI bell peppers with ultra-high resolution absorption mode Fourier transform ion cyclotron resonance mass spectrometry. Int. J. Mass Spectrom. 2021, 470, 116722. [Google Scholar] [CrossRef]
- Acquavia, M.A.; Pascale, R.; Martelli, G.; Bondoni, M.; Bianco, G. Natural Polymeric Materials: A Solution to Plastic Pollution from the Agro-Food Sector. Polymers 2021, 13, 158. [Google Scholar] [CrossRef]
- Santarsiero, A.; Onzo, A.; Pascale, R.; Acquavia, M.A.; Coviello, M.; Convertini, P.; Todisco, S.; Marsico, M.; Pifano, C.; Iannece, P.; et al. Pistacia lentiscus Hydrosol: Untargeted Metabolomic Analysis and Anti-Inflammatory Activity Mediated by NF-κ B and the Citrate Pathway. Oxid. Med. Cell. Longev. 2020, 2020, 4264815. [Google Scholar] [CrossRef] [PubMed]
- Caivano, M.; Pascale, R.; Mazzone, G.; Buchicchio, A.; Masi, S.; Bianco, G.; Caniani, D. N2O and CO2 Emissions from Secondary Settlers in WWTPs: Experimental Results on Full and Pilot Scale Plants. In Lecture Notes in Civil Engineering; Mannina, G., Ed.; Springer: Cham, Switzerland, 2017; Volume 4, pp. 412–418. [Google Scholar]
- Citti, C.; Linciano, P.; Panseri, S.; Vezzalini, F.; Forni, F.; Vandelli, M.; Cannazza, G. Cannabinoid Profiling of Hemp Seed Oil by Liquid Chromatography Coupled to High-Resolution Mass Spectrometry. Front. Plant Sci. 2019, 10, 120. [Google Scholar] [CrossRef]
- Delgado-Povedano, M.M.; Sánchez-Carnerero Callado, C.; Priego-Capote, F.; Ferreiro-Vera, C. Untargeted characterization of extracts from Cannabis sativa L. cultivars by gas and liquid chromatography coupled to mass spectrometry in high resolution mode. Talanta 2020, 208, 120384. [Google Scholar] [CrossRef] [PubMed]
- CE n. 796/2004 Regolamento CE n. 796/2004 Recante Modalità di Applicazione Della Condizionalità, Della Modulazione e del Sistema Integrato di Gestione e di Controllo di cui al Reg. (CE) n. 1782/2003. Available online: https://www.politicheagricole.it/flex/cm/pages/ServeBLOB.php/L/IT/IDPagina/2942 (accessed on 19 October 2021).
- CE n. 1164/89 Regolamento CE n. 1164/89 della Commissione del 28 Aprile 1989 Relativo Alle Modalità D’applicazione Concernenti L’aiuto per il Lino Tessile e la Canapa—Publications Office of the EU. Available online: https://op.europa.eu/en/publication-detail/-/publication/fb3afa66-f593-4145-8a14-77aba838eb8c/language-it/format-PDFA1B (accessed on 19 October 2021).
- Lelario, F.; Scrano, L.; De Franchi, S.; Bonomo, M.G.; Salzano, G.; Milan, S.; Milella, L.; Bufo, S.A. Identification and antimicrobial activity of most representative secondary metabolites from different plant species. Chem. Biol. Technol. Agric. 2018, 5, 13. [Google Scholar] [CrossRef]
- Lelario, F.; Labella, C.; Napolitano, G.; Scrano, L.; Bufo, S.A. Fragmentation study of major spirosolane-type glycoalkaloids by collision-induced dissociation linear ion trap and infrared multiphoton dissociation Fourier transform ion cyclotron resonance mass spectrometry. Rapid Commun. Mass Spectrom. 2016, 30, 2395–2406. [Google Scholar] [CrossRef] [PubMed]
- Hillig, K.; Mahlberg, P. A chemotaxonomic analysis of cannabinoid variation in Cannabis (Cannabaceae). Am. J. Bot. 2004, 91, 966–975. [Google Scholar] [CrossRef] [Green Version]
- Santos, N.A.D.; Tose, L.V.; Silva, S.R.C.D.; Murgu, M.; Kuster, R.M.; Ortiz, R.S.; Camargo, F.A.O.; Vaz, B.G.; Lacerda, V.; Romão, W. Analysis of Isomeric Cannabinoid Standards and Cannabis Products by UPLC-ESI-TWIM-MS: A Comparison with GC-MS and GC × GC-QMS. J. Braz. Chem. Soc. 2019, 30, 60–70. [Google Scholar] [CrossRef]
- Ferrer, I. Analyses of cannabinoids in hemp oils by LC/Q-TOF-MS. Compr. Anal. Chem. 2020, 90, 415–452. [Google Scholar] [CrossRef]
- Meijer, E.P.M.D.; Bagatta, M.; Carboni, A.; Crucitti, P.; Moliterni, V.M.C.; Ranalli, P.; Mandolino, G. The inheritance of chemical phenotype in Cannabis sativa L. Genetics 2003, 163, 335. [Google Scholar] [CrossRef] [PubMed]
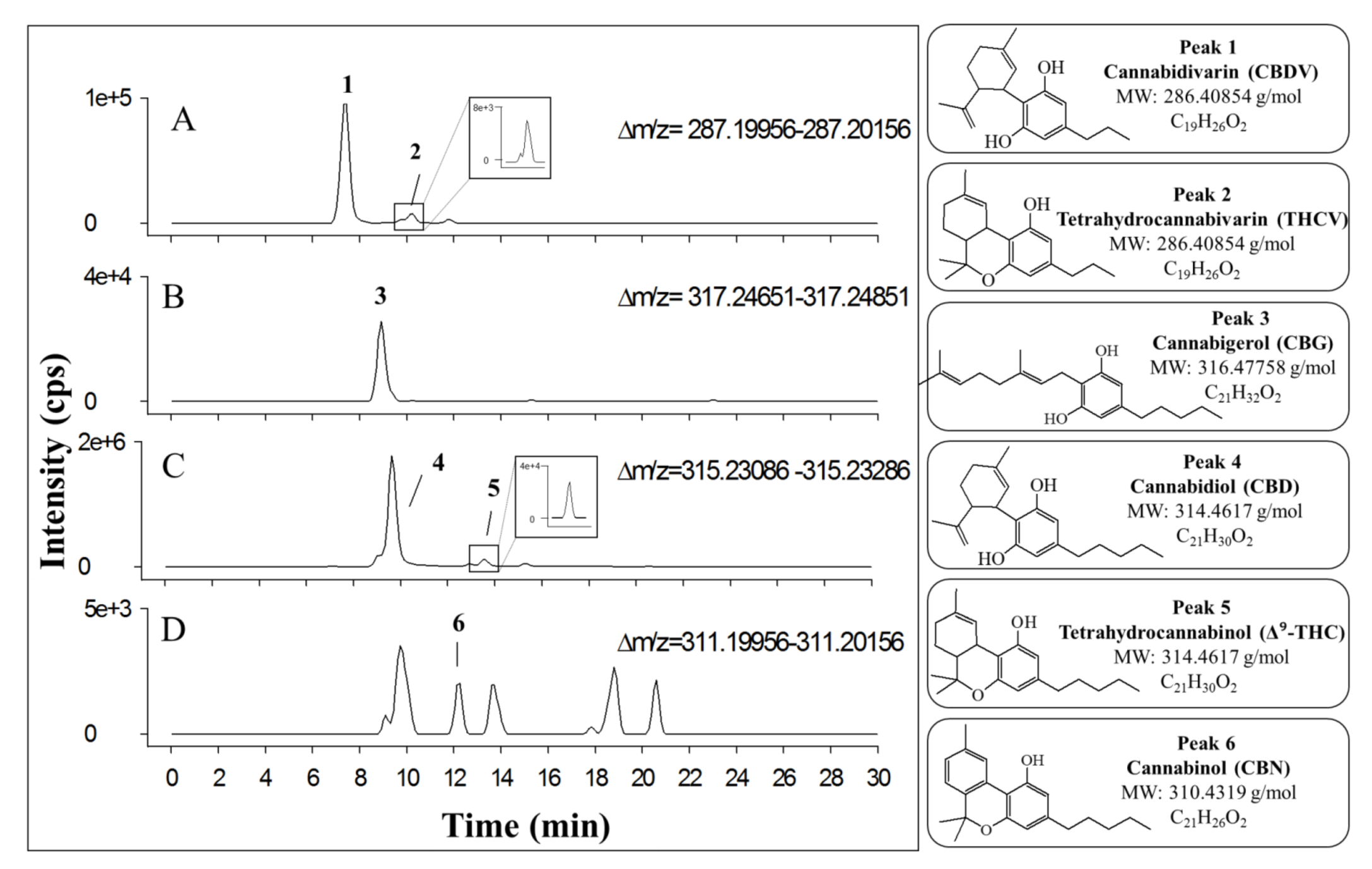

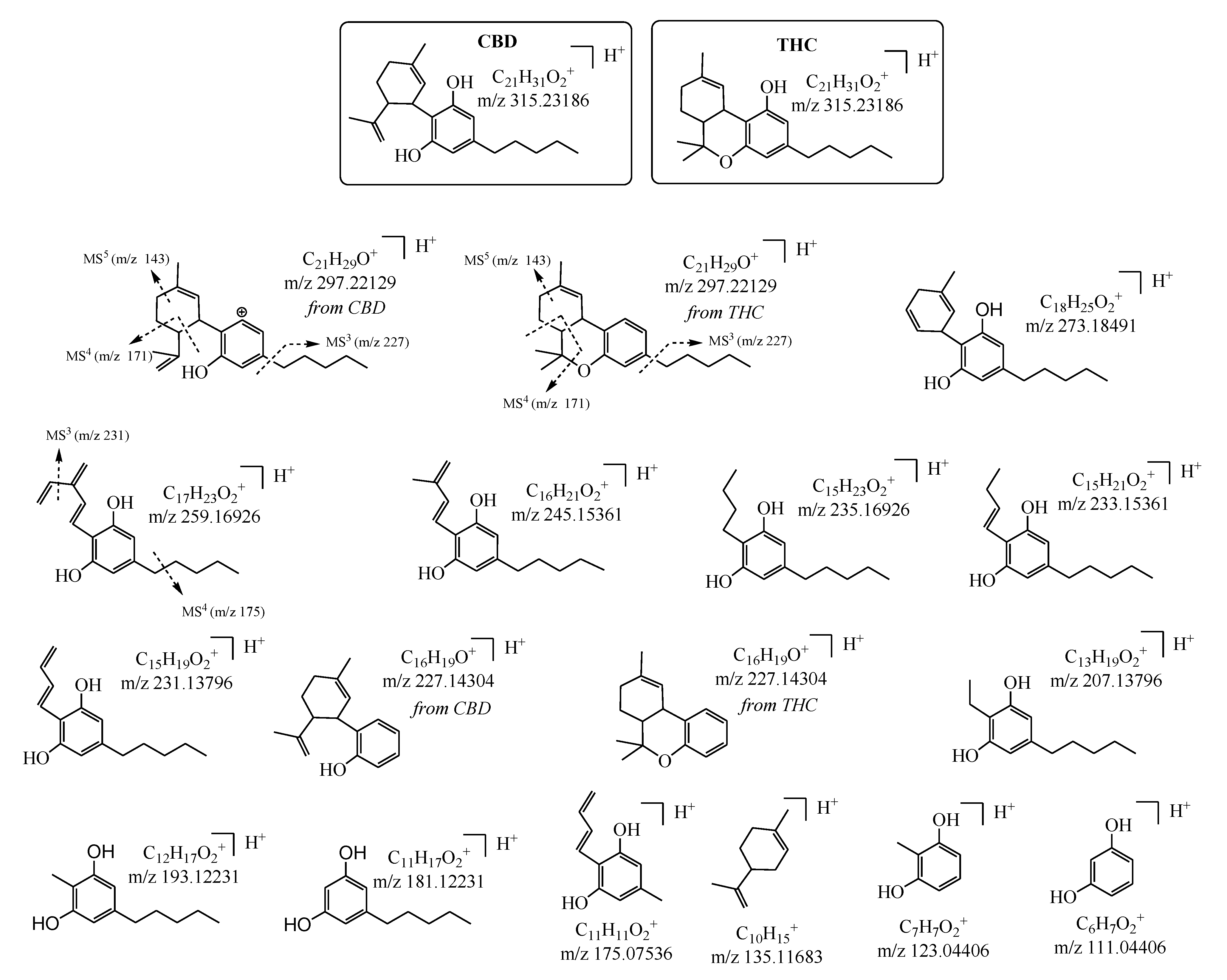

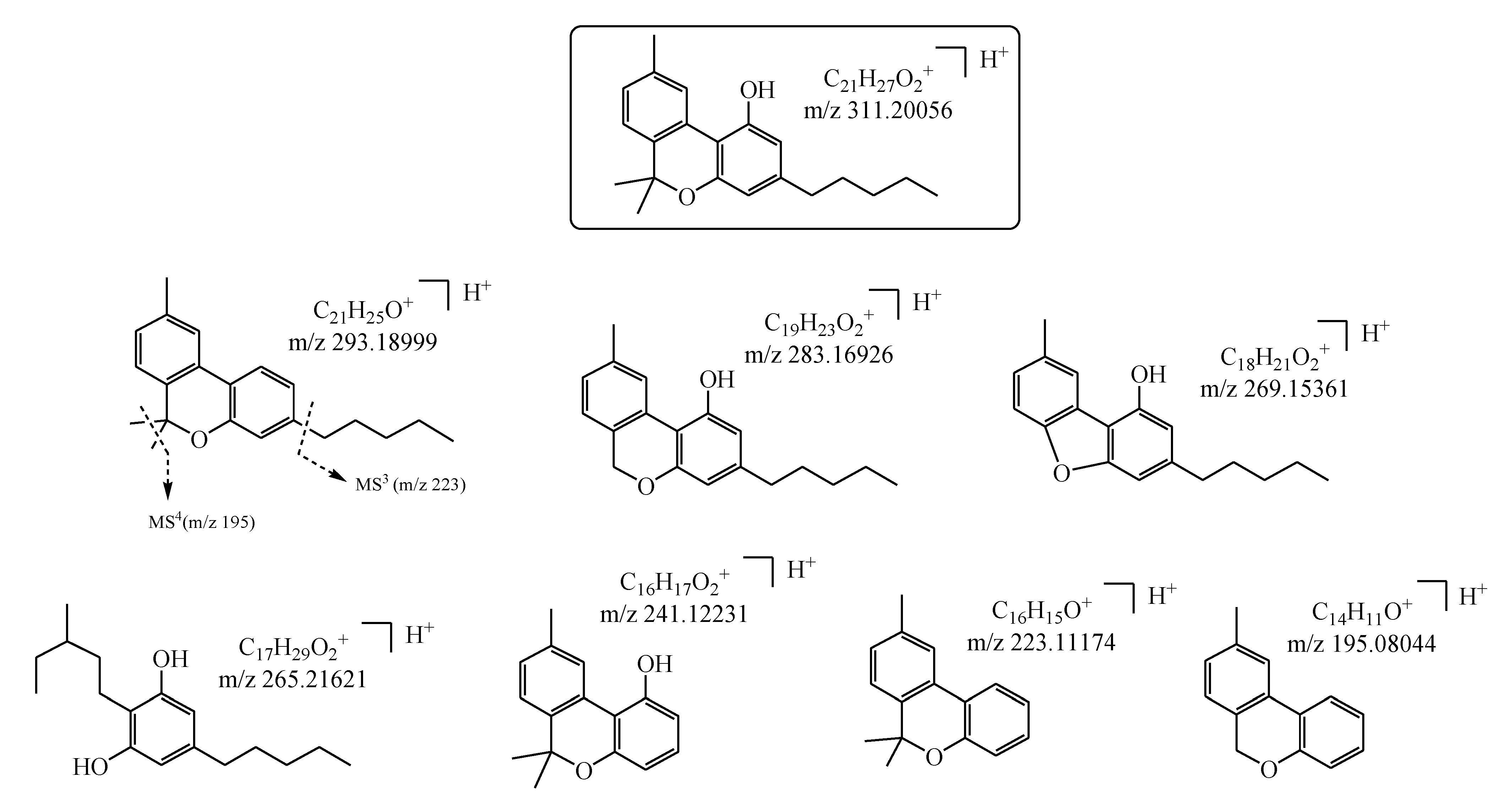
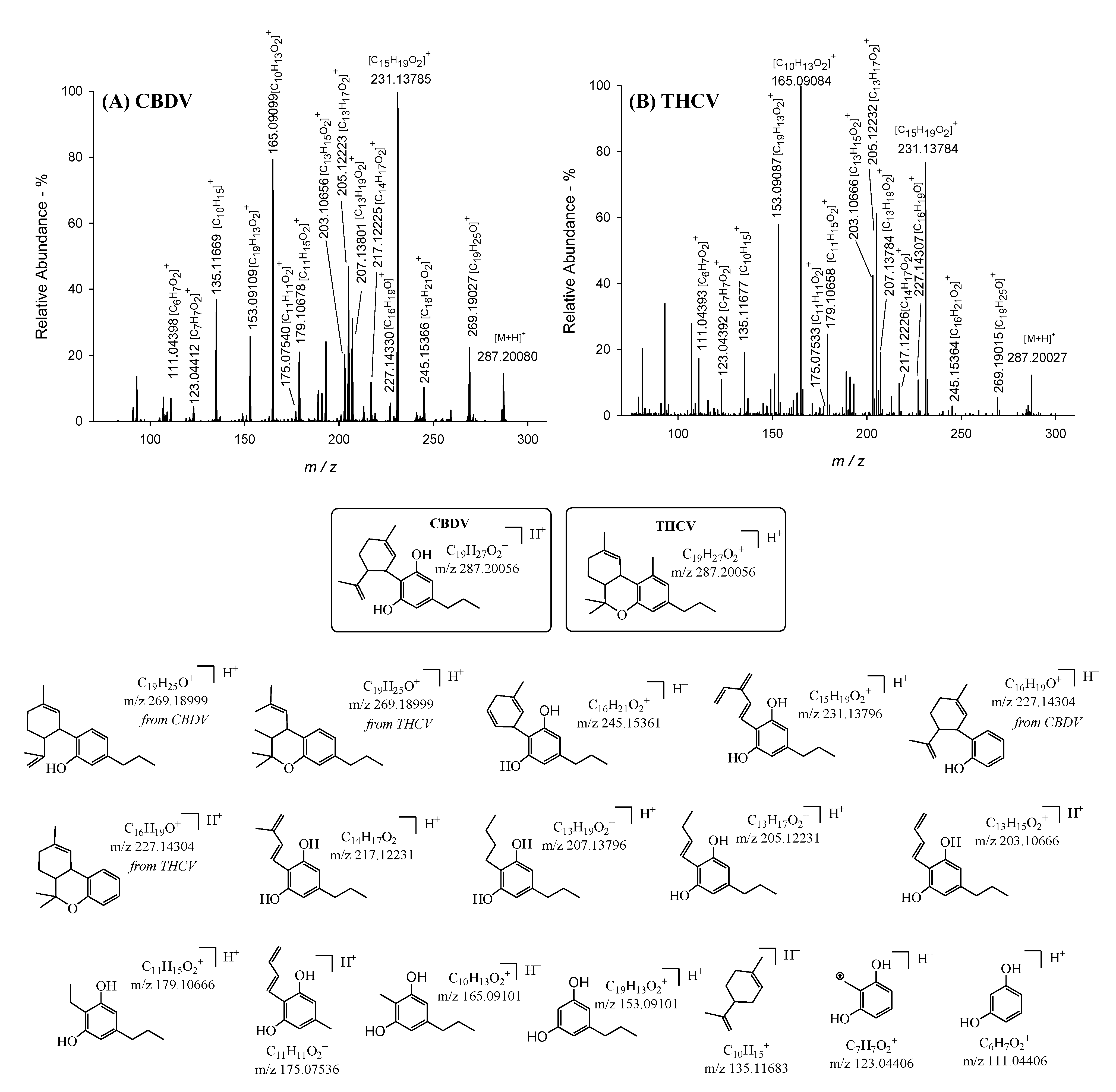
| N a | Cannabinoid | tr (min) b | Molecular Formula as [M+H]+ c | Monoisotopic Accurate Value [M+H]+ (m/z) d | Mass Error (ppm) e | Main IRMPD MS/MS Productions (Accurate m/z) d and Mass Error (ppm) e |
|---|---|---|---|---|---|---|
| 1 | CBDV | 7.4 | C19H27O2+ | 287.20080 | 0.84 | 111.04398 (0.72); 123.04412 (0.49); 135.11669 (1.04); 153.09109 (0.52); 165.09099 (0.12); 175.07540 (0.23); 179.10678 (0.68); 203.10656 (0.52); 205.12223 (0.38); 207.13801 (0.24); 217.12225 (0.28); 227.14330 (1.15); 231.13785 (0.48); 245.15366 (0.20); 269.19027 (1.04) |
| 2 | THCV | 10.3 | C19H27O2+ | 287.20027 | 1.01 | 111.04393 (1.22); 123.04392 (1.11); 135.11677 (0.44); 153.09087 (0.91); 165.09084 (1.02); 175.07533 (0.20); 179.10658 (0.44); 203.10666 (0.02); 205.12232 (0.03); 207.13784 (0.60); 217.12226 (0.23); 227.14307 (0.13); 231.13784 (0.50); 245.15364 (0.13); 269.19015 (0.61) |
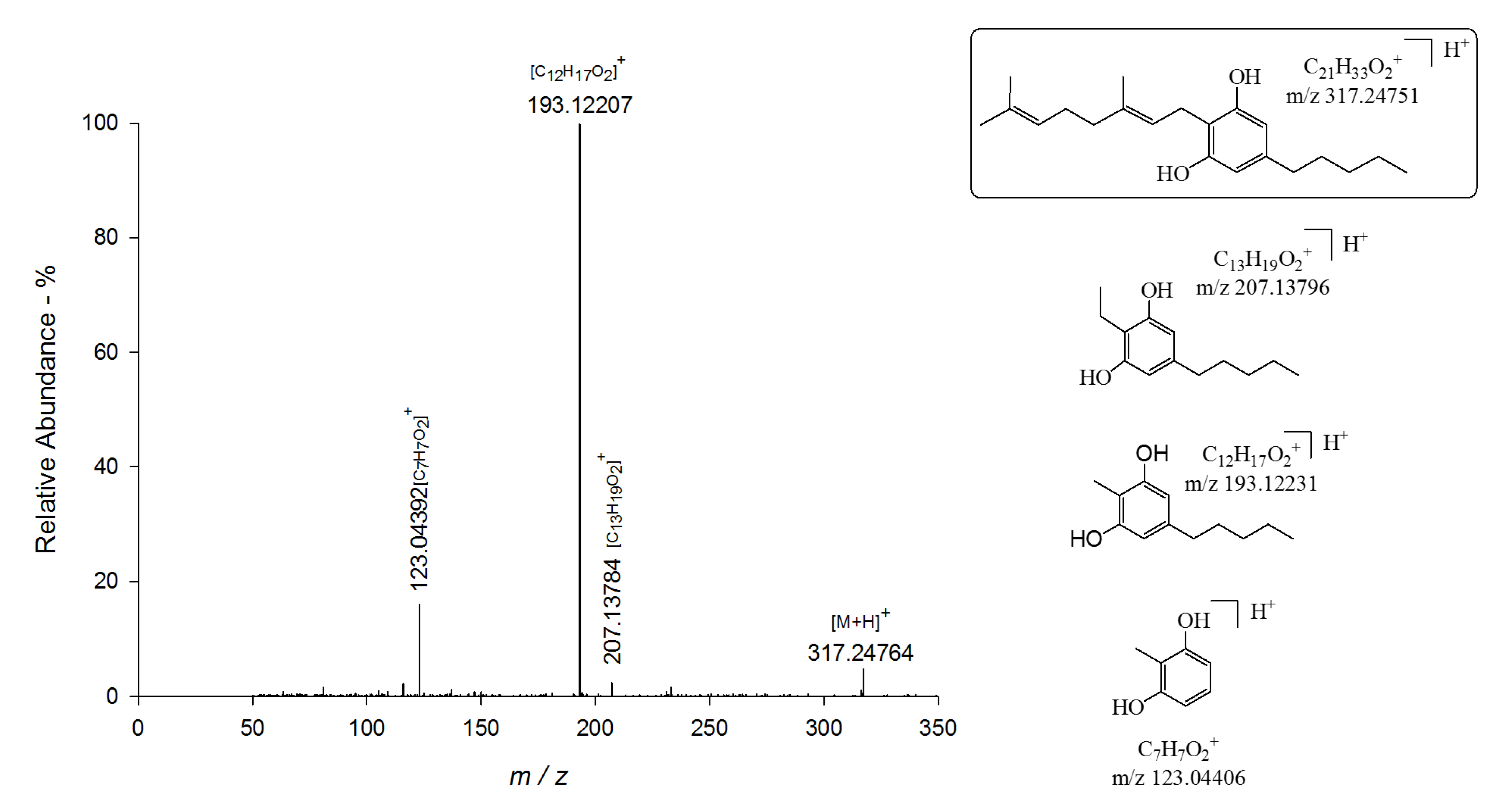
| 3 | CBG | 8.9 | C21H33O2+ | 317.24764 | 0.42 | 123.04392 (1.11); 193.12207 (1.18); 207.13784 (0.60) |
| 4 | CBD | 9.6 | C21H31O2+ | 315.23156 | 0.95 | 111.04400 (0.54); 123.04402 (0.37); 135.11677 (0.44); 175.07533 (0.20); 181.12216 (0.81); 193.12230 (0.016); 207.13784 (0.60); 227.14307 (0.14); 231.13784 (0.50); 233.15364 (0.14); 235.16906 (0.85); 245.15364 (0.13); 259.16923 (0.14); 273.18463 (1.03); 297.22138 (0.31) |
| 5 | THC | 13.6 | C21H31O2+ | 315.23184 | 0.063 | 111.04405 (0.09); 123.04396 (0.49); 135.11685 (0.15); 175.07533 (0.20); 181.12227 (0.22); 193.12223 (0.36); 207.13787 (0.43); 227.14313 (0.40); 231.13797 (0.043); 233.15363 (0.085); 235.16914 (0.51); 245.15368 (0.29); 259.16916 (0.39); 273.18491 (0.00); 297.22133 (0.13) |
| 6 | CBN | 12.3 | C21H27O2+ | 311.20071 | 0.48 | 195.08050 (0.31); 223.11191 (0.76); 241.1206 (0.87); 265.21632 (0.41); 269.15382 (0.80); 283.16900 (0.92); 293.19000 (0.034) |
| Parameter | Sample | ||
|---|---|---|---|
| Carmagnola C.S. | Carmagnola | Eletta Campana | |
| THC % | 0.053 ± 0.001 | 0.105 ± 0.002 | 0.111 ± 0.003 |
| CBD % | 0.897 ± 0.002 | 1.387 ± 0.003 | 1.991 ± 0.002 |
| CBN % | 0.009 ± 0.001 | 0.011 ± 0.001 | 0.007 ± 0.002 |
| CBD/THC | 16.925 ± 0.322 | 13.210 ± 0.253 | 17.937 ± 0.485 |
| CBN/THC | 0.170 ± 0.019 | 0.105 ± 0.010 | 0.063 ± 0.018 |
| (THC + CBN)/CBD | 0.069 ± 0.002 | 0.084 ± 0.002 | 0.059 ± 0.002 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lelario, F.; Pascale, R.; Bianco, G.; Scrano, L.; Bufo, S.A. Hemp Chemotype Definition by Cannabinoids Characterization Using LC-ESI(+)-LTQ-FTICR MS and Infrared Multiphoton Dissociation. Separations 2021, 8, 245. https://doi.org/10.3390/separations8120245
Lelario F, Pascale R, Bianco G, Scrano L, Bufo SA. Hemp Chemotype Definition by Cannabinoids Characterization Using LC-ESI(+)-LTQ-FTICR MS and Infrared Multiphoton Dissociation. Separations. 2021; 8(12):245. https://doi.org/10.3390/separations8120245
Chicago/Turabian StyleLelario, Filomena, Raffaella Pascale, Giuliana Bianco, Laura Scrano, and Sabino Aurelio Bufo. 2021. "Hemp Chemotype Definition by Cannabinoids Characterization Using LC-ESI(+)-LTQ-FTICR MS and Infrared Multiphoton Dissociation" Separations 8, no. 12: 245. https://doi.org/10.3390/separations8120245
APA StyleLelario, F., Pascale, R., Bianco, G., Scrano, L., & Bufo, S. A. (2021). Hemp Chemotype Definition by Cannabinoids Characterization Using LC-ESI(+)-LTQ-FTICR MS and Infrared Multiphoton Dissociation. Separations, 8(12), 245. https://doi.org/10.3390/separations8120245










